Abstract
Achromobacter spp. are emerging respiratory pathogens in cystic fibrosis patients. Since 2013 the genus Achromobacter includes 15 species for which innate antibiotic resistance is unknown. Previously the AxyXY-OprZ efflux system has been described to confer aminoglycoside (AG) resistance in A. xylosoxidans. Nevertheless, some Achromobacter spp. strains are susceptible to AG. This study including 49 Achromobacter isolates reveals that AG resistance is correlated with different Achromobacter spp. It is noteworthy that the axyXY-oprZ operon is detected only in AG-resistant species, including the most frequently encountered in cystic fibrosis patients: A. xylosoxidans, A. ruhlandii, A. dolens and A. insuavis.
Keywords: Achromobacter, aminoglycoside resistance, AxyXY-OprZ, cystic fibrosis, nrdA, selection pressure
Achromobacter spp. are nonfermenting Gram-negative bacilli considered as emerging pathogens in cystic fibrosis (CF) patients [1], [2]. Since the description of the type species, A. xylosoxidans [3], 14 other species have been ranked into the genus Achromobacter: A. piechaudii and A. ruhlandii [4], A. denitrificans [5], A. spanius and A. insolitus [6], A. marplatensis [7], A. animicus, A. mucicolens, A. pulmonis and A. spiritinus [8], A. insuavis, A. aegrifaciens, A. anxifer and A. dolens [9], and 6 other genogroups. These 21 species and genogroups can be distinguished by the multilocus sequence typing (MLST) scheme proposed by Spilker et al. [10]. The study demonstrated that sequencing a 765 bp internal fragment of the only nrdA gene is sufficient for correct identification [11]. Because of the actual difficulty in performing accurate species identification, most isolates are still referred by default as A. xylosoxidans, preventing the evaluation of the real epidemiology and clinical impact of each species. Moreover, the data about the mechanisms of innate antibiotic resistance are scarce [12], [13]. The AxyXY-OprZ RND efflux system confers resistance to aminoglycosides (AG) in A. xylosoxidans AXX-A since reclassified as A. insuavis (accession number AFRQ01000000). Nevertheless, AG, which take an important part in CF antimicrobial therapy, remain active against some isolates of Achromobacter spp. [14], [15].
We sought to describe the distribution of AG-resistant isolates among the different species of the genus Achromobacter and to search for the axyXY-oprZ efflux operon in AG-resistant and -susceptible isolates to assess if AG resistance is correlated with the presence of the operon.
Forty-nine Achromobacter isolates harbouring various AG resistance patterns were included in this study: 21 from CF patients' sputum, 20 from non-CF clinical samples and eight from environmental samples (Table 1). Most of them (n = 35) were collected in our laboratory; nine collection strains were purchased from the Institut Pasteur, France, including six type strains, and five were kindly provided by J. J. LiPuma (Department of Pediatrics and Communicable Diseases, University of Michigan Medical School). Isolates were identified at the genus level either by using the conventional biochemical method API 20NE (bioMérieux) or by sequencing the 16S rRNA gene. The identification to the species level was performed by sequencing the 765 bp internal nrdA fragment followed by Achromobacter PubMLST database query (http://pubmlst.org/achromobacter/). Minimal inhibitory concentrations (MICs) of tobramycin, amikacin, gentamicin and netilmicin were measured by the Etest method (bioMérieux). Mueller-Hinton agar plates were inoculated by swabbing from a 0.5 McFarland turbidity bacterial suspension, and MICs were recorded after overnight incubation at 37°C by two persons independently. The phenotype “AG-susceptible” (AG-S) was attributed to isolates susceptible to all AG and the phenotype “AG-resistant” (AG-R) to the other by using the European Committee on Antimicrobial Susceptibility Testing clinical breakpoints for Pseudomonas spp. (http://www.eucast.org/clinical_breakpoints/; version 5.0). Detection of the axyXY-oprZ operon was performed by 2 PCRs targeting the genes (a) axyY, encoding the RND transporter, and (b) oprZ, encoding the outer membrane factor. PCRs were carried out in reaction mixtures containing dNTP (0.2 mM), forward and reverse primers (0.25 μM each), Taq polymerase (Fermentas) (2.5 U) with the supplied buffer, MgCl2 (1.5 mM), dimethyl sulfoxide (5% volume) and template DNA (1 μL), adjusted with water to a final volume of 50 μL. The cycling parameters were 94°C for 10 minutes, 30 cycles of 94°C for 90 seconds, annealing primers temperature for 90 sections, 72°C for 60 seconds, and 72°C for 10 minutes. The results are summarized in Table 1.
