Abstract
Background:
Campylobacter infections may lead to serious conditions, including septicemia or other invasive forms of the disease, which require rapid and accurate laboratory diagnosis and subsequently appropriate antimicrobial therapy. The aim of this study was to compare the species distribution and antimicrobial susceptibility pattern of Campylobacter spp. strains isolated from patients and food samples.
Methods:
Biochemical identification was performed on 15 clinical and 30 food isolates of Campylobacter recovered onto Brucella agar containing 5% sheep blood. PCR was carried out to confirm the identity of Campylobacter spp. using primers for cadF, hipO, and asp genes of Campylobacter. To determine antibiotic sensitivity of isolates, Kirby-Bauer assay was carried out using 16 different antibiotic discs.
Results:
PCR assay and biochemical tests confirmed all 45 isolates as Campylobacter: 20 (44.44%) as C. jujeni, 10 (22.22%) as C. coli, and 15 (33.34%) as other Campylobacter strains. The maximum resistance was observed to cefotaxime and imipenem (each 86.49%) and the maximum sensitivity to erythromycin (48.65%).
Conclusion:
C. jujeni is dominant among isolates from clinical and food samples. In addition, tetracycline remains the first-line therapeutic agent against Campylobacter infections in Iran.
Key Words: Campylobacter, Genetic diversity, Drug resistance
INTRODUCTION
Campylobacter spp. are motile, oxidase-positive microorganisms with spiral or corkscrew appearance belonging to a group of Gram-negative microaerophilic bacteria[1]. Many Campylobacter spp. have been reported to be implicated in human diseases, such as Campylo-bacteriosis, periodontitis, diarrhea etc.[2,3]. However, C. jejuni and C. coli are the most common isolates from human pathological samples, including enteritis[1,4]. In addition, C. fetus is also seen as an opportunistic pathogen in human[5].
Most estimates of incidence in developing countries are from laboratory-based surveillance of pathogens responsible for diarrhea. Isolation rates of Campylo-bacter in developing countries range from 5 to 20%[6,7]. Although in most of the cases Campylobacter infections are self-limiting, some serious conditions may happen, such as diarrhea, cramping, abdominal pain, and fever within two to five days after exposure to the organism. Diarrhea may be bloody and can be accompanied by nausea and vomiting. In persons with compromised immune systems, Campylobacter occasionally spreads to the bloodstream and causes a serious life-threatening infection[6]. Some people develop arthritis and others may develop a rare disease called Guillain-Barré syndrome that affects nerves of the body and begins several weeks after the diarrheal illness[8]. Accordingly, rapid and accurate identification of the causing strains and the selection of the most efficient therapeutics are required[9]. However, increasing antimicrobial resistance and different patterns of antibiotic susceptibility among different clinical and environmental isolates of Campylobacter have been frequently reported by some investigations[10,11]. For example, in the late 1980s, resistance to quinolones was increased in Asia and Europe, following the introduction and indiscriminate use of these drugs in livestock[12]. Interestingly, despite the widespread use of erythromycin, resistance of Campylobacter to this antibiotic has remained low in industrialized countries[8].
Biochemical and molecular methods are used for identification of Campylobacter spp. and strains. Many methods used to identify Campylobacter spp. are based on classic phenotypes of these bacteria, e.g. morphology, growth temperature, biochemical and serological reactions, and tolerance to higher temperatures[13-15]. Moreover, commercial kits such as enzyme immunoassay and ProSpecT Campylobacter Microplate Assay are available for direct and rapid identification of antigens from C. jejuni and C. coli in stool[16,17]. The aim of this study was to characterize Campylobacter spp. strains isolated from food and clinical samples using biochemical methods and PCR assay and to determine their antibiotic susceptibility patterns by disc diffusion test.
MATERIALS AND METHODS
Bacterial strains
The current study included 15 clinical and 30 food Campylobacter spp. isolates that were obtained during a 14-month period (June 2004- July 2005).The food isolates were collected from different areas of Tehran,
Iran, including shopping centers and retails. Clinical isolates were obtained from patients referred to one major hospital in Tehran. The isolates were stocked in skim milk medium containing 15% glycerol and preserved at -20°C until use.
Growth conditions and biochemical tests
Campylobacter strains were cultivated on Brucella agar containing 5% sheep blood, vancomycin, polymyxin B and trimethoprim (pH 7.2±0.2) at 42°C for 48 h. The cultivation was performed under microaerobic conditions provided by gas replacement method[18]. Gram staining, catalase and urease production, nitrate reduction[19], hippurate hydro-lysis[20], and indoxyl acetate hydrolysis[21] tests were used for biochemical identification and bio-typing of the strains.
