Abstract
In this study, we investigate whether miR-128 is capable of regulating the apoptosis and proliferation of human U251 glioma cells by downregulating RhoE. The expression of miR-128 was assessed by quantitative polymerase chain reaction in normal brain tissue and glioma samples. A significant downregulation of the expression of miR-128 was detected in glioma in contrast to normal brain tissue. Following the transfection of pre-miR-128 and anti-miR-128 into U251 cells, the high expression of miR-128 could inhibit proliferation and induce apoptosis in U251 cells, and those effects could be restored by miR-128 knockdown. To analyze the regulation mechanism of miR-128, TargetScan, miRanda and PicTar were used to ascertain whether RhoE was a potential target gene. Next, luciferase activity assay and western blot analysis confirmed that RhoE was a direct and specific target gene of miR-128. The advanced effects of pre-miR-128 on the apoptosis and proliferation of U251 cells were reversed by the upregulation of RhoE expression. In summary, aberrantly expressed miR-128 regulates apoptosis and proliferation in human glioma U251 cells partly by directly targeting RhoE. This finding may offer a new potential therapeutic strategy for the treatment of glioma.
Keywords: glioma, miR-128, RhoE, U251 cell
Introduction
Glioma is the most common primary tumor in the nervous system, and has the highest mortality rate among endocranial tumors due to its malignant proliferation and invasion characteristics (1). Gliomas are rarely curable. Treatment for glioma includes surgery, chemotherapy or radiation therapy, depending on the patient. Surgery is the most significant initial approach, and chemotherapy and radiotherapy following the initial surgical resection is effective in preventing recurrence and metastasis (2). However, chemotherapy is only effective for ~50% of individuals treated for glioma. For this reason, the prognosis remains poor, particularly for patients with high-grade glioma, even when prompt and comprehensive treatment is administered. The median survival time for adults with an anaplastic astrocytoma is 2–3 years, and for those with more aggressive glioblastomas, the median survival time drops off to 12–14.6 months, with a two-year median survival rate of 30% (3).
microRNAs (miRNAs) are small endogenous noncoding RNAs. They exhibit an abnormal expression and function in various types of malignancies, and act as different types of tumor-related gene (4,5). miR-128 is an anti-oncogene which is expressed at low levels in certain malignant tumors, including glioma (6,7). miR-128 specifically inhibits a number of target genes related to tumor cell characteristics, including apoptosis and proliferation (8,9). Evidence suggests that miR-128 is an apoptosis-induced factor and functions at least in part by targeting a host of apoptosis-related genes, including Bmi-1, EGFR and E2F3 (10). miR-128 may silence the expression of certain anti-apoptotic genes and induce apoptosis in cancer cells.
Our study identified that miR-128 was expressed at lower levels in glioma samples than in controls. Upregulation of miR-128 inhibited U251 cell proliferation by targeting the RhoE gene at the translational level. Our novel findings suggest that abnormal expression of miR-128 is pivotal for the apoptosis and proliferation of human glioma cells. Further related research may aid the development of new therapeutic strategies against glioma.
Materials and methods
Clinical specimens
A total of 48 glioma and corresponding paracancerous tissues were provided by the Department of Neurosurgery at Shengjing Hospital of China Medical University between November 2012 and April 2014. The study was approved by the hospital's ethics committees, and we obtained consent from all patients prior to surgery. All specimens were obtained from patients through surgical resection and then histologically proven to be gliomas. None of the patients had undergone radiation or chemotherapy before surgery. Following surgery, the tissue samples were stored at −80°C, and the pathological information was obtained shortly after.
Quantitative polymerase chain reaction (qPCR)
After total RNA was extracted from tissue and cell samples, cDNA was synthesized and used to detect the mRNA expression with a miRNA detection kit (Invitrogen Life Technologies, Carlsbad, CA, USA) (11). The samples were normalized to 18S and the CT values from 18 to 30 were calculated by the 2−ΔΔCT method using the Applied Biosystems 7500 system (Applied Biosystems; Thermo Fisher Scientific, Waltham, MA, USA).
Transfection
Twenty-four hours before transfection, an appropriate concentration (~80%) of resuspended U251 cells were seeded on six-well plates. The miRNAs or expression vectors (Life Technologies, Grand Island, NY, USA) were transfected into U251 cells with Lipofectamine™ 2000 reagent (Invitrogen Life Technologies). The final concentration of miRNAs was 60 nM. The transfected cells were incubated for 4 h, and normal media were added. After 48 h, the cells were harvested for further studies.