Table 1.
Achromobacter isolates and main results
| nrdA identification | Isolate | Origin | MIC (mg/L) |
AG S/R | PCR axyY | PCR oprZ | |||
|---|---|---|---|---|---|---|---|---|---|
| TOB | AMK | GEN | NET | ||||||
| A. aegrifaciens | ACH-CF-D59 | CF sputuma | >256 | >256 | >256 | >256 | R | + | + |
| A. aegrifaciens | ACH-CF-802 | CF sputuma | 64 | 48 | 16 | 32 | R | + | + |
| A. aegrifaciens | ACH-ENV-2 | Hospital hand-washing sinka | 12 | 8 | 3 | 8 | R | + | + |
| A. aegrifaciens | ACH-CF-766 | CF sputuma | 192 | 48 | 12 | 64 | R | + | + |
| A. animicus | ACH-CF-864 | CF sputuma | 1.5 | 4 | 1 | 1 | S | − | − |
| A. animicus | ACH-NCF-33 | Cathetera | 1.5 | 6 | 2 | 2 | S | − | − |
| A. animicus | ACH-CF-D63 | CF sputuma | 1.5 | 4 | 1.5 | 1 | S | − | − |
| A. animicus | ACH-CF-D64 | CF sputuma | 2 | 8 | 2 | 1.5 | S | − | − |
| A. animicus | ACH-CF-D65 | CF sputuma | 1 | 3 | 1.5 | 1.5 | S | − | − |
| A. animicus | ACH-CF-711 | CF sputuma | 2 | 4 | 1.5 | 1.5 | S | − | − |
| A. denitrificans | CIP-77.15T | Soil | 32 | 256 | 64 | 64 | R | + | + |
| A. dolens | AU18822 | CF sputum | >256 | 64 | >256 | 12 | R | + | + |
| A. dolens | AU20310 | CF sputum | >256 | >256 | >256 | 128 | R | + | + |
| A. genogroup 12 | ACH-ENV-3 | Dialysis watera | 3 | 32 | 8 | 8 | R | + | + |
| Novel species | ACH-CF-583 | CF sputuma | 24 | >256 | 96 | >256 | R | + | + |
| A. insolitus | CIP-108202T | Leg wound | 256 | 32 | 48 | >256 | R | + | + |
| A. insuavis | ACH-CF-476 | CF sputuma | >256 | >256 | >256 | >256 | R | + | + |
| A. insuavis | ACH-CF-777 | CF sputuma | 96 | >256 | >256 | >256 | R | + | + |
| A. insuavis | AXX-A | Ear swaba | 16 | 256 | 24 | 64 | R | + | + |
| A. insuavis | CIP-102062 | Blood | 12 | 256 | 16 | 24 | R | + | + |
| A. marplatensis | ACH-ENV-4 | Lakea | 12 | 256 | 24 | 32 | R | + | + |
| A. mucicolens | ACH-NCF-34 | Tracheal aspiratea | 1.5 | 6 | 1.5 | 1.5 | S | − | − |
| A. mucicolens | ACH-NCF-35 | Tracheal aspiratea | 2 | 8 | 2 | 1.5 | S | − | − |
| A. mucicolens | ACH-NCF-36 | Blooda | 2 | 6 | 2 | 2 | S | − | − |
| A. mucicolens | ACH-CF-510 | CF sputuma | 2 | 8 | 2 | 2 | S | − | − |
