Summary
Acute myeloid leukemia (AML) is characterized by recurrent mutations that affect the epigenetic regulatory machinery and signaling molecules, leading to a block in hematopoietic differentiation. Constitutive signaling from mutated growth factor receptors is a major driver of leukemic growth, but how aberrant signaling affects the epigenome in AML is less understood. Furthermore, AML cells undergo extensive clonal evolution, and the mutations in signaling genes are often secondary events. To elucidate how chronic growth factor signaling alters the transcriptional network in AML, we performed a system-wide multi-omics study of primary cells from patients suffering from AML with internal tandem duplications in the FLT3 transmembrane domain (FLT3-ITD). This strategy revealed cooperation between the MAP kinase (MAPK) inducible transcription factor AP-1 and RUNX1 as a major driver of a common, FLT3-ITD-specific gene expression and chromatin signature, demonstrating a major impact of MAPK signaling pathways in shaping the epigenome of FLT3-ITD AML.
Graphical Abstract
Highlights
-
•
FLT3-ITD signaling is associated with a common gene expression signature
-
•
FLT3-ITD-specific gene expression is associated with a common chromatin signature
-
•
FLT3-ITD AML displays chronic activation of the inducible transcription factor AP-1
-
•
AP-1 cooperates with RUNX1 to shape the epigenome of FLT3-ITD AML
Cauchy et al. identify a specific gene expression and regulatory signature associated with aberrant signaling in acute myeloid leukemia with FLT3-ITD mutations. In FLT3-ITD AML, the inducible transcription factor AP-1 is chronically activated and cooperates with RUNX1, shaping the epigenome to transactivate specific target genes.
Introduction
In cancer, genes encoding signaling molecules and transcription factors (TFs) can become mutated, thereby leading to changes in homeostatic gene regulatory networks, and a block in the normal program of terminal differentiation. This is also true for acute myeloid leukemia (AML). AML is a heterogeneous disease that is caused by multiple mutations that affect normal blood cell development from hematopoietic stem cells (HSCs) and prime developing cells to become malignant. However, during the progression of the leukemic state, pre-leukemic and leukemic cells undergo clonal evolution creating heterogeneous cell populations with different functional properties. In recent years it has become clear that the majority of initial mutations occur in genes encoding transcriptional and epigenetic regulators (Corces-Zimmerman et al., 2014, Corces-Zimmerman and Majeti, 2014). A few studies have examined in a system-wide fashion how leukemic TFs reprogram chromatin and establish specific epigenetic signatures and gene expression programs (Prange et al., 2014). An example for this notion is the case of AML with t(8;21) chromosomal translocation, which gives rise to the fusion protein RUNX1/ETO. This aberrant TF competes with RUNX1 for its binding motifs, leading to epigenetic reprogramming with a block in myeloid differentiation and the concomitant activation of a precursor-like AML-specific transcriptional network (Martens et al., 2012, Ptasinska et al., 2012, Ptasinska et al., 2014). However, in the case of karyotypically normal (KN) AML, it has proved to be much harder to identify consistent patterns of deregulation associated with specific mutations. This difficulty is due to (1) the heterogeneous nature of the combinations of underlying mutations and (2) the fact that AML cells often span a range of differentiation states. There is the added complexity that mutations in genes encoding signaling molecules tend to appear late in the process of leukemogenesis, making it harder to definitively link them to consistent target gene alteration (Corces-Zimmerman et al., 2014). Some of the most frequent secondary mutations found in AML are in tyrosine kinase receptors such as FLT3 and KIT, which are rendered constitutively active (Masson and Rönnstrand, 2009). These receptors normally control the regulated growth and survival of myeloid progenitor cells. Mutations deregulate this process, thus effectively cutting off cancer cells from normal growth regulating controls. However, we currently know very little about how mutations in signaling molecule genes impact on gene expression and whether they also establish a common epigenetic signature.
To address this question, we focused on the identification of changes in the chromatin landscape and patterns of gene expression that are driven by constitutive activation of intracellular signaling pathways and concentrated on KN-AML with internal tandem duplications (ITDs) in the FLT3 transmembrane domain as a paradigm. FLT3-ITD mutations are found in ∼25% of KN-AML and lead to constitutive activation of the downstream mitogen-activated protein kinase (MAPK), AKT, and signal transducer and activator of transcription (STAT) signaling pathways linked to FLT3 (Stirewalt and Radich, 2003, Thiede et al., 2002). FLT3-ITD frequently acts as a partner in AML with other mutations such as those in transcription factors, epigenetic regulators, and Nucleophosmin (NPM1). Tyrosine kinase receptor signaling has a direct influence on multiple signaling pathways (Gu et al., 2011, Scholl et al., 2008) and the activities of many TFs (Goyama et al., 2014, Yordy and Muise-Helmericks, 2000), but could also signal to chromatin directly (Badeaux and Shi, 2013, Dawson et al., 2009, Ray et al., 2013). Our goal was therefore to examine whether the presence of the FLT3-ITD leads to the establishment of a common epigenetic signature. To this end we mapped open regions of chromatin that exist as DNase I hypersensitive sites (DHSs) as this identifies active regulatory elements (Cockerill, 2011). Using complementary genome-wide analyses of gene expression and regulation in primary AML cells, we show that the FLT3-ITD mutation is associated with extensive changes in the epigenetic landscape and report a FLT3-ITD-specific gene expression signature that is associated with FLT3-ITD-specific DHSs. The integration of these global data reveals a cooperation between the MAPK inducible transcription factor AP-1 and RUNX1 as two of the main drivers of a FLT3-ITD-specific open chromatin signature.
Results
System-wide Genomic and Epigenomic Analysis of Karyotypically Normal AML
In order to define the specific genomic targets of aberrant signaling in KN AML with the FLT3-ITD mutation, we purified undifferentiated CD34 or CD117 positive cells, or obtained a mononuclear fraction comprising greater than 92% undifferentiated blast cells, either from peripheral blood or from bone marrow of AML patients. This protocol avoided confounding issues associated with contamination by differentiated cells expressing markers unrelated to leukemogenesis. Profiles from AML cells were compared to those of the CD34+ progenitor population of mobilized peripheral blood stem cells (PBSCs). To identify specific genetic and epigenetic signatures, we performed a comprehensive set of genome-wide analyses that included (1) global mRNA microarray analysis to identify aberrantly regulated genes, (2) a DNA sequence screen for exonic mutations within 55 candidate myeloid oncogenes and tumor suppressor genes to uncover pathways responsible for aberrant gene expression patterns (Table S1), (3) global DNase sequencing (DNase-seq) mapping of DHS patterns to identify aberrantly activated (and absent) cis-regulatory elements, (4) identification of regions within DHSs protected from DNase I digestion (digital footprints), and the underlying specific motifs bound by TFs, (5) chromatin immunoprecipitation sequencing (ChIP-seq) assays to detect specific proteins bound at DHSs, and (6) DNA methylation array analysis of 450,000 CG sites to detect epigenetically silenced loci. These analyses included 19 KN AML samples, of which 9 were from FLT3-ITD+ patients. Because the exon sequencing screen was unable to detect all of the FLT3-ITD mutations, which vary greatly in size and location, we confirmed the predicted FLT3-ITD status by PCR analyses (Figure 1A), which indicated that the ITD had occurred on both alleles in three of the samples (ITD3, ITD5, and ITD7), while ITD4 had two different forms of the ITD mutation. Data File S1 describes all of the mutations in all our samples, which were found in 21 different genes, plus their allele frequencies. Most of these mutations and FLT3 PCR products were detected at levels close to 50% or 100%, indicating that these samples were clonal and that some mutations such as those in JAK2 were bi-allelic. Of the nine ITD+ patients, one sample had no other defined mutations (ITD1), and this became the prototype ITD+ sample anchoring this study. Three of the patients with wild-type (WT) FLT3 had mutations in genes encoding other signaling molecules linked to FLT3 signaling pathways, which included NRAS, PTN11, SOCS1, and JAK2, which might therefore share some common features with FLT3-ITD+ AML. SOCS1 mutations are also known to cooperate with FLT3-ITD in AML (Reddy et al., 2012).
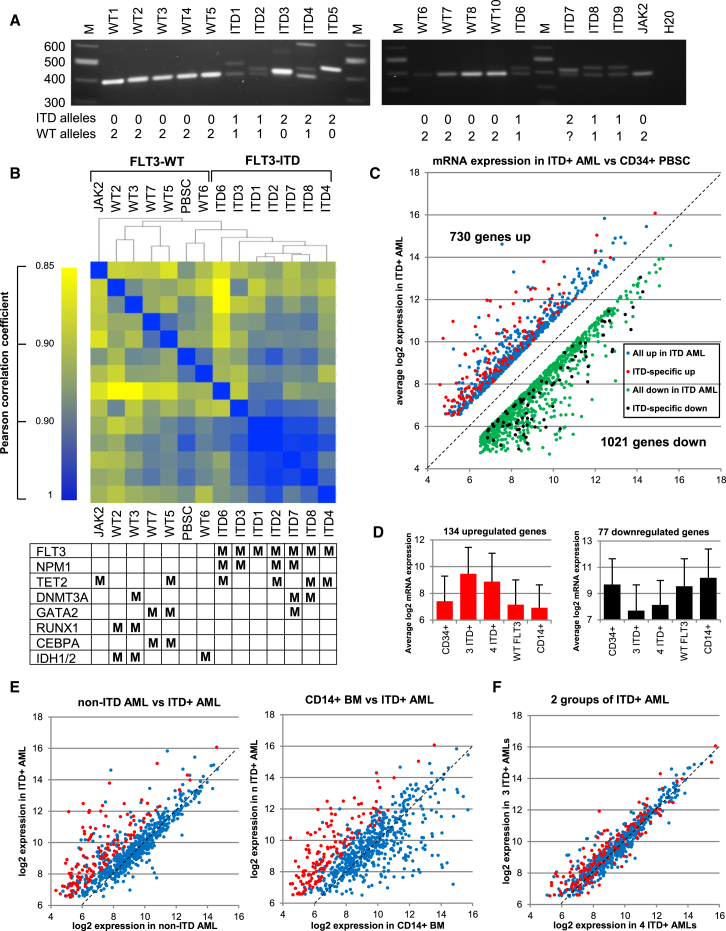
Figure 1.