Molecular characterization by PCR
PCR was carried out to detect Campylobacter species[22]. DNA was extracted using boiling method. Briefly, a clone of the bacteria was suspended in 200 μl sterile distilled water, boiled for 10 min and incubated at -20°C for 10 min. It was then centrifuged at 24148 ×g at room temperature for 10 min and the supernatant was used as template DNA in PCR reaction.
PCR amplification was performed using primers specific for different genes, Campylobacter adhesion genes: fibronectin gene (cadF) for detection of Campylobacter genus, hippuricase or benzoylglycine amidohydrolase gene (hipO) for C. jejuni, and aspartokinase or aspartate kinase gene (asp) for C. coli (Table 1). For PCR, 0.3 μl dNTP (25 mM), 0.2 μl Taq polymerase (5 unit/μl), 0.6 μl MgCl2 (50 mM), 5 μl DNA template, 0.25 μl primer-F (100 pM), 0.25 μl primer-R (100 pM), and 2.5 μl PCR buffer (10×) were mixed and brought to a volume of 25 μl using distilled water.
Table 1.
Characteristics of the primers used for detection of Campylobacter isolates by PCR
| Gene | Primer | Sequence | Length (nt) | Band (bp) |
|---|---|---|---|---|
| cadF | Forward Reverse |
5'-TTGAAGGTAATTTAGATATG-3' 5'-CTAATACCTAAAGTTGAAAC-3' |
20 20 |
400 |
| hipO | Forward Reverse |
5'-GAAGAGGGTTTGGGTGGTG-3' 5'-AGCTAGCTTCGCATAATAACTTG-3' |
19 23 |
735 |
| asp | Forward Reverse |
5'-GGTATGATTTCTACAAAGCGAG-3' 5'-ATAAAAGACTATCGTCGCGTG-3' |
22 21 |
500 |
bp, base pair; nt, nucleotide
The final volume was subjected to PCR amplification within a thermal cycler (Eppendorf, Germany) with the initial denaturation at 95°C for 5 min (for all three genes), followed by 30 amplification cycles. The amplification cycles included denaturation at 95°C for 45 seconds (for all three genes), annealing at 43°C (cadF), 55°C (hipO) and 52°C (asp) for 1 min, extension at 72°C for 1 min (for all three genes), and the final extension at 72°C for 5 min.
Afterwards, 8 μl PCR amplicons together with 2 μl loading buffer were run on 1% agarose gel in 1× Tris-Borate-EDTA buffer at 95 V for 2 h. Finally, the electrophoresis gel was stained in ethidium bromide solution (10 μg/ml), and visualized at wavelength of 590 nm under an ultraviolet trans-illuminator (Medox Biotech, India).
Antibiotic susceptibility test
Antibiotic susceptibility assay was performed using Kirby-Bauer disk diffusion test[23]. For this purpose, 16 antibiotic disks were chosen (μg/disc), including erythromycin (15), azithromycin (15), gentamicin (10), ampicillin (30), tetracycline (5), imipenem (10), ciprofloxacin (5), nalidixic acid (30), kanamycin (5), chloramphenicol (30 μg/disc), streptomycin (30), cefotaxime (30), trimethoprim (30), cefepime (30), tobramycin (10), and amikacin (30), which all were purchased from MAST Company, UK. The susceptibility of the bacteria to each antibiotic was determined according to the latest guidelines published by the Clinical and Laboratory Standards Institute.
RESULTS
Biochemical identification of isolates
Results of Gram staining, nitrate reduction, catalase and urease production, and indoxyl acetate hydrolysis tests revealed that all 45 suspected isolates belonged to
Campylobacter spp. Among them, the hippurate hydrolysis test identified 20 isolates (44.44%) as C. jujeni and 10 isolates (22.22%) as C. coli. The remaining 15 (33.34%) Campylobacter spp. isolates belonged to other species.