| A. piechaudii | CIP-60.75T | Pharynx | 1.5 | 6 | 3 | 3 | S | − | − |
| A. ruhlandii | CIP-77.26T | Soil | 8 | 24 | 12 | 16 | R | + | + |
| A. ruhlandii | AU19877 | CF sputum | 16 | >256 | 48 | 64 | R | + | + |
| A. ruhlandii | AU19911 | CF sputum | 3 | 48 | 6 | 12 | R | + | + |
| A. ruhlandii | AU19929 | CF sputum | >256 | >256 | >256 | >256 | R | + | + |
| A. spanius | CIP-108199T | Blood | 1.5 | 6 | 4 | 4 | S | − | − |
| A. spanius | ACH-NCF-37 | Foot wounda | 1 | 4 | 1 | 1.5 | S | − | − |
| A. spanius | ACH-CF-746 | CF sputuma | 2 | 6 | 2 | 2 | S | − | − |
| A. xylosoxidans | ACH-CF-809 | CF sputuma | 128 | >256 | 256 | >256 | R | + | + |
| A. xylosoxidans | ACH-NCF-39 | Insertion-site skin swaba | 24 | >256 | 64 | 128 | R | + | + |
| A. xylosoxidans | ACH-NCF-18 | Tracheal aspiratea | 64 | >256 | 128 | 256 | R | + | + |
| A. xylosoxidans | CIP-71.32T | Ear discharge | 192 | >256 | >256 | >256 | R | + | + |
| A. xylosoxidans | CIP-101902 | Pleural fluid | >256 | >256 | >256 | >256 | R | + | + |
| A. xylosoxidans | CIP-102236 | Sputum | 48 | >256 | 256 | >256 | R | + | + |
| A. xylosoxidans | ACH-NCF-41 | Sputuma | 32 | >256 | 64 | 128 | R | + | + |
| A. xylosoxidans | ACH-NCF-42 | Tracheal aspiratea | 8 | 256 | 24 | 64 | R | + | + |
| A. xylosoxidans | ACH-ENV-1 | Dental instrumenta | 192 | >256 | >256 | >256 | R | + | + |
| A. xylosoxidans | ACH-NCF-13 | Bronchial aspiratea | 64 | >256 | 128 | >256 | R | + | + |
| A. xylosoxidans | ACH-CF-805 | CF sputuma | 24 | >256 | 48 | 192 | R | + | + |
| A. xylosoxidans | ACH-NCF-11 | Sputuma | 16 | 256 | 32 | 48 | R | + | + |
| A. xylosoxidans | ACH-CF-842 | CF sputuma | 8 | 192 | 24 | 32 | R | + | + |
| A. xylosoxidans | ACH-NCF-40 | Blooda | 32 | >256 | 96 | 192 | R | + | + |
| A. xylosoxidans | ACH-ENV-5 | Rivera | 16 | >256 | 24 | 64 | R | + | + |
| A. xylosoxidans | ACH-ENV-6 | Domestic hand-washing sinka | 32 | >256 | 48 | 128 | R | + | + |
AG, aminoglycoside; AMK, amikacin; CF, cystic fibrosis; GEN, gentamicin; MIC, minimum inhibitory concentration; NET, netilmicin; R, resistant.; S, susceptible; TOB, tobramycin.
Isolates collected in our laboratory.