FLT3-ITD+ AML Displays a Characteristic mRNA Expression Profile
(A) PCR amplification of the transmembrane coding region of the FLT3 gene used to identify ITD mutations. On the left are the sizes of the DNA marker bands (M). Below are the estimated numbers of normal and mutated FLT3 alleles.
(B) Hierarchical clustering of Pearson correlation coefficients of mRNA values. For each sample the mutations present in the most commonly mutated genes are indicated as M in the table underneath.
(C–F) Identification of genes that differ by at least 2-fold in a comparison of the average log2 mRNA microarray values for a core group of three ITD+ AML samples (ITD1, ITD2, and ITD3) to the equivalent values determined from either the average of two independent PBSC samples (C), the average of four WT FLT3 AML samples (WT2, WT3, WT5, and WT7), for CD14+ bone marrow cells (E), or a second group of ITD+ AML samples (ITD4, ITD6, ITD7, and ITD8) used here for validation of the ITD+ pattern (F). The 134 genes that are consistently expressed at 2-fold higher levels in ITD+ AML in each of the three comparisons in (C) and (E) are shown in red, and the remaining genes that are upregulated in at least (C) are shown in blue. In (C) we also highlight 77 genes with values that are consistently 2-fold lower in ITD+ AML compared to each of PBSCs, WT FLT3 AML, and CD14+ cells in black, and the remaining genes that are also downregulated at least in (B) are shown in green. (D) Average log2 mRNA values for the specific groups of 134 upregulated and 77 downregulated genes in each of the 5 groups used in this analysis, with the SD for mRNA expression shown as error bars.
AML with FLT3-ITD Displays a Specific Gene Expression Profile
We used microarray analysis to define mRNA levels for ∼22,000 genes for AML samples and in 2 independent samples of CD34+ PBSCs. Hierarchical clustering of the Pearson correlation coefficients was then used to identify similarities and differences between these AMLs (Figure 1B). This analysis included seven FLT3-ITD+ AMLs, five AMLs with wild-type (WT) FLT3 (and no other known receptor mutations), and one AML with an activating JAK2 mutation (and WT FLT3). The seven ITD+ AMLs formed a discrete cluster with high correlation coefficients, while the five WT FLT3 AMLs clustered more closely with the PBSCs, and the JAK2 mutated AML had a pattern distinctly different to all the other samples. The parallel mutation analysis revealed that mutations in the epigenetic regulators DNMT3A and TET2 were divided between the FLT3-ITD+ and WT FLT3 groups, and mutations in the genes encoding the TFs RUNX1, GATA2, and C/EBPα occurred predominantly in the WT FLT3 AMLs. NPM1 mutations, which frequently partner FLT3-ITD, were restricted to the FLT3-ITD+ AML cluster, but could not alone account for the observed patterns as they did not independently cluster together.
To identify a set of genes specifically deregulated in FLT3-ITD+ AML, we focused on the most related samples from each group and independently analyzed expression data from three core ITD+ AMLs (ITD1, ITD2, and ITD3) and four core WT FLT3 AMLs (WT2, WT3, WT5, and WT7), for which we also had accompanying DHS data. The three ITD+ AMLs included two with mutated NPM1 (ITD2 and ITD3) plus ITD1 with no other defined mutations (Data File S1). We compared the average log2 mRNA microarray values with those for CD34+ PBSCs. As an additional control and to assess the stage of the differentiation block, we also compared these values with those from normal bone-marrow derived CD14+ myeloid cells. We used this second criterion to assist in the identification of artifacts arising from high levels of gene expression within minor subsets of more differentiated monocytic cells.
Within the core group of 3 ITD+ AMLs, we identified 730 upregulated genes with average log2 values at least 1.0 above PBSCs, (and absolute log2 values of at least 6.5 to select genes at least 1 log above background), plus 1,021 downregulated genes with values at least 1.0 below that of PBSCs (blue and green dots, Figure 1C). However, only 134 of the upregulated genes and 77 of the downregulated genes maintained the same specific pattern of deregulation relative to both the WT FLT3 AML subset and the CD14+ve cells (Figures 1C–1E, represented by red and black dots). Note that some of the 730 genes upregulated in ITD+ cells were expressed at very high levels in CD14+ cells, meaning that the elevated values may have arisen from a minor contamination with more differentiated cells. The 134 ITD-specific genes were on average expressed at levels 4-fold higher than in PBSCs, WT FLT3 AML, and CD14+ve cells (Figure 1D). Conversely, the 77 downregulated genes were on average expressed at levels 4-fold lower than the other three cell types. We confirmed the generality of these observations by demonstrating that the gene expression profile for the genes upregulated in the core ITD+ group was essentially identical to the average mRNA profile observed for an additional group of four other ITD+ AMLs (ITD4, ITD6, ITD7, and ITD8) (Figure 1F). These analyses highlighted the need for more than one reference cell type when assessing expression patterns to identify true FLT3-ITD responsive genes. Our data also suggest that the block in differentiation in the FLT3-ITD+ AML occurs further down the myeloid differentiation pathway as indicated by the presence of the many genes expressed at levels similar to those in more mature CD14+ve cells (and the WT FLT3 AML), but higher than in PBSCs.
The average values for mRNA expression for the above two groups of deregulated genes are listed in Data File S2. They include upregulated genes that could influence the development of AML cells such as genes encoding (1) the transcriptional regulators FOXC1, PRDM16 (MEL1), and NFIX, (2) growth factor receptor genes such as IL2RA, IL3RA, TNFSF9, TNFRSF4, and TNFRSF18, (3) additional genes influencing growth and survival such as CCNA1 (Cyclin A), PTP4A3 (PRL3), and IGFBP2, (4) genes controlling differentiation such as the HOXB2 to HOXB6 cluster, and PBX3, (5) genes controlling homing such as the chemokine genes CCL1 and CCL5, and (6) genes for proteases that are known to remodel tissues and enhance migration such as CTSG, ADAMTS14, MMP15, and (7) a gene cluster that includes the protease genes ELANE, PRTN3, and AZU1 as well as LPPR3. The upregulated class of genes also contained other potentially important genes regulating signaling and migration such as AK2 (adenylate cyclase), DSC2 (a protein involved in cell adhesion), and the gene encoding the macrophage scavenger receptor SCARA3, which may be related to the fact that FLT3-ITD cells increase ROS production (Sallmyr et al., 2008). FOXC1 encodes a pioneer-type transcription factor (Zaret and Carroll, 2011). PRDM16 is related to the cancer-promoting histone methyl transferase gene EVI1, and both EVI1 and PRDM16 are activated via RAS mutations in models of myeloid leukemia (Wolf et al., 2013).
The expression patterns of seven of these upregulated genes, plus TBP as a control, are shown in Figure S1A for all of the samples analyzed. Within the core cluster of three ITD+ AMLs, FOXC1 was the only FOX gene significantly upregulated in ITD+ AML compared to the cluster of four WT FLT3 AMLs, while FOXO1 expression was downregulated (Figure S1B). Most notable of the 77 downregulated genes was a group of 10 HLA class II genes that were consistently downregulated in ITD+ AML and not in WT FLT3 AML (Figure S1C).
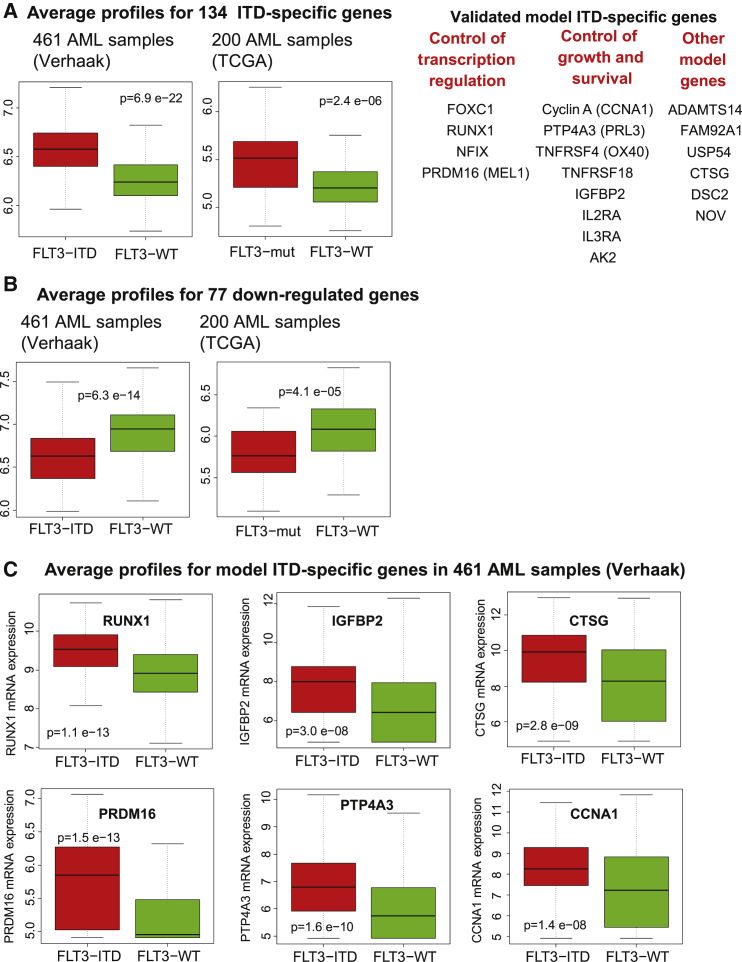
To validate the conclusions of our mRNA expression analysis, we analyzed data from two published studies of much larger cohorts of AML samples (Figure 2). For the ITD target genes identified above, we calculated the average mRNA levels for data from (1) a study of 461 AML patients comparing FLT3 ITD+AML samples with WT FLT3 AML samples (Verhaak et al., 2009) and (2) a study of 200 patients from The Cancer Genome Atlas (TCGA) Research Network comparing all samples with any FLT3 mutations with WT FLT3 AML samples (Cancer Genome Atlas Research Network, 2013). This confirmed that on average the above 134 upregulated genes and 77 downregulated genes displayed the same behavior in these much larger studies (Figures 2A and 2B). Several genes controlling gene expression or cell growth were confirmed as upregulated ITD target genes (Figure 2A), including RUNX1, IGFBP2, PRDM16, PTP4A3, and CCNA1 (Figure 2C).