Molecular characterization by PCR
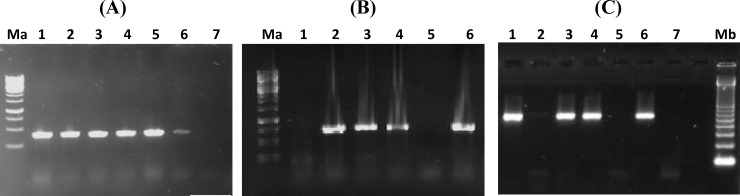
The result of PCR assay confirmed the biochemical test results for detection of Campylobacter spp. and related species. Using cadF primers, all isolates revealed a 400-bp band on electrophoresis, which indicated the presences of Campylobacter spp. (Fig. 1A). Amplification with hipO primers revealed 20 C. jujeni among the isolates (Fig. 1B). In addition, a 500-bp band of asp gene was detected in 10 isolates corresponding to the presence of C. coli (Fig. 1C).
Fig. 1.
(A) PCR amplification of cadF (400 bp): Lane1, positive control (C. jejuni ATCC 29428), lanes 2-6, patient or food samples with positive results; lane7, negative control. (B) PCR amplification of hipO (735 bp): Lane1, negative control, lane 2, positive control (C. jejuni ATCC 29428), lanes 3-6, patient or food samples. (C) PCR amplification of asp (500 bp): lane1, positive control (C. coli ATCC 43478); lanes 2-6, patient or food samples; lane 7, negative control. Ma, 1 kb DNA ladder (Fermentas, USA) and Mb, 100 bp DNA ladder (Fermentas, USA). Negative control is DNA from Escherichia coli ATCC 25922
Antibiotic sensitivity test
Antibiotic sensitivity testing was performed on clinical and food isolates of Campylobacter spp. Disregarding the source of isolates, maximum resistance was observed to cefotaxime and imipenem, each seen among 86.49% of the isolates. In addition, the maximum sensitivity belonged to erythromycin, observed among 48.65% of the isolates. Among the strains isolated from food samples, the maximum resistance was seen to imipenem (90.9%), and the maximum sensitivity to chloramphenicol (40.9%). Moreover, among the isolates from clinical samples, the maximum resistance belonged to cefotaxime and trimethoprim (each, 93.34%), and the maximum sensitivity to tobramycin (46.66%). Comparisons of the susceptibility data among Campylobacter strains isolated from clinical and food samples were performed by SPSS software (version 19), and no significant difference was observed between the two groups (P>0.05).
DISCUSSION
Due to the problems associated with the application of biochemical and biological methods for identification of Campylobacter strains, there are overwhelming intricacies about characterization and epidemiological studies of these bacteria. In the present study, we used both biochemical and molecular methods for detection of Campylobacter isolates from food and clinical samples. Although our results revealed no difference between the sensitivity of PCR assay and that of biochemical tests for species identification of Campylobacter isolates, many advantages have been suggested for molecular approaches in making differentiation between various strains of the bacteria. First, the accuracy and speed of molecular diagnostics are higher than biochemical methods with no need to long incubation at different temperatures. Second, molecular techniques are easier and less costly than biochemical methods, and they are more suitable for epidemiological studies.
Using PCR assay, we found that among 45 studied isolates, almost 45% were C. jujeni and 23% C. coli, and the remaining belonged to other Campylobacter species. Given the clinical significance of C. jujeni and C. coli, our overall focus was on these two species. These findings are in accordance with previous studies that have shown the dominance of C. jujeni over C. coli[24-26]. For instance, Denis et al.[24]. observed that among 513 isolates from chickens, 61.5% were C. jujeni and 38.5% C. coli. In addition, Fitzgerald et al.[25] in an investigation on isolates from farm and clinical environments suggested that the higher frequency of C. jujeni over the C. coli may be due to the extensive colonization of the C. jujeni in a vast range of hosts living as commensalism. Moreover, Manfreda et al.[27] found that C. jujeni is dominant among isolates in cold seasons, while C. coli is more frequent in warm seasons as a thermo-tolerant species; however, in the current study, the overall dominance is attributed to the C. jujeni, regardless of its seasonality.