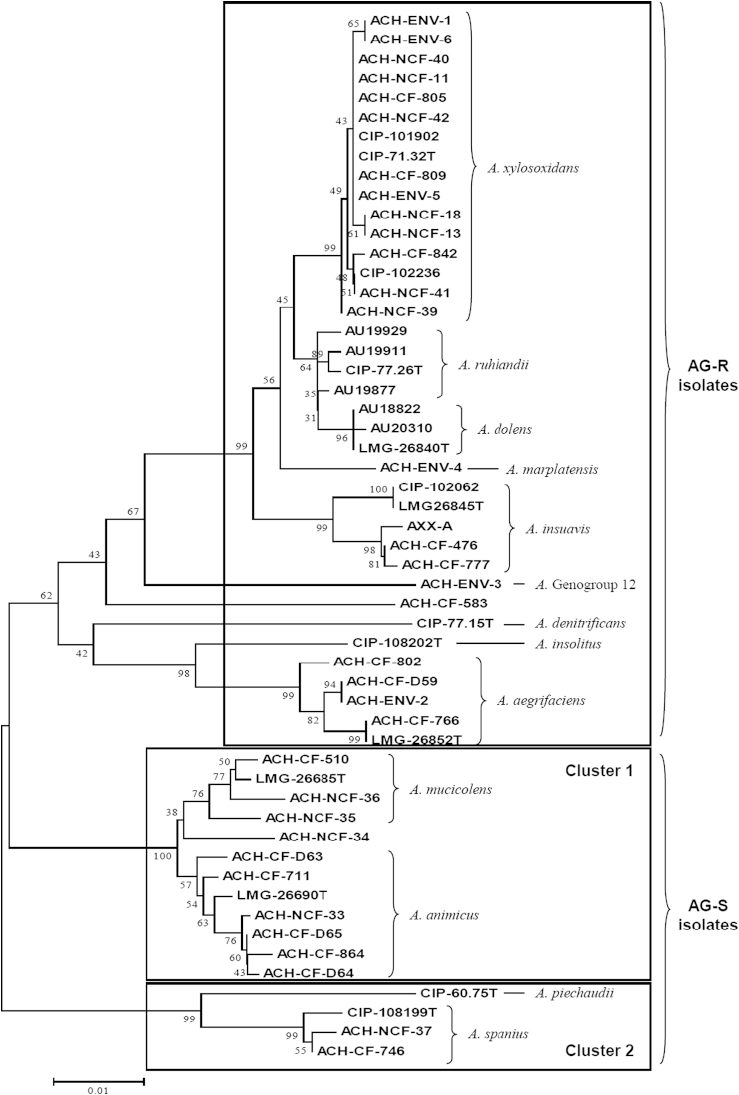
The nrdA sequences analysis allowed identification of 48 of the 49 isolates. nrdA sequence of ACH-CF-583 harboured 39 nucleotide differences, with its closest match in database (genogroup 19) (Fig. 1) indicating that this isolate belonged to a novel genogroup or a novel species. Fourteen of the 49 studied isolates were categorized as AG-S. Interestingly, all isolates belonging to a same species harboured the same AG resistance pattern. In the resistant species, the level of resistance was sometimes variable among the isolates. Nevertheless, none of these isolates had been categorized as susceptible for all four AG molecules. A variable expression level of the efflux operon might account for these differences as already observed for Pseudomonas aeruginosa [16]. Distribution of the AG-S and AG-R isolates according to species identification is represented in a dendrogram (Fig. 2) generated from the nrdA sequences using the neighbour-joining method with 1000 bootstrap replications (MEGA6). We also included in the dendrogram the nrdA sequences of the type strains (LMG) of the recently described novel species. AG-S isolates were divided up into two clusters of species including A. animicus and A. mucicolens (cluster 1), and A. spanius and A. piechaudii (cluster 2). These two clusters were supported by high bootstrap values. All other isolates were AG-R and did not belong to cluster 1 or 2. They included the 16 A. xylosoxidans isolates and all isolates from the species A. aegrifaciens, A. denitrificans, A. dolens, A. genogroup 12, A. insolitus, A. insuavis, A. marplatensis and A. ruhlandii. A similar tree topology was obtained by using the maximum likelihood and the maximum parsimony methods (data not shown). There was a perfect correlation between the AG resistance profile and the presence of the AxyXY-OprZ efflux system. Indeed, the operon was detected in all AG-R isolates and not in the AG-S ones. An additional PCR was performed in all AG-S isolates with primers designed in flanking sequences of the axyXY-oprZ operon and confirmed the absence of the whole operon. Because the GC content of the efflux operon is similar to that of whole genome, we hypothesized that a deletion occurred in the course of evolution. These findings indicate that susceptibility or resistance to AG might be a phenotypic trait correlated with Achromobacter species evolution.