Flow cytometry detection
Cells (5×106) were harvested, and an apoptosis detection kit (Biosea, Beijing, China) was used to examine the apoptosis rate in accordance with the manufacturer's instructions. Next, the cells were read by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) (Ex, 488 nm; Em, 635 nm), and the obtained numerical values were analyzed with CellQuest 3.0 software (BD Biosciences). Annexin V-positive cells were considered as apoptotic cells. The cells were counted by a dual-color flow cytometric method.
Cell proliferation assay
Cells were seeded in a 96-well plate with 2×104 cells per well. Twenty microliters of 0.5 mg/ml MTT solution (Sigma-Aldrich, St. Louis, MO, USA) was added to each well, and the 96-well plate was incubated at 37°C. The media were replaced after 4 h, and 0.2 ml DMSO was added to each well. The 96-well plate was incubated for 30 min, and results were read on an enzyme-labeled instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at a 570 nm wavelength. The obtained numerical values were used to construct the cell growth curve.
Target prediction
The prediction of the RhoE 3′-untranslated region (UTR) as a miRNA binding target was determined using Targetscan (www.targetscan.org), miRanda (www.mirbase.org) and PicTar (pictar.mdc-berlin.de). miRNAs that were simultaneously predicted by all three programs were selected for this study.
Vector construction and luciferase reporter assay
The luciferase reporter vectors were constructed by Invitrogen Life Technologies. The RhoE 3′-UTR, which included miR-128 seed-binding sites, was cloned into the pGL3-promotor vector to construct the wide-type (WT) vector, and the predicted seed zone with miR-128 was replaced by nonsense sequences in the mutation-type (MuT) vector.
In the luciferase reporter assay, 100 ng RhoE 3′UTR luciferase construct and 400 ng microRNA were cotransfected into U251 cells, and the Dual-Glo luciferase assay (Promega Corporation, Madison, WI, USA) was applied to quantitate the relative luciferase activity, which was computed by the luciferase activity of firefly to renilla. In order to normalize the transfection efficiency, the β-galactosidase expression vector was used in every transfection.
Western blot analysis
Cells were harvested and protein was extracted. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis and antibody hybridization were performed as described previously (12). The ECL analysis system (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was used for detection in accordance with the manufacturer's instructions. Western blot quantification was performed using Fluor Chen 2.0 image processing and analysis software (Bio-Rad Laboratories, Inc.). GAPDH was selected as the reference protein.
Statistical analysis
All experiments were carried out three times. All numerical data were presented as the means ± standard deviation and analyzed with SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance was used to analyze the statistical significance. P<0.05 and P<0.001 were considered to indicate a statistically significant difference.
Results
miR-128 is downregulated in glioma specimens
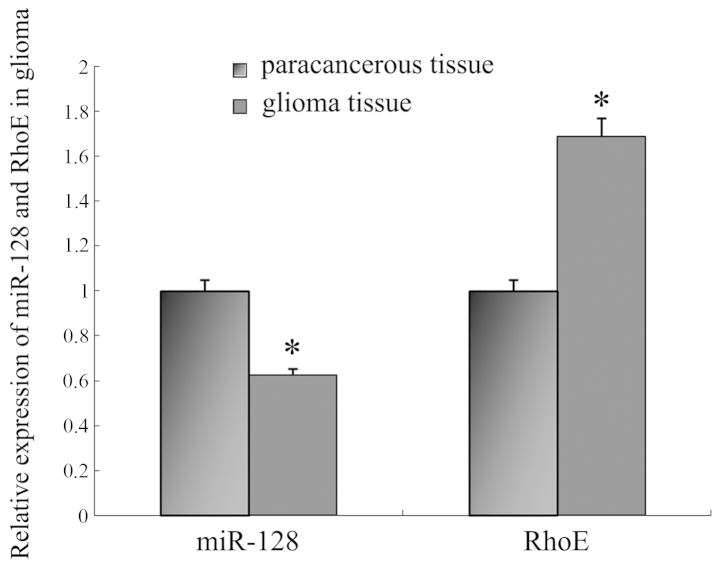
In order to research the potential effect of miR-128 in glioma, the expression of miR-128 was firstly analyzed in 48 glioma and corresponding paracancerous tissue samples. Compared with the matching paracancerous tissues, miR-128 expression in glioma was notably decreased (P<0.001; Fig. 1). Furthermore, the expression of miR-128 was negatively correlated with the pathological grade of glioma (P<0.01; Table I).