In the present study, disc diffusion test was performed to determine the resistance pattern of isolates to antibiotics belonging to aminoglycosides, quinolones, beta-lactams, sulfonamides, and cephalo-sporins family of antibiotics according to the globally accepted standard criteria. We observed that the maximum resistance belonged to cefotaxime and imipenem and then to nalidixic acid, trimethoprim, ampicillin, ciprofloxacin, tetracycline, streptomycin, and kanamycin, respectively. We also found that the maximum sensitivity was to erythromycin and then to chloramphenicol, gentamicin, azithromycin, and cefepime, respectively. Senok and colleagues[28] also indicated a high degree of erythromycin sensitivity and ciprofloxacin resistance among C. jejuni isolates of human and poultry origin. Our study shows a high resistance to nalidixic acid and ciprofloxacin and low resistance to azithromycin and gentamicin. Antibacterial susceptibility test by Lehtopolku and coworkers[29] on 1808 isolates isolated between 2003-2005 also showed high resistance to nalidixic acid (41.4% C. jejuni and 83.3 C. coli strains) and ciprofloxacin (42.4% C. jujeni and 83.3 C. coli strains) as well as low resistance to azithromycin (5% C. jejuni and 38.9 C. coli strains) and gentamicin (0.9% C. jejuni and 0 C. coli strains). In addition, Dadi and Asrat[30] found the maximum susceptibility to erythromycin, chloramphenicol, amoxicillin, and the maximum resistance to ampicillin, gentamicin, tetracycline, streptomycin and kanamycin. Our findings are consistent with those of Dadi and Asrat[30] with the exception of higher susceptibility to gentamicin that was observed in our study.
Moreover, Oza and colleagues[31] reported the lowest resistance to ciprofloxacin (3%), which is in contrary with our findings. High resistance to ciprofloxacin in the present study may be due to the fact that fluoroquinolones such as ciprofloxacin are frequently used for treatment of campylobacteriosis because of their broad spectrum of activity against enteric pathogens[32]. Furthermore, Oza and colleagues[31] observed the susceptibility to erythromycin, gentamicin, and chloramphenicol in more than 99% of human or poultry isolates of Campylobacter spp., whereas maximum antimicrobial resistance was seen for ampicillin, nalidixic acid and tetracycline, respectively. Thus, our findings together with the previous reports support the continued use of erythromycin and chloramphenicol as first-line therapy for enteritis of Campylobacter spp. in Iran and other countries encountering campylobacteriosis.
In the present investigation, the maximum resistance was observed to cefotaxime and imipenem (each 86.49%). In an earlier study by Tajada and colleagues[33], all strains were susceptible to imipenem. Likewise, in another study among clinical C. coli and C. jejuni strains, imipenem was highly effective against multidrug resistance campylobacter[29]. Also, in a study in Kuwait during 2002-2010, 97 C. jejuni isolates were investigated, and no resistance to imipenem was observed[34]. High resistance to imipenem, which was observed among our isolates, makes further observation and tracking necessary for finding the exact molecular basis of such high resistance.
Tetracycline resistance was high among our isolates (78.38%). Albert[34] and Gallay et al.[35] showed that resistance to tetracycline has a tendency to increase from 2003-2010 and 1999-2004, respectively. The high rate of tetracycline resistance may be due to tetO, which is a plasmid encoding gene introduced to be responsible for tetracycline resistance in Campylo-bacter spp.[36].
In conclusion, we found that the isolation frequency of C. jujeni in both clinical and food specimens is higher than that of C. coli. Our findings indicated that the Campylobacter has the highest resistance to ciprofloxacin, nalidixic acid, and tetracycline as well as the least resistance to erythromycin and chlor-amphenicol, which still suggest the two latter as being first-line therapeutic agents against Campylobacter infections in Iranian clinics.
ACKNOWLEDGMENTS
We thank the Research Council of Tarbiat Modares University, Tehran, Iran for financially supporting the project.
CONFLICT OF INTEREST. None declared.