Figure 2.
FLT3-ITD Consistently Deregulates the Same Genes in AML
(A–C) Average mRNA microarray values for FLT3-ITD target genes in 2 datasets obtained from 461 (Verhaak et al., 2009) or 200 (Cancer Genome Atlas Research Network, 2013) AML samples.
(A) Average profiles for 134 upregulated genes, with a subset of validated genes listed on the right.
(B) Average profiles for 77 downregulated genes.
(C) Profiles for individual upregulated genes.
AML with FLT3-ITD Displays a Distinct Chromatin Signature
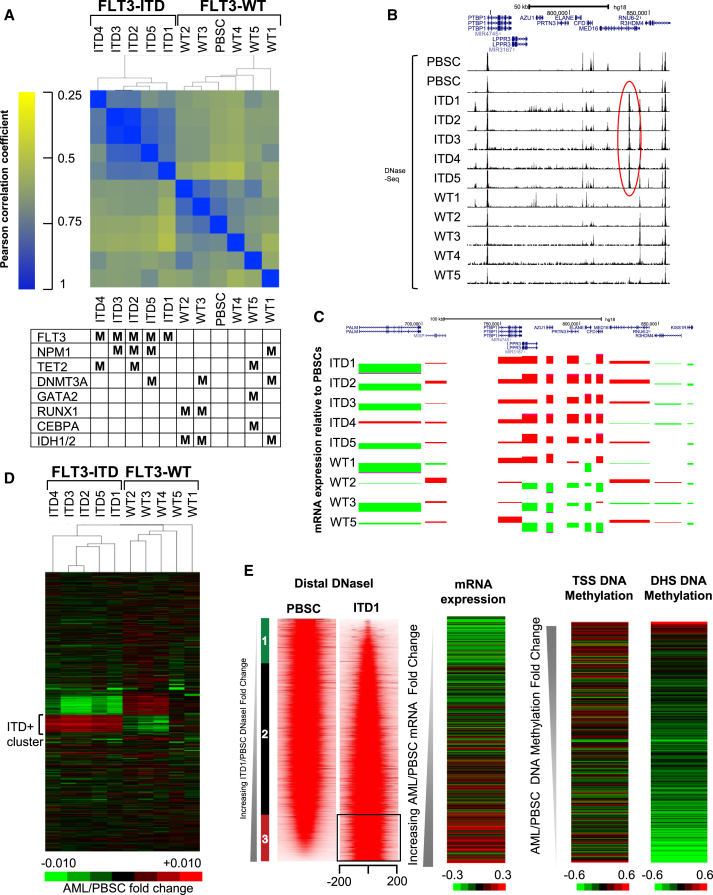
Tumor-specific DHSs patterns provide vital clues regarding the nature of TFs involved in tumorigenesis (Kreher et al., 2014). To identify DHSs specifically enriched in the ITD+ AMLs, we performed DNase-seq on a matched set of five ITD+ AMLs, five AMLs with WT FLT3, and two independent samples of CD34+ PBSCs. In a global evaluation of all the DHSs detected, we first identified all peaks for each dataset and determined the sequence tag counts for 400-bp regions centered on each peak. We then divided the peaks into the two separate groups of distal DHSs and promoter associated DHSs. Because we found that the greatest differences between the ITD+ and WT FLT3 AMLs were seen for the distal DHSs, we focused most of our subsequent more detailed analyses on this group. Previous global studies of transcriptional networks have similarly found that tissue-specific patterns are more associated with distal rather than proximal cis-elements (Heinz et al., 2010). For the initial comparisons, we performed hierarchical correlation clustering of the DNase-seq peak data (Figure 3A). As seen for the mRNA microarray values, the ITD+ samples clustered together as a discrete group distinct from both the WT FLT AMLs and the PBSCs. Parallel analysis of mutations in these samples again suggested that the ITD mutation had the greatest influence in defining the overall patterns of distal DHSs (Figure 3A, bottom). Interestingly, within the WT FLT3 group the two AMLs with mutated RUNX1 genes clustered together, as did the two samples with both GATA2 and CEBPA mutations.
Figure 3.
FLT3-ITD+ AML Has a Characteristic Chromatin Signature
(A) Hierarchical clustering of Pearson correlation coefficients of DHS peaks. For each sample the mutations present in the most commonly mutated genes are indicated as M in the table underneath.
(B and C) UCSC Genome Browser views of the DNase-seq patterns (B), and relative mRNA values (C) of genes surrounding an ITD+ AML-specific DHS in MED16 marked by a red oval, and a cluster of adjacent genes upregulated in ITD+ AML. mRNA values are shown as a ratio of the values detected in the AML samples compared to PBSCs (red: up, green: down).
(D) Heatmap depicting hierarchical clustering of the relative DNase-seq signals seen in each distal DHS peak in each AML sample relative to PBSCs.
(E) Profiles of the DNase-seq signals within each 400-bp window centered on each peak for PBSC and ITD1, with peaks shown in the order of increasing DNase-seq tag count signal for ITD1 relative to PBSC. This analysis includes the union of all peaks present in either ITD1 or in PBSC. Shown to the right of the DNase-seq profiles are the relative mRNA expression values for genes with the nearest transcription start sites (TSS) in ITD1 relative to PBSC, and the DNA methylation signals for the nearest TSS and for the DHS in ITD1 relative to PBSCs.
Figure 3B depicts the genome browser view of a gene cluster that includes the myeloid genes LPPR3, AZU1, PRTN3, ELANE, and CFD. Four of these closely linked genes were included in the group of 134 highly ITD-specific mRNAs listed in Data File S2. This region includes an ITD-specific DHS within an intron of MED16, which was essentially absent in both CD34+ PBSCs and in AMLs with WT FLT3. Figure 3C reveals a strong trend for specific upregulation of expression of the whole cluster of five genes in the ITD+ samples (shown in red), with a modest upregulation of MED16, which harbors the specific DHS, and no upregulation of the flanking genes. With the exception of AML sample WT1, which carries a NRAS mutation, these upregulated genes were actually downregulated in AML with WT FLT3 (shown in green).
To visualize the overall specific DHS pattern in FLT3-ITD AML, we calculated the ratios for all of the distal DNase-seq peaks for each of the AML samples depicted in Figure 3A relative to the same regions, using a PBSC DNase-seq dataset that was sequenced at high read depth. These ratios were then used for hierarchical clustering of the values, which are represented as a heatmap in Figure 3D. This analysis identified a cluster of ∼2,000 DHSs that were consistently enriched in the ITD+ samples and under-enriched in the WT FLT3 samples. Conversely, a separate cluster of DHSs was consistently downregulated in ITD+ AML and upregulated in the WT FLT3 samples, indicating that FLT3-ITD has both positive and negative effects on DHSs.
We next identified the full complement of FLT3-ITD-specific DHS peaks by calculating ratios of DNase-seq tag counts for all distal peaks detected in each AML sample and/or the PBSCs. Peaks were ranked in order of increasing DNase-seq tag count ratios for the AML sample relative to CD34+ PBSCs, and the DNase-seq data were plotted in order of increasing ratio as density maps spanning each 400-bp window. In the case of the prototype ITD+ AML sample ITD1 (which had no other defined mutations), we identified a total of 19,551 distal peaks present either in the ITD1 or PBSC dataset. The DNase-seq density maps are shown side-by-side in Figure 3E for both PBSCs and ITD1, revealing that most DHSs spanned ∼200–250 bp and identifying a group of 3,728 distal DHSs that were at least 2-fold more intense in ITD1 (boxed region in Figure 3E). Parallel pair-wise analyses were also performed comparing the distal peaks in ITD2 and in ITD3, and the promoter-associated DHSs in ITD1, ITD2, and ITD3, with the equivalent regions in PBSCs (Figures S2A–S2C). In each case we identified the subsets of DHSs that were at least 2-fold more intense in the AML sample than in PBSCs. The profiles of the average tag counts for the distal DHSs are plotted in Figure S2D and reveal similar patterns of deregulation in each of ITD1, ITD2, and ITD3. The parallel analyses of the promoter regions showed a less pronounced upregulation of a small subset proximal DHSs in each of the ITD+ AML samples (Figure S2C).
To examine how the presence of DHSs impacted on the expression of nearby genes, we plotted the relative AML/PBSC mRNA expression levels for the genes nearest to each DHS in the three core FLT3-ITD AML samples relative to PBSCs (Figures 3E, S2A, and S2B). These heatmaps revealed a trend for upregulation of the genes nearest to ITD-specific DHSs. In parallel, we determined ratios for the level of DNA methylation for CG elements covered by the Illumina 450K platform, and we plotted heatmaps of this data for regions spanning both the DHS site itself and the transcription start site of the nearest gene for ITD1 and ITD2. This revealed a trend for DNA demethylation of both the ITD-specific DHSs and the adjacent transcription start sites (TSSs) and for increased DNA methylation in the DHSs that were absent (Figures 2E and S2A). However, the overall changes in DNA methylation were less pronounced than the chromatin changes, suggesting that these may be secondary events.