Figure 1.
Quantitative polymerase chain reaction analysis of the expression of miR-128 and RhoE in brain glioma tissue. Compared with paracancerous tissue, the expression of miR-128 was lower in brain glioma tissue (P<0.05), and RhoE demonstrated significant upregulation (P<0.05). *P<0.05 vs. paracancerous tissue.
Table I.
Correlation between miR-128 expression in brain glioma tissue and pathological differentiation.
| Differentiation (grade) | Number of cases | miR-128 expression relative quantification (gliomas/paracancerous tissues) | P-value |
|---|---|---|---|
| Medium/well differentiated (grade I–II) | 21 | 0.673±0.057 | <0.05 |
| Anaplastic glioma (grade III) | 15 | 0.632±0.062 | |
| Glioblastoma (grade IV) | 12 | 0.528±0.048 |
miR-128 functions as an anti-oncogene in glioma cells
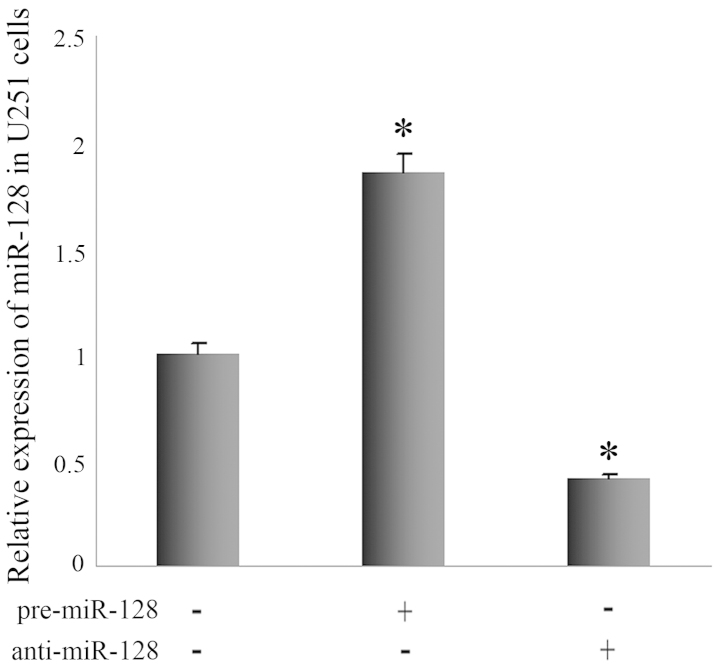
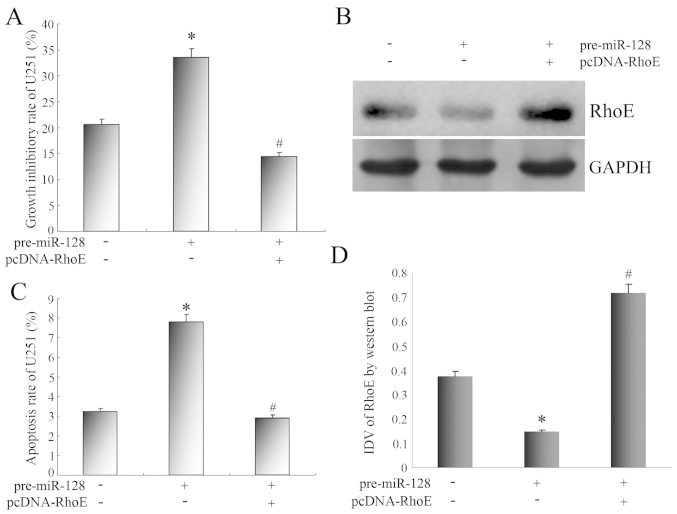
We investigated whether miR-128 influences apoptosis and proliferation using U251 cells. Pre-miR-128 was transfected into U251 cells to increase miR-128 expression (Fig. 2). In comparison with control U251 cells, the growth inhibitory rate of U251 cells with miR-128 overexpression was markedly increased by MTT assay (Fig. 3A). Flow cytometry revealed that the apoptosis rate of U251 cells with miR-128 overexpression was 7.83%, and that of control U251 cells was 3.18%. The statistical analysis revealed that the apoptosis rate increased significantly (Fig. 3C).