Fig. 1.
ACH-CF-583 nrdA sequence alignment with its closest match (genogroup 19) in Achromobacter PubMLST database. Query: ACH-CF-583; Ref: closest match in database.
Fig. 2.
Neighbour-joining dendrogram based on nrdA sequence. Numbers at nodes indicate bootstrap values. Scale bar indicates number of substitutions per site. AG-resistant (AG-R) isolates are in upper box; AG-susceptible (AG-S) isolates are in boxes ‘cluster 1’ and ‘cluster 2.’ AG resistance was not determined for the LMG strains.
To date, and to our knowledge, only four studies including an appropriate Achromobacter identification method report distribution of the different Achromobacter species in clinical samples. They indicate that A. xylosoxidans is the species the most frequently recovered from CF patients [11], [17], [18], [19]. Other species are also widely prevalent: A. ruhlandii, A. dolens and A. insuavis. It is noteworthy that all isolates belonging to these four species have been categorized as AG-R in the present work. One can therefore wonder whether these AG-R species are more pathogenic than the AG-S ones, which might explain their high prevalence. One might also hypothesize that these species have emerged under the selection pressure of AG treatment frequently prescribed to those in the CF population.
In conclusion, AG resistance is the first phenotypic characteristic correlated with the different species of the genus Achromobacter. More studies including accurate species level identification are required in order to improve knowledge about the epidemiology and virulence of these pathogens. They might also help to elucidate whether the use of inhaled AG promotes selection of Achromobacter that belong to the species harbouring the AxyXY-OprZ efflux system.
Acknowledgements
We thank J. J. LiPuma for providing isolates and T. Spilker for his help in analysing nrdA sequences.
Conflict of interest
None declared.
References
- 1.Amoureux L., Bador J., Siebor E., Taillefumier N., Fanton A., Neuwirth C. Epidemiology and resistance of Achromobacter xylosoxidans from cystic fibrosis patients in Dijon, Burgundy: first French data. J Cyst Fibros. 2013;12:170–176. doi: 10.1016/j.jcf.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Emerson J., McNamara S., Buccat A.M., Worrell K., Burns J.L. Changes in cystic fibrosis sputum microbiology in the United States between 1995 and 2008. Pediatr Pulmonol. 2010;45:363–370. doi: 10.1002/ppul.21198. [DOI] [PubMed] [Google Scholar]
- 3.Yabuuchi E., Oyama A. Achromobacter xylosoxidans n. sp. from human ear discharge. Jpn J Microbiol. 1971;15:477–481. doi: 10.1111/j.1348-0421.1971.tb00607.x. [DOI] [PubMed] [Google Scholar]
- 4.Yabuuchi E., Kawamura Y., Kosako Y., Ezaki T. Emendation of genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. nov., Achromobacter piechaudii (Kiredjian et al.) comb. nov., and Achromobacter xylosoxidans subsp. denitrificans (Rüger and Tan) comb. nov. Microbiol Immunol. 1998;42:429–438. doi: 10.1111/j.1348-0421.1998.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 5.Coenye T., Vancanneyt M., Cnockaert M.C., Falsen E., Swings J., Vandamme P. Kerstersia gyiorum gen. nov., sp. nov., a novel Alcaligenes faecalis–like organism isolated from human clinical samples, and reclassification of Alcaligenes denitrificans Rüger and Tan 1983 as Achromobacter denitrificans comb. nov. Int J Syst Evol Microbiol. 2003;53:1825–1831. doi: 10.1099/ijs.0.02609-0. [DOI] [PubMed] [Google Scholar]