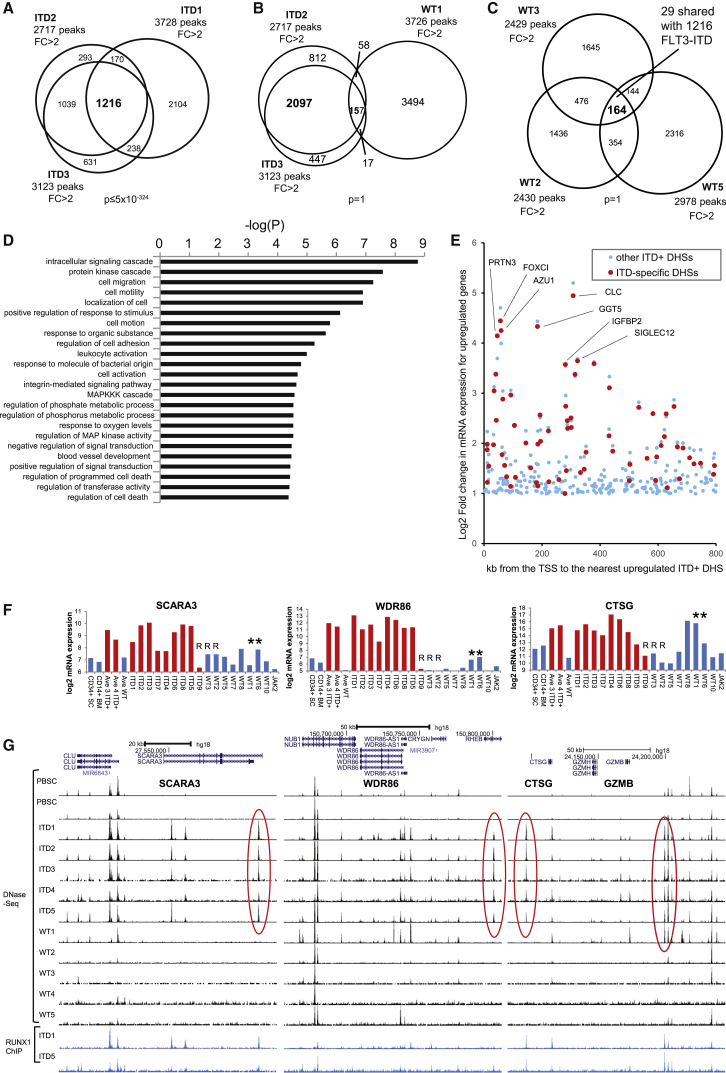
Using the ranking defined in Figure 3E, we also carried out direct side-by-side comparisons of the complete set of 19,551 distal ITD1/PBSC DHSs with each of the above datasets, plus the data for CD14+ BM cells, and for 3 previously published DHS datasets derived from normal CD34+ and CD14+ cells from the NIH Epigenome Roadmap consortium (Bernstein et al., 2010) plotted in the same order (Figure S2E). This confirmed that (1) the DHS pattern for PBSCs resembled independently derived equivalent CD34+ cell datasets, and (2) a common subset of DHSs was upregulated in each of the ITD+ samples, and (3) the DHS patterns present in the two more mature CD14+ cell datasets and the WT FLT3 AML samples more closely resembled PBSCs than ITD+ AML (Figure S2E). To better define these profiles, we used the relative DNase I signals to divide the DHSs into the three groups of (1) 2-fold downregulated, (2) less than 2-fold change, and (3) 2-fold upregulated (as depicted to the left of Figures 3E and S2E). The average DNase I profiles for each group are shown in Figure S2F. This clearly demonstrates that other ITD+ AMLs shared the same core of upregulated DHSs (group 3), whereas the patterns for the WT FLT AMLs more closely resembled the patterns for CD34+ and CD14+ cells. Last, but not least, we defined a discrete subset of reproducibly upregulated ITD-specific DHSs by determining the overlap between the FLT3-ITD-specific DHSs of each of ITD1, ITD2, and ITD3, and we found that 1,216 of these DHSs were shared between each group with high significance (Figure 4A). The locations of these DHSs are defined in Data File S3. ITD2 and ITD3, which both have NPM1 mutations, were the most similar with 83% of the top ITD2 group being part of the top ITD3 group. However, the NPM1 mutation is unlikely to be a major driver of this pattern because the upregulated DHSs from AML sample WT1 (Figure 4B) and WT8 (data not shown), which each carry an NPM1 mutation but WT FLT3, showed non-significant overlap with the ITD+ group. An equivalent analysis of the DHSs that are 2-fold upregulated in 3 AML samples with WT FLT3 (WT2, WT3, and WT5) also showed non-significant overlap with each other and with just 29 of the164 overlapping DHSs shared with the 1,216 ITD-specific DHSs (Figure 4C).
Figure 4.
FLT3-ITD Mutations Are Associated with a Specific Subset of DHSs
(A–C) Venn diagrams depicting the overlaps between populations of DHSs that are 2-fold upregulated in AML samples compared to PBSCs (FC > 2).
(A) Intersections between ITD1, ITD2, and ITD3.
(B) Intersections between ITD1, ITD2, and WT1, which has a NPM1 mutation.
(C) Intersections between WT2, WT3, and WT5.
(D) Gene ontology analysis of the genes closest to the 1,216 ITD-specific DHSs defined in (A).
(E) Plot of the relationship between the log2 mRNA fold change for genes upregulated in ITD+ AML versus the distance to the nearest ITD-specific DHS for all of the 134 ITD-specific genes (red), and the remainder of the 703 upregulated genes (blue) for genes located within 800 kb of an ITD-specific DHS.
(F and G) Log2 mRNA microarray values (as in Figure S1) (F) and UCSC Genome Browser views for DNase-seq and RUNX1 ChIP-seq data (G) for the ITD-specific genes SCARA3, WDR86, and CTSG/GZMB. ITD-specific DHSs are enclosed by red ovals.
ITD-Specific DHSs Are Associated with ITD-Specific Genes and an Activation Signature
Gene ontology analysis of the genes located closest to the 1,216 distal ITD-specific DHSs identified a strong correlation with genes linked to cell signaling and activation (Figure 4D). To directly link the ITD-specific epigenetic signature with the gene expression profile, we calculated the distance of each ITD-specific gene promoter region from the nearest upregulated DHS for both the highly ITD-specific 134 genes (shown in red) and for the additional genes included in the 730 upregulated genes (shown in blue), and we depicted this relationship graphically in Figure 4E. Interestingly, this revealed an increasing degree of upregulation at the mRNA level for the genes closest to an ITD-specific DHS. Examples of individual ITD-specific genes close to ITD-specific DHSs are shown for SCARA3, WDR86, and CTSG/GZMB in Figures 4F and 4G and for FOXC1, DSC2, and AK2 in Figures S1, S3A, and S3B. More complex patterns were seen for the HOXB cluster, where many DHSs were upregulated in ITD+ AML (Figure S3A), in parallel with the cluster of HOXB2 to HOXB6, and for a DHS in the C10orf128 locus, where both C10orf128 locus and VSTM4 were included in the group of 134 ITD-specific mRNAs (Figure S3B). A different pattern was seen for ID1, a previously defined ITD target gene (Tam et al., 2008), which contains an ITD-specific DHS at –2 kb and was upregulated in ITD+ AML compared to most WT FLT3 AMLs, but not compared to PBSC or normal CD14+ cells (Figure S3B). Interestingly, ID1 is also regulated by a downstream STAT5-dependent enhancer at +5 kb (Tam et al., 2008, Wood et al., 2009). STAT5 is FLT3-ITD inducible (Mizuki et al., 2000). To examine the role of STAT5 in the regulation of this and other genes, we therefore measured genome-wide binding sites for this factor by ChIP-seq. This experiment demonstrates that the +5 kb element indeed binds STAT5 in the MV4-11 FLT3-ITD cell line (Figure S3B). However, this DHS was not FLT3-ITD-specific.
Overall, the ITD-specific genes were upregulated in either all or most ITD+ samples (Figures 4F and S1A). However, ITD9 represented a notable exception to this pattern. Interestingly, this patient carried a RUNX1 frameshift mutation, suggesting an important role of RUNX1 in activating FLT3-ITD-specific genes. Furthermore, the three samples carrying RUNX1 mutations (labeled R) tended to have similar expression profiles (WT2, ITD9, and WT3 in Figures 4, S1, and S3). Conversely, we also noted that among these loci there were also numerous examples of DHSs that were preferentially enhanced in both the ITD+ AMLs and in AML samples WT1 and/or WT6, which carry mutations in signaling molecules (labeled with asterisks). AML sample WT1 carries an activating NRAS G12S mutation, which is a powerful activator of the MAPK pathway. Hence, activating RAS mutations with a concomitant upregulation of MAPK signaling may activate a subset of genes and DHSs that we defined here as FLT3-ITD targets.
Taken together, our analysis demonstrates that in spite of the presence of a wide variety of mutations, the presence of a chronically active FLT3-ITD correlates with a common core epigenetic signature that is associated with a distinct gene expression pattern. In addition, our mutation data suggested a role for both RUNX1 and MAPK signaling in activating FLT3-ITD-specific DHSs.