References
- 1.Moore JE, Corcoran D, Dooley JS, Fanning S, Lucey B, Matsuda M, McDowell DA, Mégraud F, Millar BC, O'Mahony R, O'Riordan L, O'Rourke M, Rao JR, Rooney PJ, Sails A, Whyte P. Campylobacter. Veterinary research. 2005;36(3):351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- 2.Humphrey T, O'Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. International journal of food microbiology. 2007;117(3):237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Zeigler M, Claar C, Rice D, Davis J, Frazier T, Turner A, Kelly C, Capps J, Kent A, Hubbard V, Ritenour C, Tuscano C, Shultz ZQ, Leaumont CF. Outbreak of campylobacteriosis associated with a long-distance obstacle adventure race-Nevada, October 2012. Morbidity and mortality weekly report. 2014;63(17):375–378. [PMC free article] [PubMed] [Google Scholar]
- 4.Vellinga A, Van Loock F. The dioxin crisis as experiment to determine poultry-relat Campylobacter enteritis. Emerginginfectious diseases. 2002;8(1):19–22. doi: 10.3201/eid0801.010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauerwein RW, Bisseling J, Horrevorts AM. Septic abortion associated with Campylobacter fetus subspecies fetus infection: case report and review of the literature. Infection. 1993;21(5):331–333. doi: 10.1007/BF01712458. [DOI] [PubMed] [Google Scholar]
- 6.Oberhelman RA, Taylor DN. Campylobacter infections in developing countries. Campylobacter. 2nd ed. Washington, DC: ASM Press; [Google Scholar]
- 7.Ghorbanalizadgan M, Bakhshi B, Lili AK, Najar-Peerayeh S, Nikmanesh B. Molecular survey of Campylobacter jejuni and Campylobacter coli virulence and diversity. Iranian biomedical journal. 2014;18(3):158–164. doi: 10.6091/ibj.1359.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coker AO, Isokpehi RD, Thomas BN, Amisu KO, Obi CL. Human campylobacteriosis in developing countries. Emerging infectious diseases. 2002;8(3):237–244. doi: 10.3201/eid0803.010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eiland LS, Jenkins LS. Optimal treatment of campylobacter dysentery. The journal of pediatric pharmacology and therapeutics. 2008;13(3):170–174. doi: 10.5863/1551-6776-13.3.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deckert AE, Reid-Smith RJ, Tamblyn SE, Morrell L, Seliske P, Jamieson FB, Irwin R, Dewey CE, Boerlin P, Mcewen SA. Antimicrobial resistance and antimicrobial use associated with laboratory-confirmed cases of campylobacter infection in two health units in Ontario. TheCanadian journal of infectious diseases and medical microbiology. 2013;24(1):e16–21. doi: 10.1155/2013/176494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez-Boto D, Garcia-Pena FJ, Abad-Moreno JC, Echeita MA. Antimicrobial susceptibilities of Campylobacter jejuni and Campylobacter coli strains isolated from two early stages of poultry production. Microbial drug resistance. 2013;19(4):323–330. doi: 10.1089/mdr.2012.0160. [DOI] [PubMed] [Google Scholar]
- 12.Endtz HP, Ruijs GJ, van Klingeren B, Jansen WH, van der Reyden T, Mouton RP. Quinolone resistance in campylo-bacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. The journal of antimicrobial chemotherapy. 1991;27(2):199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- 13.Oza AN, Thwaites RT, Wareing DR, Bolton FJ, Frost JA. Detection of heat-stable antigens of Campylobacter jejuni and C. coli by direct agglutination and passive hemagglutination. The journal of clinical microbiology. 2002;40(3):996–1000. doi: 10.1128/JCM.40.3.996-1000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Procop GW. Gastrointestinal infections. Infection disease clinics North America. 2001;15(4):1073–1108. doi: 10.1016/s0891-5520(05)70187-2. [DOI] [PubMed] [Google Scholar]
- 15.Totten PA, Patton CM, Tenover FC, Barrett TJ, Stamm WE, Steigerwalt AG, Lin Jy, Holmes KK, Brenner DJ. Prevalence and characterization of hippurate-negative Campylobacter jejuni in King County, Washington. The journal of clinical microbiology. 1987;25(9):1747–1752. doi: 10.1128/jcm.25.9.1747-1752.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindiyeh M, Jense S, Hohmann S, Benett H, Edwards C, Aldeen W, Croft A, Daly J, Mottice S, Carroll KC. Rapid detection of Campylobacter jejuni in stool specimens by an enzyme immunoassay and surveillance for Campylobacter upsaliensis in the greater salt lake city area. The journal of clinical microbiology. 2000;38(8):3076–3079. doi: 10.1128/jcm.38.8.3076-3079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dediste A, Vandenberg O, Vlaes L, Ebraert A, Douat N, Bahwere P, Butzler JP. Evaluation of the ProSpecT microplate assay for detection of campylobacter: a routine laboratory perspective. Clinical microbiology and infection. 2003;9(11):1085–1090. doi: 10.1046/j.1469-0691.2003.00705.x. [DOI] [PubMed] [Google Scholar]
- 18.Arimi SM, Fricker CR, Park RW. Occurrence of 'thermophilic' campylobacters in sewage and their removal by treatment processes. Epidemiology and infection. 1988;101(2):279–286. doi: 10.1017/s0950268800054194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sellars MJ, Hall SJ, Kelly DJ. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. The journal of bacteriology. 2002;184(15):4187–4196. doi: 10.1128/JB.184.15.4187-4196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris GK, el Sherbeeny MR, Patton CM, Kodaka H, Lombard GL, Edmonds P, Hollis DG, Brenner DJ. Comparison of four hippurate hydrolysis methods for identification of thermophilic campylobacter spp. The journal of clinical microbiology. 1985;22(5):714–718. doi: 10.1128/jcm.22.5.714-718.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodge DS, Borczyk A, Wat LL. Evaluation of the indoxyl acetate hydrolysis test for the differentiation of campylobacters. The journal of clinical microbiology. 1990;28(6):1482–1483. doi: 10.1128/jcm.28.6.1482-1483.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Amri A, Senok AC, Ismaeel AY, Al-Mahmeed AE, Botta GA. Multiplex PCR for direct identification of campylobacter spp. in human and chicken stools. The journal of medical microbiology. 2007;56(10):1350–1355. doi: 10.1099/jmm.0.47220-0. [DOI] [PubMed] [Google Scholar]
- 23. http://shop.clsi.org/ microbiology-documents/ M02-M100-PK.html.