Figure 2.
Quantitative polymerase chain reaction analysis of the expression of miR-128 in U251 cells following transfection with pre-miR-128 or anti-miR-128. Pre-miR-128 increased the miR-128 expression level efficiently, and anti-miR-128 demonstrated a contradictory regulative effect. *P<0.05 vs. control U251 cells.
Figure 3.
(A) Impact of RhoE gene expression changes on the growth inhibitory rate of U251 cells. (B) Western blot analysis for RhoE expression of U251 cells treated with pre-miR-128 and pcDNA-RhoE transfection. GAPDH was used as a reference control. (C) Impact of RhoE gene expression changes on the apoptosis rate of U251 cells. (D) Quantitative analysis of the relative protein levels of RhoE normalized to those of GAPDH. *P<0.05 vs. control U251 cells, #P<0.05 vs. U251 cells transfected with pre-miR-128. IDV, integrated density values (calculated by Fluor Chen 2.0 computerized image analysis system and normalized with those of GAPDH).
RhoE is a direct and specific target of miR-128 in U251 cells
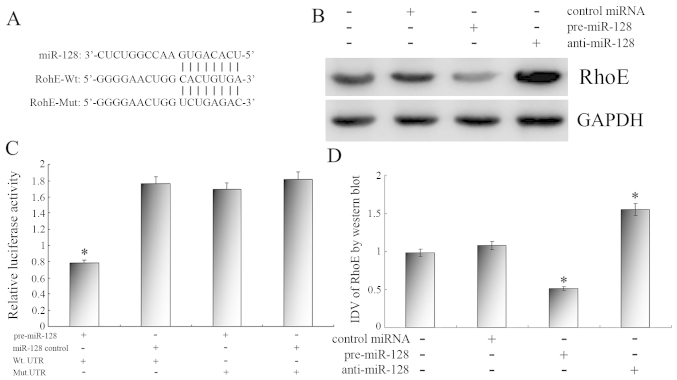
We then investigated the mechanisms of how miR-128 affects the apoptosis and proliferation phenotype in U251 cells. An online search of the target genes of miR-128 by TargetScan, miRBase and PicTar indicated that RhoE gene was a possible target of miR-128 (Fig. 4A).
Figure 4.
(A) The 3′-untranslated region (UTR) of RhoE is a target of miR-128 predicted by TargetScan, miRanda and PicTar. (B) Representative image of the protein level of RhoE. GAPDH was used as a reference control. (C) Reporter assay results with each bar representing values from three independent experiments. The transfection efficiency was normalized by cotransfected renilla luciferase and the light units were calculated by relative luciferase activity of firefly to renilla. (D) Quantitative analysis of the relative protein levels of RhoE normalized to those of GAPDH. Data are the means ± standard deviation of three independent experiments. *P<0.05.
qPCR was used to confirm that the expression of the RhoE gene was upregulated significantly in glioma tissues compared with matching paracancerous tissues (Fig. 1). Furthermore, Pearson's correlation analysis revealed a negative correlation between the expression of miR-128 and RhoE.
The luciferase reporter assay was performed to identify the direct miR128-RhoE interaction. Pre-miR-128 and various reporter vectors were cotransfected into U251 cells respectively. As shown in Fig. 4C, the relative luciferase activity was much lower in U251 cells cotransfected with WT vector and pre-miR-128 than in other transfected U251 cells. These results indicate that miR-128 could specifically bind to the seed zone of RhoE 3′UTR to inhibit its expression, while the MuT vector which changed the pivotal binding sequence could not be combined with miR-128 to decrease the relative luciferase activity. RhoE is a specific and direct target gene of miR-128.
Next, we confirmed the regulatory effects of miR-128 on endogenous RhoE expression. The results revealed that pre-miR-128 increased the miR-128 expression level efficiently, and anti-miR-128 demonstrated a contradictory regulative effect (Fig. 2). Western blot analysis revealed that the RhoE protein level was significant suppressed with miR-128 enhancement and was upregulated significantly in U251 cells transfected with anti-miR-128 (Fig. 4B and D), whereas the qPCR results demonstrated that endogenous RhoE expression was not significantly altered in U251 cells (data not shown). Together, these results confirm that miR-128 negatively regulates endogenous RhoE expression at the post-translational level.