FLT3-ITD-Specific DHSs Are Associated with Specific DNA Binding Motifs for RUNX1 and AP-1, but Not STAT5
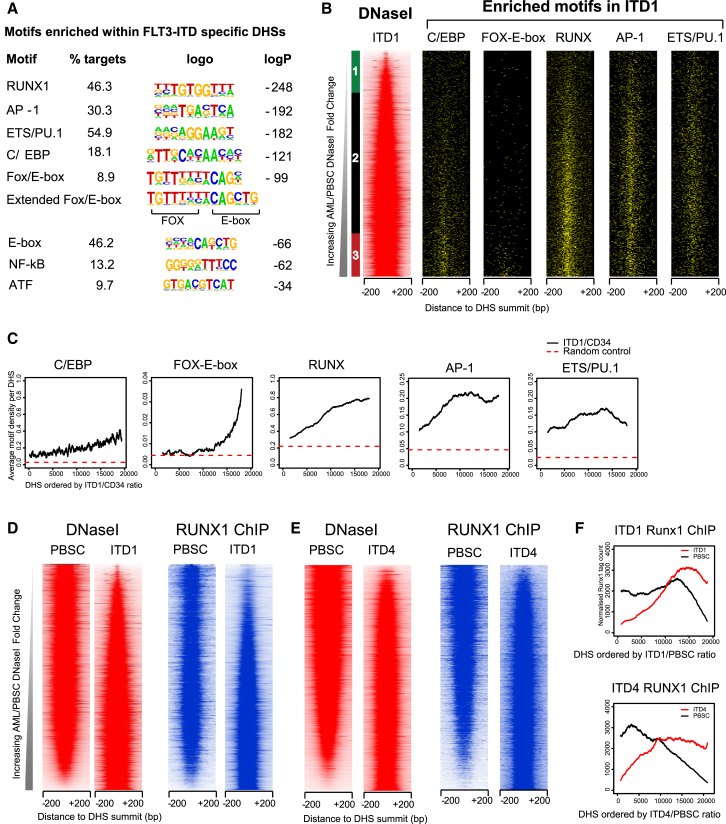
Having identified a FLT3-ITD+ AML-specific epigenetic signature, we investigated its underlying molecular basis by performing an unbiased search for enriched sequence motifs within active chromatin regions using HOMER (Heinz et al., 2010) (Figure 5A). This analysis revealed that RUNX, AP-1, ETS, E-box, and C/EBP motifs were the most common binding motifs present in the top 1,216 ITD-specific DHSs, each being found in 18%–55% of all sites. Interestingly, we also identified a motif for Forkhead (FOX) family proteins, which was part of a composite FOX/E-box motif that might recruit complexes containing both FOX and HLH family TFs such as TAL1/SCL and LYL1, which function together with RUNX, ETS, and GATA family TFs in the combinatorial regulation blood stem cell development (Wilson et al., 2010). We also saw an enrichment of motifs for the inducible TFs NF-κB and ATF, but with less statistical significance. In order to validate the above results, we plotted the coordinates of each motif found back on to the coordinates of the DHSs, plotted in the same order of increasing DHS ratio as above (Figure 5B). We also plotted the rolling average of the numbers of each motif found per DHS across the same series of DHSs (Figure 5C), plus the average distributions of the motifs with each of the previously defined three groups of distal ITD1/PBSC DHSs (Figure S4A). All five motifs were present in the ITD-specific DHSs at levels substantially higher than the predicted random level (indicated by the dashed lines in Figure 5C). Most of these motifs aligned with the midpoints of the DHSs, suggesting that the two features are functionally related. Both the FOX/E-box and the C/EBP motifs were preferentially enriched in the ITD-specific DHSs, represented as group 3 in Figure S4A. ITD-specific DHSs were also highly enriched in motifs widely used in normal myeloid cells (Heinz et al., 2013). RUNX, ETS, and AP-1 motifs were highly abundant in both the ITD-specific DHSs and the DHSs that were shared between the AML and the PBSCs. This suggests that the FLT3-ITD signature involves a substantial degree of redeployment of TFs driving the normal myeloid program.
Figure 5.
ITD-Specific DHSs Have a Specific Motif Signature and Bind RUNX1
(A) Result of de novo motif search of 1,216 ITD-specific DHSs using HOMER.
(B and C) Alignment of ITD-specific DHS motifs with distal DHSs present in either ITD1 or PBSCs (B) with the rolling averages of motif densities plotted underneath (C).
(D–F) Analysis of RUNX1 ChIP-seq data comparing ITD1 (D), ITD4 (E), and PBSCs (F) plotted alongside the ITD1 and ITD4 DHS data. (F) Depiction of the rolling averages of the ChIP signals.
We also looked directly for the presence of binding motifs of other TFs linked to common signaling pathways that were not among the motifs found by HOMER, such as the FLT3-ITD inducible TF STAT5 (Mizuki et al., 2000). However, we did not see any significant enrichment for STAT motifs in ITD-specific DHSs or for motifs linked to other signaling pathways, such as interferon response factors (IRFs), which are common mediators of receptor signaling, or NFAT, which is a mediator of Ca2+ signaling (Figures S4A and S4B). These three classes of motifs were present at low abundance and mostly showed an anti-correlation with the ITD-specific DHSs. The average densities of each motif across each the three groups of DHS are depicted in Figure S4A. Because the TF GATA2 is important for blood stem cell development (de Pater et al., 2013, Wilson et al., 2010), we also analyzed the distribution of GATA motifs. These motifs were most enriched in the PBSC-specific group 1 but were depleted in the ITD-specific group 3 DHSs, providing another indication that ITD1 represents a more mature myeloid cell.
To evaluate the role of STAT5 in shaping the FLT3-ITD-specific epigenetic signature, we integrated the DHS data with the STAT5 ChIP-data from FLT3-ITD+ MV4-11 cells. This analysis revealed STAT5 peaks in the HOXB, AK2, VSTM4/c10orf128, and ID1 loci, but not at the above-defined ITD-specific DHSs (Figure S3). Moreover, when STAT5 peaks were plotted alongside the ITD1/PBSC DHS comparison, they were found to be broadly distributed throughout the genome and predominantly in the shared DHS group (Figure S4B). Furthermore, just 249 of the 9,572 STAT5 peaks detected were present within the group of 1,216 ITD-specific DHSs (Figure S4C). These data suggest that STAT5 is binding predominantly at pre-existing DHSs and only plays a minor role in maintaining the FLT3-ITD-specific DHSs. A de novo motif analysis of the STAT5 ChIP peaks identified a STAT consensus motif in 16% of these peaks and further suggested that much of the STAT binding is in association with DHSs containing ETS, RUNX, C/EBP, and AP-1 motifs (Figure S4D).
A parallel analysis of DHSs and motifs associated with 380 ITD-specific promoter-associated DHSs revealed a similar ITD-specific epigenetic and binding motif signature (Figures S4E and S4F). Gene ontology analysis of the genes associated with this group of DHSs also revealed a cell activation signature similar to that seen for genes associated with the distal ITD-specific DHSs (Figure S4G).
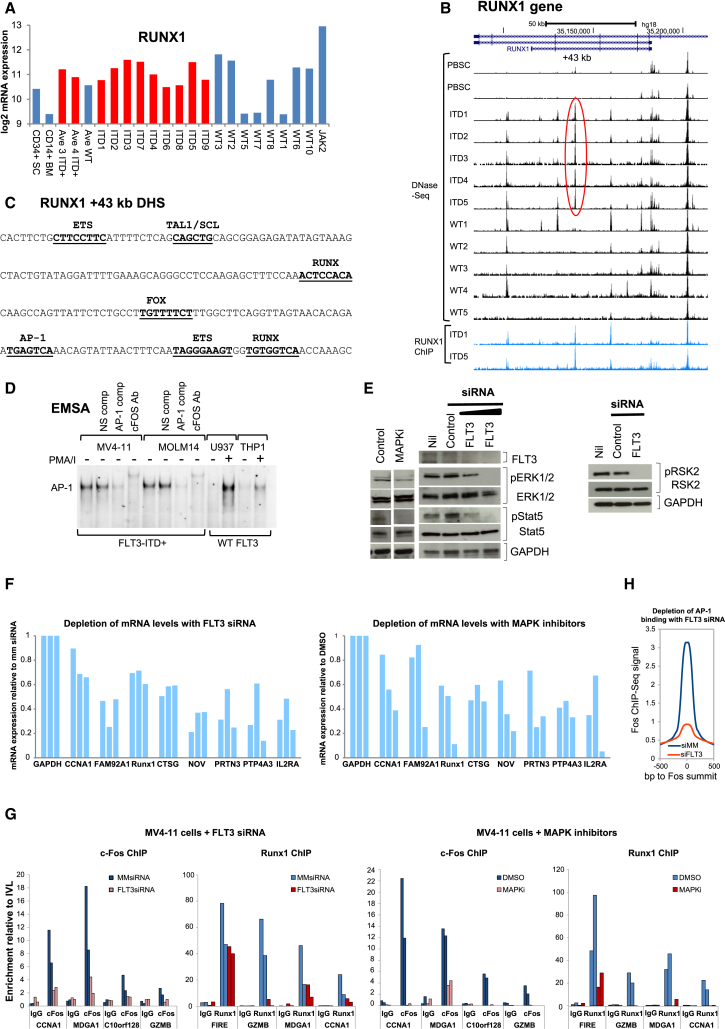
RUNX1, AP-1, FOX/E-box, and C/EBP Motifs Are Occupied in ITD-Specific DHSs
Our motif analysis pointed to a major role for RUNX and AP-1 in activating FLT3-ITD-specific DHSs. However, the presence of enriched motifs within specific DHSs does not necessarily mean that they are occupied. To verify occupancy by RUNX1, we performed ChIP-seq assays on ITD1, ITD4, and PBSCs (Figures 5D and 5E). The average profiles of RUNX1 binding (Figure 5F) closely mirrored the distribution of ITD-specific DHSs (Figure S2D) and RUNX motifs (Figure 5C). To account for the increase in RUNX1-bound sites in the ITD+ cells, we examined RUNX1 expression levels. RUNX1 expression was elevated in most of the ITD+ AMLs and in the AML with JAK2 mutations compared to normal CD34+ and CD14+ cells, but only in some of the WT FLT3 AMLs (Figure 6A; manual PCR validation shown in Figure S5A). A parallel PCR analysis confirmed that FOXC1 mRNA was also often upregulated in ITD+ AML (Figure S5B), with both RUNX1 and FOXC1 being significantly increased in each of the core ITD+ group of ITD1, ITD2, and ITD3. The upregulation of RUNX1 expression may be in part due to the presence of an ITD-specific DHS within the RUNX1 gene 43 kb downstream of the downstream promoter P2 (Figure 6B), which contains seven motifs linked to ITD-specific DHSs (RUNX, ETS, FOX, AP-1, and E-box) (Figure 6C). These motifs were also seen in the ITD-specific DHSs associated with FOXC1, DSC2, AK2, and ID1 (Figure S3) and in the SCARA3, CTSG, MDGA1, MED16, GZMB, VSTM4/c10orf128, and CCNA1 ITD-specific DHSs (Figure S5C). Notable among the 1,216 ITD+ DHSs was a DHS near C8orf87 containing 5 FOX motifs, one of which was the exact composite FOX/E-box motif defined in Figure 5A. This DHS was 221 kb away from the highly ITD-specific gene FAM92A1 (Figure S5D). In this case, the gene was again not expressed in AML samples with WT FLT3 unless they carried other mutations in the signaling molecules NRAS or SOCS1 (ITD1 and ITD6, marked by asterisks), or JAK2, and was also not upregulated in samples carrying RUNX1 mutations.