- 24.Denis M, Rose V, Huneau-Salaun A, Balaine L, Salvat G. Diversity of pulsed-field gel electrophoresis profiles of Campylobacter jejuni and Campylobacter coli from broiler chickens in France. Poultry science. 2008;87(8):1662–1671. doi: 10.3382/ps.2008-00010. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald C, Stanley K, Andrew S, Jones K. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Applied and environmental microbiology. 2001;67(4):1429–1436. doi: 10.1128/AEM.67.4.1429-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivoal K, Ragimbeau C, Salvat G, Colin P, Ermel G. Genomic diversity of Campylobacter coli and Campylobacter jejuni isolates recovered from free-range broiler farms and comparison with isolates of various origins. Applied and environmental microbiology. 2005;71(10):6216–6227. doi: 10.1128/AEM.71.10.6216-6227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manfreda G, De Cesare A, Bondioli V, Stern NJ, Franchini A. Enumeration and identity of Campylobacter spp. in Italian broilers. Poultry science. 2006;85(3):556–562. doi: 10.1093/ps/85.3.556. [DOI] [PubMed] [Google Scholar]
- 28.Senok A, Yousif A, Mazi W, Sharaf E, Bindayna K, Elnima el A, Botta G. Pattern of antibiotic susceptibility in Campylobacter jejuni isolates of human and poultry origin. the japanese journal of infectious disease. 2007;60(1):1–4. [PubMed] [Google Scholar]
- 29.Lehtopolku M, Nakari UM, Kotilainen P, Huovinen P, Siitonen A, Hakanen AJ. Antimicrobial susceptibilities of multidrug-resistant Campylobacter jejuni and C. coli strains: in vitro activities of 20 antimicrobial agents. Antimicroial agents and chemotherapy. 2010;54(3):1232–6. doi: 10.1128/AAC.00898-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dadi L, Asrat D. Prevalence and antimicrobial susceptibility profiles of thermotolerant Campylobacter strains in retail raw meat products in Ethiopia. Ethiopian journal of health development. 2008;22(2):195–200. [Google Scholar]
- 31.Oza AN, McKenna JP, McDowell SW, Menzies FD, Neill SD. Antimicrobial susceptibility of Campylobacter spp. isolated from broiler chickens in Northern Ireland. the journal of antimicrobial and chemotherapy. 2003;52(2):220–223. doi: 10.1093/jac/dkg333. [DOI] [PubMed] [Google Scholar]
- 32.Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clinical infectious diseases. 2001;32(8):1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 33.Tajada P, Gomez-Graces JL, Alos JI, Balas D, Cogollos R. Antimicrobial susceptibilities of Campylobacter jejuni and Campylobacter coli to 12 beta-lactam agents and combinations with beta-lactamase inhibitors. Antimicrobial agents and chemotherapy. 1996;40(8):1924–1925. doi: 10.1128/aac.40.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albert MJ. In vitro susceptibility of Campylobacter jejuni from Kuwait to tigecycline & other antimicrobial agents. Indian journal of medical research. 2013;137(1):187–190. [PMC free article] [PubMed] [Google Scholar]
- 35.Gallay A, Prouzet-Mauleon V, Kempf I, Lehours P, Labadi L, Camou C, Denis M, de Valk H, Desenclos JC, Mégraud F. Campylobacter antimicrobial drug resistance among humans, broiler chickens, and pigs, France. Emerging infectious diseases journal. 2007;13(2):259–266. doi: 10.3201/eid1302.060587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wardak S, Szych J, Zasada AA, Gierczyński R. Antibiotic resistance of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. Antimicrobial agents and chemotherapy. 2007;51(3):1123–115. doi: 10.1128/AAC.01187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]