- 6.Coenye T., Vancanneyt M., Falsen E., Swings J., Vandamme P. Achromobacter insolitus sp. nov. and Achromobacter spanius sp. nov., from human clinical samples. Int J Syst Evol Microbiol. 2003;53:1819–1824. doi: 10.1099/ijs.0.02698-0. [DOI] [PubMed] [Google Scholar]
- 7.Gomila M., Tvrzová L., Teshim A. Achromobacter marplatensis sp. nov., isolated from a pentachlorophenol-contaminated soil. Int J Syst Evol Microbiol. 2011;61:2231–2237. doi: 10.1099/ijs.0.025304-0. [DOI] [PubMed] [Google Scholar]
- 8.Vandamme P., Moore E.R., Cnockaert M. Achromobacter animicus sp. nov., Achromobacter mucicolens sp. nov., Achromobacter pulmonis sp. nov. and Achromobacter spiritinus sp. nov., from human clinical samples. Syst Appl Microbiol. 2013;36:1–10. doi: 10.1016/j.syapm.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Vandamme P., Moore E.R., Cnockaert M. Classification of Achromobacter genogroups 2, 5, 7 and 14 as Achromobacter insuavis sp. nov., Achromobacter aegrifaciens sp. nov., Achromobacter anxifer sp. nov. and Achromobacter dolens sp. nov., respectively. Syst Appl Microbiol. 2013;36:474–482. doi: 10.1016/j.syapm.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Spilker T., Vandamme P., LiPuma J.J. A multilocus sequence typing scheme implies population structure and reveals several putative novel Achromobacter species. J Clin Microbiol. 2012;50:3010–3015. doi: 10.1128/JCM.00814-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spilker T., Vandamme P., LiPuma J.J. Identification and distribution of Achromobacter species in cystic fibrosis. J Cyst Fibros. 2013;12:298–301. doi: 10.1016/j.jcf.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Bador J., Amoureux L., Duez J.M. First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob Agents Chemother. 2011;55:4912–4914. doi: 10.1128/AAC.00341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bador J., Amoureux L., Blanc E., Neuwirth C. Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob Agents Chemother. 2013;57:603–605. doi: 10.1128/AAC.01243-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saiman L., Chen Y., Tabibi S. Identification and antimicrobial susceptibility of Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. J Clin Microbiol. 2001;39:3942–3945. doi: 10.1128/JCM.39.11.3942-3945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., Ridderberg W., Hansen C.R. Early treatment with inhaled antibiotics postpones next occurrence of Achromobacter in cystic fibrosis. J Cyst Fibros. 2013;12:638–643. doi: 10.1016/j.jcf.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Vogne C., Aires J.R., Bailly C., Hocquet D., Plesiat P. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother. 2004;48:1676–1680. doi: 10.1128/AAC.48.5.1676-1680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridderberg W., Wang M., Nørskov-Lauritsen N. Multilocus sequence analysis of isolates of Achromobacter from patients with cystic fibrosis reveals infecting species other than Achromobacter xylosoxidans. J Clin Microbiol. 2012;50:2688–2694. doi: 10.1128/JCM.00728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrado L., Brañas P., Orellana M.Á. Molecular characterization of Achromobacter isolates from cystic fibrosis and non–cystic fibrosis patients in Madrid, Spain. J Clin Microbiol. 2013;51:1927–1930. doi: 10.1128/JCM.00494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupont C., Michon A.L., Jumas-Bilak E., Nørskov-Lauritsen N., Chiron R., Marchandin H. Intrapatient diversity of Achromobacter spp. involved in chronic colonization of cystic fibrosis airways. Infect Genet Evol. 2015;32:214–223. doi: 10.1016/j.meegid.2015.03.012. [DOI] [PubMed] [Google Scholar]