Figure 6.
AP-1 and RUNX Pathways Are Activated in ITD+ AML
(A) Log2 RUNX1 mRNA microarray values.
(B) UCSC Genome Browser view for RUNX1 DNase-seq.
(C) Sequence of the ITD-specific RUNX1 +43-kb DHS with the indicated sequence motifs underlined, which underlay the ITD-specific DHS signature.
(D) EMSA using nuclear extracts from the indicated cell lines grown in the presence and absence of stimulation for 2 hr with phorbol 12-myristate 13-acetate and calcium ionophore A23187 (PMA/I) to induce AP-1 activity. Some assays include AP-1 or non-specific (NS) DNA competitors or FOS antibodies.
(E) Western blot analyses extracts from ITD+ MV4-11 cells treated with FLT3 or mismatch (MM) control siRNA, using the indicated antibodies.
(F) RT-QPCR analysis of ITD target gene mRNA expression after treatment of MV4-11 cells with either siRNA against FLT3 (left) or with the MAPK pathway inhibitors PD98059, SP600125, and SB202190 directed against MEK1/2, JNK, and p38, respectively (right), from three independent experiments for each gene. Values were calculated relative to GAPDH.
(G) ChIP analyses of FOS and RUNX1 binding to ITD-specific DHSs, with normal IgG used as a control. Data are expressed relative to FOS or RUNX1 binding to a region in the inactive IVL locus.
(H) Average FOS ChIP peak profiles obtained from MV4-11 cells treated with either FLT3 or MM siRNA.
AP-1 is highly relevant to this study because it normally functions as a tightly regulated MAPK-inducible factor known to play major roles in the control of cell growth, survival, and cancer (Shaulian, 2010, Shaulian and Karin, 2002). To test for enhanced AP-1 activity, we used electrophoretic mobility shift assays (EMSAs) to examine the levels of nuclear AP-1 DNA-binding activities in both FLT-ITD+ and WT FLT3 myeloid cell lines (Figure 6D). Both of the ITD+ cell lines MV4-11 and MOLM14 had high levels of constitutive AP-1 activity, equivalent to the induced levels of AP-1 seen in the WT FLT3 cell lines U937 and THP1. Because AP-1 is a MAPK-inducible factor, we also used western blots of MV4-11 cell proteins to look for evidence of FLT3-dependent activation of MAPK signaling pathways (Figure 6E). This analysis revealed FLT3-dependent phosphorylation of both ERK1/2 and RSK2 (which are downstream of RAS signaling), which was suppressed by small interfering RNA (siRNA) directed against FLT3. This confirmed the FLT3-dependent activation not only of MAPK and the STAT pathways but also of STAT5. Furthermore, quantitative RT-PCR analyses revealed suppression of expression of several previously defined ITD-specific genes in MV4-11 cells after treatment with either FLT3 siRNA or a combination of inhibitors directed against the MEK, JNK, and p38 MAPK pathways (Figure 6F). We used ChIP to confirm that one partner of the AP-1 complex, FOS, was bound to four ITD-specific DHSs in MV4-11 cells (Figure 6G) that each contain AP-1 motifs (Figure S5C) and that RUNX1 was also bound to three of these DHSs and to the CSF1R FIRE element (Ptasinska et al., 2014) used here as a control for RUNX1 binding. In parallel we demonstrated that either FLT3 siRNA or MAPK inhibitors were sufficient to substantially diminish binding of FOS and RUNX1 to these ITD-specific DHSs, to a greater extent than seen at the FIRE RUNX site (Figure 6G). Last, but not least, we performed a ChIP-seq analysis for FOS in MV4-11 cells treated with FLT3 siRNA, which demonstrated a global decrease in the binding of FOS (Figure 6H). In summary, our data demonstrate that constitutive FLT3-ITD signaling leads to chronic activation of AP-1 via the MAPK pathway, which together with RUNX1 leads to chromatin remodeling and the activation of specific genes.
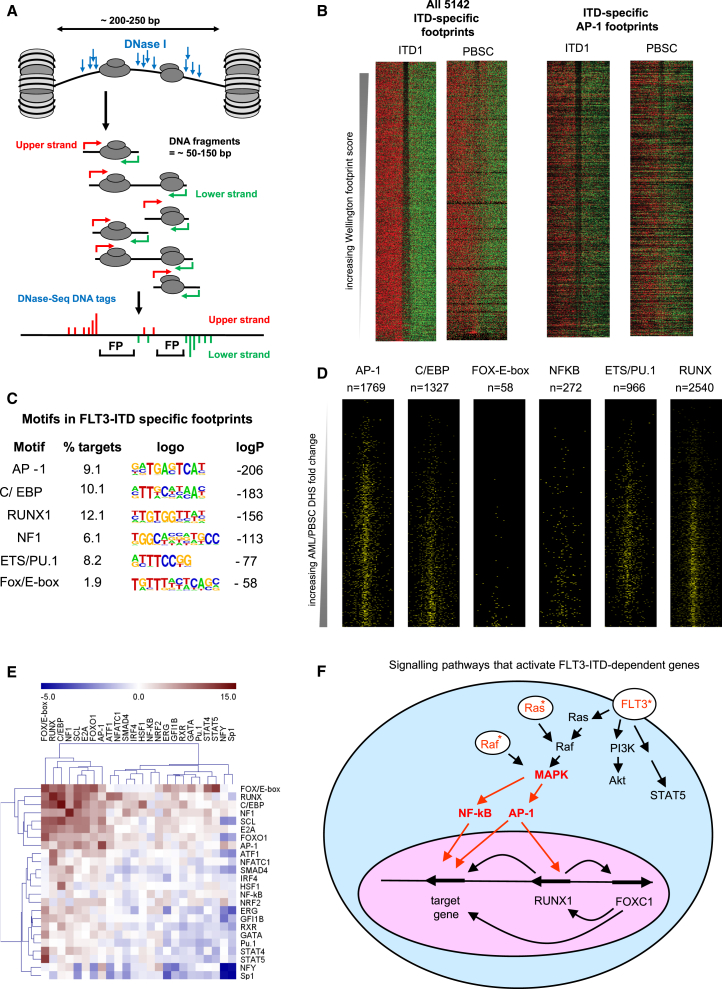
We next wanted to understand the role of other TFs in shaping ITD-specific DHS and how they would work together. To find additional evidence for the binding of these factors, we employed our recently developed Wellington algorithm that uses DNase-seq data to perform genome-wide in silico DNase I footprinting of regulatory motifs (Piper et al., 2013). This methodology determines statistically whether a given DNA sequence is protected from DNase I digestion, thereby indicating that it is occupied by a TF, as modeled in Figure 7A. Figure 7B depicts the density of upper (red) and lower (green) strand DNase I cleavage sites detected in ITD1 and PBSCs at all 5,142 FLT3-ITD-specific predicted footprints and at the specific subset carrying AP-1 motifs. The black gap between the red and the green signal indicates that the AP-1 motifs were occupied at a high frequency in ITD+ cells, but not in PBSCs. This analysis provided convincing evidence that AP-1 motifs are preferentially occupied in the FLT3-ITD AML sample, even though ITD1 and PBSCs share many DHSs that contain AP-1 motifs. These ITD-specific footprints also included 63 STAT5 motifs and 226 STAT4 motifs (some of which will be the same motif), but none of these were associated with any of the 134 or 77 FLT3-ITD-specific upregulated or downregulated genes.
Figure 7.
ITD-Specific DHS Motifs Are Occupied in ITD+ AML
(A) Model depicting the generation of DNase I footprints at DHSs.
(B) DNase I cleavage patterns within ITD1-specific footprints predicted by Wellington. Upper strand cut sites are shown in red and lower strand cut sites in green within a 200-bp window centered on each footprint (gap), for all 5,142 ITD-specific footprints, and for those containing AP-1 sites.
(C) Analysis of overrepresented binding motifs within each footprint using HOMER.
(D) Profiles of motifs in ITD-specific footprints plotted on the same DHS axes as used in Figure 2E.
(E) Analysis of statistically significant co-localization of the indicated footprinted motifs within 50 bp of each other using bootstrapping analysis (Z score).
(F) Model of an ITD-specific transcriptional network based on our data.
A de novo search for motifs within the 5,142 footprints revealed essentially the same signature as that of the 1,216 ITD-specific DHSs (Figure 7C), but with an additional motif for Nuclear Factor 1 (NF1) that may be linked to the ITD-specific upregulation of NF1X, which contains an ITD-specific DHS occupied by RUNX1 (data not shown). Figure 7D depicts the locations of the footprinted ITD-specific motifs on the ITD1/PBSC DHS coordinates displayed above in Figures 3E and 5B, demonstrating a close alignment with the DHS peak summits and with the FOX/E-box, C/EBP, and NF-κB motifs each being preferentially occupied in the ITD-specific DHSs. Occupied AP-1 motifs were equally distributed between ITD1-specific DHSs and DHSs shared with PBSCs, indicating constitutive binding of AP-1 to the shared sites in FLT3-ITD AML. We confirmed the existence of footprints at many of the predicted protected motifs by plotting the frequency of DNase I cleavage on the upper and lower strand in ITD1, alongside the footprint probability profiles and the motifs in Figure S6A. The parallel analysis of footprints in ITD2 showed similar results (not shown). To confirm the ability of our algorithm to detect STAT footprints, we show a footprint spanning the STAT motif, plus a second footprint spanning ETS and AP-1 motifs within the ID1 +5 kb DHS in ITD1 (Figure S6B). Overall, these results highlight (1) that gene activation in ITD+ AML involves FLT3-ITD-driven constitutive binding of normally inducible factors to pre-existing DHSs and (2) that RUNX1 and AP-1 are associated with both pre-existing and ITD-specific DHSs.
To examine, whether TFs binding to ITD-specific DHSs function in cooperation, we performed a bootstrapping analysis to identify which specific footprinted motifs were co-localized with 50 bp of each other (Figure 7E). This revealed a strong co-association of the occupied FOX, E-box, C/EBP, RUNX1, and AP-1 motifs within the same DHSs. Curiously, the ETS motifs were not preferentially footprinted in the ITD-specific DHSs, and they did not co-localize with the other ITD-specific motifs, which may tie in with the finding that PU.1 activity is downregulated in FLT3-ITD (Gerloff et al., 2015). In summary, these analyses demonstrate that the establishment of FLT3-ITD-specific DHSs is mediated by the interaction of a limited set of constitutive and inducible transcription factors.
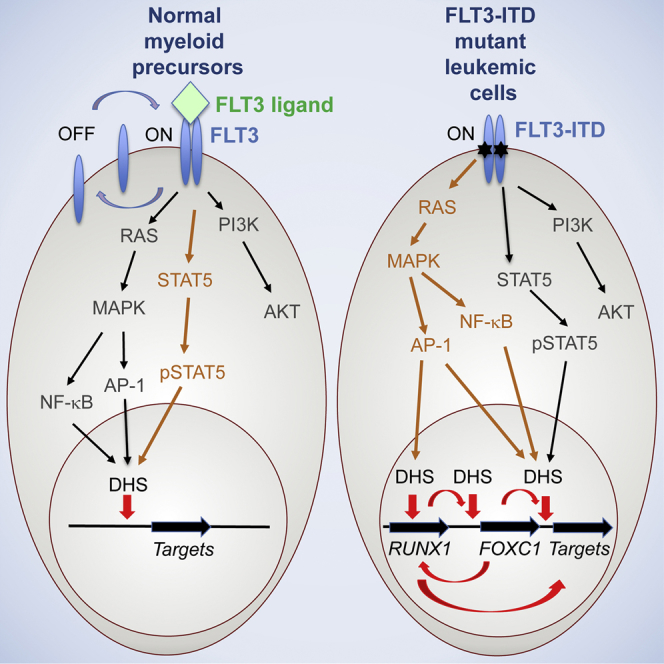
Discussion
A two-hit model of leukemogenesis is commonly accepted whereby a mutation in a transcriptional or epigenetic regulator gene impacting on gene expression and differentiation (class II mutation) cooperates with a mutation in a gene such as a signaling molecule regulating growth (class I mutation) (Renneville et al., 2008). Here we show at the systems level, in primary cells from patients, that this distinction is becoming blurred, with signaling having wide-ranging impacts on gene expression. Our study uncovers a significant role for chronic signaling by a mutated FLT3 growth factor receptor to the nucleus and demonstrates a profound impact of aberrant signaling on transcription factor binding and FLT3-ITD-specific gene expression. Our findings are consistent with the model depicted in Figure 7F in which aberrant FLT3-signaling activates both STAT and MAPK signaling. The latter, via the activation of inducible transcription factors such as AP-1/ATF family members and NF-κB, upregulates genes such as RUNX1 and FOXC1, which in cooperation with other factors, including C/EBP, ETS, and E-box family members, activate specific cis-regulatory elements driving the expression of many target genes. A similar cooperation between RUNX1 and AP-1 in response to MAPK signaling has been seen during megakaryocyte differentiation (Pencovich et al., 2011). Moreover, our mutation data indicate that alternate mutation of another member of the MAPK pathway (NRAS) may upregulate related sets of cis-regulatory elements. Furthermore, the mutation of RUNX1 disturbs the FLT3-ITD-specific expression pattern and may also influence the expression of the FOXC1 gene, which is not highly expressed in the patient containing mutant RUNX1 (ITD9) (Figure S1A). FOXC1 is itself linked to an ITD-specific DHS that contains three RUNX consensus motifs and that binds RUNX1 in ITD1 (Figure S3A).
A surprise finding from our study was the absence of enriched STAT motifs in our global analysis of ITD-specific DHSs in spite of overwhelming evidence (also from this study) that FLT3-ITD feeds into the STAT pathway and is required for enhanced translocation of STAT5 into the nucleus and leukemic survival (Chatterjee et al., 2014, Gu et al., 2011, Masson and Rönnstrand, 2009, Mizuki et al., 2000, Zhang et al., 2000). Furthermore, the data from MV4-11 cells suggested that STAT5 is bound predominantly at pre-existing DHSs that are shared with CD34+ cells, not STAT5-dependent DHSs, and thus is not involved in opening up additional FLT3-ITD-dependent DHSs. It is also possible that STAT5 binding to its targets may be only be required for the upregulation of a limited set of survival genes (such as MYC or BCLXL) (Chatterjee et al., 2014) and so is simply not present at most ITD-specific DHSs. In contrast, AP-1 may interact with many more ITD-specific targets. It is known that AP-1 regulates the inflammatory response and is involved in activating gene expression in multiple types of cancers with an inflammatory phenotype such as breast cancer, melanoma, and Hodgkin’s lymphoma (Giancotti, 2006, Kappelmann et al., 2014, Kreher et al., 2014). We previously showed that the chronic activation of regulators of inflammatory response genes such as JUN and IRF5 activates a Hodgkin’s lymphoma-specific gene expression program characterized by cytokine and chemokine expression, which during disease progression is progressively upregulated by chronic autocrine and paracrine stimulation (Kreher et al., 2014). It is therefore possible that STAT5 interaction with DNA is required for the initiation of the leukemic phenotype, but during leukemia development in patients it becomes less important due to the progressive establishment of a FLT3-ITD-specific transcriptional network that is driven by chronic inflammation and MAPK responsive transcription factors. This feature is consistent with our finding of the FLT3-ITD-specific upregulation of inflammatory response genes, such as chemokine genes, growth factor receptor genes, and genes involved in leukocyte activation and response to bacterial stimuli.
The FLT3-ITD mutation comes with a bad prognosis, and attempts to cure this type of AML using FLT3 inhibitors have met with only limited success (Grundler et al., 2003, Levis et al., 2011). Our system-wide studies of primary cells highlighting a common MAPK-responsive transcriptional network are therefore of utmost importance for cancer therapy, which may be applicable to ITD+ AML. Efforts to evaluate the role of different genes within this network with respect to the maintenance of the leukemogenic phenotype are currently underway.
Experimental Procedures
Detailed methods are in the Supplemental Information.
Patient Samples
AML cells and PBSCs were purified by density gradient centrifugation. Where the blasts were less than 92%, cells were further purified using CD34 or CD117 antibody coupled beads.
DNase-Seq and ChIP-Seq
Global analyses of DHSs and RUNX1 binding, and data analyses, were performed as in Ptasinska et al. (2014).
Mutations
Screening of DNA for mutations in the 55 AML-associated genes listed in Data File S1 was performed by WMRGL, Birmingham Women’s Hospital.
DNA Methylation
Analyses of DNA methylation were performed by Gen-Probe using the Illumina 450K platform.
Author Contributions
P.N.C., C.B., and P.C. wrote the paper. P.N.C., C.B., P.C., S.R.J., A.P., J.Z.-C., M.R.I., S.A.A., J.P., M.C., M.H., D.R.W., and S.O. designed and performed the experiments and analyzed the results. P.N.C., C.B., S.A., S.J.C., and M.J.G. coordinated and analyzed the mutation analyses. M.R., J.L., M.J.G., and S.J.R. obtained patent samples and analyzed patient data.
Acknowledgments
We would like to thank A. Jack, C. Craddock, A. Lubenko, R. Tooze, J. Burrow, J. Jesson, P. Jenkin, and D. Hollyman for help obtaining samples, D. Tenen and M. Wu for help with DNA sequencing, K. Keeshan, P. Twiss, and A. Rettino for help with mutation analyses, and S. Kissane for microarray analyses. This research was funded by a Specialist Programme Grant from Leukaemia Lymphoma Research to P.N.C. and C.B., the Kay Kendall Leukemia Fund, and an EPSRC studentship for J.P.
Published: July 23, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, six figures, one table, and three data files and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.06.069.
Contributor Information
Constanze Bonifer, Email: c.bonifer@bham.ac.uk.
Peter N. Cockerill, Email: p.n.cockerill@bham.ac.uk.
Accession Numbers
The DNase-seq, ChIP-seq, and mRNA array data reported in this study are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) as a superseries under the accession number GEO:GSE64874.
Supplemental Information
List of all the individual gene mutations identified from the mutation screen, including the DNA and amino acid changes, the proportion of sequences mutated (allele bias), the number of wild type and mutated sequences detected (allele depth), the method used for the mutation detection (Pindel or GATK), the nature of the two alleles (0 = normal, 1 = mutated), the lengths of insertions (INS) and deletions (DEL), and whether the mutation is previously recorded in the COSMIC database.
Alphabetical list of 134 upregulated genes and 77 downregulated genes in FLT3-ITD+ AML. Data are based on values obtained in Figure 1 for the relative differences in the average log2 mRNA array values for the core group of ITD+ AML samples (ITD1, ITD2, and ITD3) relative to the other four populations defined in Figure 1. Also listed are the actual average log2 mRNA array values for each of the five populations.
Chromosomal coordinates of 1,216 FLT-ITD+ AML-specific DHSs in the hg18 build of the human genome sequence. DHSs are listed alphabetically according the name of the nearest gene and indicate the distance to the nearest transcription start site (TSS).
References
- Badeaux A.I., Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 2013;14:211–224. [PubMed] [Google Scholar]
- Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Ghosh J., Ramdas B., Mali R.S., Martin H., Kobayashi M., Vemula S., Canela V.H., Waskow E.R., Visconte V. Regulation of Stat5 by FAK and PAK1 in oncogenic FLT3- and KIT-driven leukemogenesis. Cell Rep. 2014;9:1333–1348. doi: 10.1016/j.celrep.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P.N. Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J. 2011;278:2182–2210. doi: 10.1111/j.1742-4658.2011.08128.x. [DOI] [PubMed] [Google Scholar]
- Corces-Zimmerman M.R., Majeti R. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. 2014;28:2276–2282. doi: 10.1038/leu.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman M.R., Hong W.J., Weissman I.L., Medeiros B.C., Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., Bannister A.J., Göttgens B., Foster S.D., Bartke T., Green A.R., Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater E., Kaimakis P., Vink C.S., Yokomizo T., Yamada-Inagawa T., van der Linden R., Kartalaei P.S., Camper S.A., Speck N., Dzierzak E. Gata2 is required for HSC generation and survival. J. Exp. Med. 2013;210:2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerloff D., Grundler R., Wurm A.A., Brauer-Hartmann D., Katzerke C., Hartmann J.U., Madan V., Muller-Tidow C., Duyster J., Tenen D.G. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29:535–547. doi: 10.1038/leu.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti V. Breast cancer markers. Cancer Lett. 2006;243:145–159. doi: 10.1016/j.canlet.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Goyama S., Huang G., Kurokawa M., Mulloy J.C. Posttranslational modifications of RUNX1 as potential anticancer targets. Oncogene. 2014;34:3483–3492. doi: 10.1038/onc.2014.305. [DOI] [PubMed] [Google Scholar]
- Grundler R., Thiede C., Miething C., Steudel C., Peschel C., Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102:646–651. doi: 10.1182/blood-2002-11-3441. [DOI] [PubMed] [Google Scholar]
- Gu T.L., Nardone J., Wang Y., Loriaux M., Villén J., Beausoleil S., Tucker M., Kornhauser J., Ren J., MacNeill J. Survey of activated FLT3 signaling in leukemia. PLoS ONE. 2011;6:e19169. doi: 10.1371/journal.pone.0019169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Romanoski C.E., Benner C., Allison K.A., Kaikkonen M.U., Orozco L.D., Glass C.K. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappelmann M., Bosserhoff A., Kuphal S. AP-1/c-Jun transcription factors: regulation and function in malignant melanoma. Eur. J. Cell Biol. 2014;93:76–81. doi: 10.1016/j.ejcb.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Kreher S., Bouhlel M.A., Cauchy P., Lamprecht B., Li S., Grau M., Hummel F., Köchert K., Anagnostopoulos I., Jöhrens K. Mapping of transcription factor motifs in active chromatin identifies IRF5 as key regulator in classical Hodgkin lymphoma. Proc. Natl. Acad. Sci. USA. 2014;111:E4513–E4522. doi: 10.1073/pnas.1406985111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis M., Ravandi F., Wang E.S., Baer M.R., Perl A., Coutre S., Erba H., Stuart R.K., Baccarani M., Cripe L.D. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood. 2011;117:3294–3301. doi: 10.1182/blood-2010-08-301796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.H., Mandoli A., Simmer F., Wierenga B.J., Saeed S., Singh A.A., Altucci L., Vellenga E., Stunnenberg H.G. ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood. 2012;120:4038–4048. doi: 10.1182/blood-2012-05-429050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson K., Rönnstrand L. Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell. Signal. 2009;21:1717–1726. doi: 10.1016/j.cellsig.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Mizuki M., Fenski R., Halfter H., Matsumura I., Schmidt R., Müller C., Grüning W., Kratz-Albers K., Serve S., Steur C. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- Pencovich N., Jaschek R., Tanay A., Groner Y. Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood. 2011;117:e1–e14. doi: 10.1182/blood-2010-07-295113. [DOI] [PubMed] [Google Scholar]
- Piper J., Elze M.C., Cauchy P., Cockerill P.N., Bonifer C., Ott S. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prange K.H., Singh A.A., Martens J.H. The genome-wide molecular signature of transcription factors in leukemia. Exp. Hematol. 2014;42:637–650. doi: 10.1016/j.exphem.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Ptasinska A., Assi S.A., Mannari D., James S.R., Williamson D., Dunne J., Hoogenkamp M., Wu M., Care M., McNeill H. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012;26:1829–1841. doi: 10.1038/leu.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptasinska A., Assi S.A., Martinez-Soria N., Imperato M.R., Piper J., Cauchy P., Pickin A., James S.R., Hoogenkamp M., Williamson D. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 2014;8:1974–1988. doi: 10.1016/j.celrep.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Kwon S.Y., Ptasinska A., Bonifer C. Chronic growth factor receptor signaling and lineage inappropriate gene expression in AML: the polycomb connection. Cell Cycle. 2013;12:2159–2160. doi: 10.4161/cc.25386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P.N., Sargin B., Choudhary C., Stein S., Grez M., Müller-Tidow C., Berdel W.E., Serve H., Brandts C.H., Study Alliance Leukemia (SAL) SOCS1 cooperates with FLT3-ITD in the development of myeloproliferative disease by promoting the escape from external cytokine control. Blood. 2012;120:1691–1702. doi: 10.1182/blood-2010-08-301416. [DOI] [PubMed] [Google Scholar]
- Renneville A., Roumier C., Biggio V., Nibourel O., Boissel N., Fenaux P., Preudhomme C. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22:915–931. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- Sallmyr A., Fan J., Datta K., Kim K.T., Grosu D., Shapiro P., Small D., Rassool F. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111:3173–3182. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- Scholl C., Gilliland D.G., Fröhling S. Deregulation of signaling pathways in acute myeloid leukemia. Semin. Oncol. 2008;35:336–345. doi: 10.1053/j.seminoncol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Shaulian E. AP-1—The Jun proteins: oncogenes or tumor suppressors in disguise? Cell. Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Shaulian E., Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Stirewalt D.L., Radich J.P. The role of FLT3 in haematopoietic malignancies. Nat. Rev. Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- Tam W.F., Gu T.L., Chen J., Lee B.H., Bullinger L., Fröhling S., Wang A., Monti S., Golub T.R., Gilliland D.G. Id1 is a common downstream target of oncogenic tyrosine kinases in leukemic cells. Blood. 2008;112:1981–1992. doi: 10.1182/blood-2007-07-103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiede C., Steudel C., Mohr B., Schaich M., Schäkel U., Platzbecker U., Wermke M., Bornhäuser M., Ritter M., Neubauer A. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- Verhaak R.G., Wouters B.J., Erpelinck C.A., Abbas S., Beverloo H.B., Lugthart S., Löwenberg B., Delwel R., Valk P.J. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wolf S., Rudolph C., Morgan M., Büsche G., Salguero G., Stripecke R., Schlegelberger B., Baum C., Modlich U. Selection for Evi1 activation in myelomonocytic leukemia induced by hyperactive signaling through wild-type NRas. Oncogene. 2013;32:3028–3038. doi: 10.1038/onc.2012.329. [DOI] [PubMed] [Google Scholar]
- Wood A.D., Chen E., Donaldson I.J., Hattangadi S., Burke K.A., Dawson M.A., Miranda-Saavedra D., Lodish H.F., Green A.R., Göttgens B. ID1 promotes expansion and survival of primary erythroid cells and is a target of JAK2V617F-STAT5 signaling. Blood. 2009;114:1820–1830. doi: 10.1182/blood-2009-02-206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy J.S., Muise-Helmericks R.C. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- Zaret K.S., Carroll J.S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Fukuda S., Lee Y., Hangoc G., Cooper S., Spolski R., Leonard W.J., Broxmeyer H.E. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J. Exp. Med. 2000;192:719–728. doi: 10.1084/jem.192.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all the individual gene mutations identified from the mutation screen, including the DNA and amino acid changes, the proportion of sequences mutated (allele bias), the number of wild type and mutated sequences detected (allele depth), the method used for the mutation detection (Pindel or GATK), the nature of the two alleles (0 = normal, 1 = mutated), the lengths of insertions (INS) and deletions (DEL), and whether the mutation is previously recorded in the COSMIC database.
Alphabetical list of 134 upregulated genes and 77 downregulated genes in FLT3-ITD+ AML. Data are based on values obtained in Figure 1 for the relative differences in the average log2 mRNA array values for the core group of ITD+ AML samples (ITD1, ITD2, and ITD3) relative to the other four populations defined in Figure 1. Also listed are the actual average log2 mRNA array values for each of the five populations.
Chromosomal coordinates of 1,216 FLT-ITD+ AML-specific DHSs in the hg18 build of the human genome sequence. DHSs are listed alphabetically according the name of the nearest gene and indicate the distance to the nearest transcription start site (TSS).