RhoE expression mediates the effect of miR-128 on U251 cell apoptosis and proliferation
Given the fact that miR-128 regulated the apoptosis and proliferation of U251 cells, and RhoE was the target gene of miR-128, we studied the physiological role of miR-128-target RhoE in U251 cells following the upregulation of RhoE expression. As shown in Fig. 3B and D, the transfection of the RhoE expression vector significantly upregulated the protein expression of RhoE, which was expressed at low levels in U251 cells transfected with pre-miR-128.
After the expression of RhoE protein was upregulated, the growth inhibitory rate was markedly reduced (Fig. 3A), and the apoptosis rate of U251 cells decreased from 7.83 to 2.87%, which was statistically significant (Fig. 3C).
Combined with our substantial evidence that miR-128 inversely regulated RhoE expression, we confirm that miR-128-controlled apoptosis and proliferation of U251 cells is mediated to a large extent through RhoE downregulation.
Discussion
It is well known that glioma cells have significant proliferation ability and a low apoptosis rate, as well as a powerful resistance to chemotherapy, radiation and biological treatment (13–15). These characteristics contribute to the malignant growth of tumors and may be a key reason for the recurrence of glioma (16).
It is known that aberrant miRNA expression regulates critical biological behavior, including apoptosis and proliferation, which may promote glioma cell development and lead to a poor prognosis (17,18). Our study revealed that miR-128 was also expressed at low levels in brain glioma. Overexpression of miR-128 could increase the apoptosis rate of U251 cells and suppress proliferation. Thus, we suggest that the expression and functional changes of miR-128 are crucial in the genesis and development of glioma.
Since miRNAs exert their roles by targeting different genes, for example miR-10b and E-cadherin, and miR-320a and ITGB3 (19,10), we speculate that miR-128 regulated a single gene to modify the regulation network and trigger cell apoptosis and proliferation in U251 cells in our study. Computational algorithms are effective tools to predict and validate the miRNA gene targets. Through analysis using TargetScan, miRanda and PicTar, a number of key candidate targets for miR-128 were predicted. Among these potential targets, RhoE is the most significant.
RhoE, also known as RND3, belongs to the Ras superfamily and is a small Rho GTPase. It has been regarded as a significant effector of cell biological characteristics, including cell cycle, apoptosis and migration (20,21). RhoE was originally identified as an endogenous ROCK1 inhibitor, and binds to ROCK1 to inhibit downstream signaling (22). Previous studies have emphasized the critical role of RhoE in a number of cancer types. The expression of RhoE decreased in lung and hepatocellular carcinoma (21,23). However, RhoE is upregulated in certain cancers, included glioma (24). These differences suggest that Rnd3 has specific roles in different cell lineages.
Our results obtained from gain-of-function approaches confirmed that RhoE is a direct target gene of miR-128. Firstly, the RhoE gene was upregulated in glioma tissues, and its expression demonstrated a negative correlation with the expression of miR-128. Second, overexpression of miR-128 notably reduces the relative luciferase activity of the WT vector containing RhoE 3′UTR. Third, mutation at the miR-128 target site in the 3′UTR of RhoE significant decreased the miR-128 regulation effect. Fourth, overexpression of miR-128 inhibits the expression of RhoE protein at the post-translational level, and knockdown of miR-128 demonstrated the opposite effect. In summary, we conclude that RhoE is a direct and pivotal target gene of miR-128.
Our studies have identified that transfection of the RhoE expression vector upregulated the expression of RhoE, which demonstrated a low expression in U251 cells transfected with pre-miR-128, and strongly influenced the apoptosis and proliferation of U251 cells. All of these results indicate that miR-128 inhibited RhoE protein expression, and the apoptosis and proliferation regulated by miR-128 may be reversed by the overexpression of RhoE. Accordingly, the verification of RhoE as a target gene of miR-128 offers a probable reason as to why the low expression of miR-128 functions as a tumor-suppressing gene in U251 cells. However, further ongoing research is essential to identify the detailed molecular mechanism of miR-128 in the genesis and development of glioma.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grants 81172408 and 81301862).
References
- 1.Kuhnt D, Becker A, Ganslandt O, Bauer M, Buchfelder M, Nimsky C. Correlation of the extent of tumor volume resection and patient survival in surgery of glioblastoma multiforme with high-field intraoperative MRI guidance. Neuro Oncol. 2011;13:1339–1348. doi: 10.1093/neuonc/nor133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mrugala MM. Advances and challenges in the treatment of glioblastoma: a clinician's perspective. Discov Med. 2013;15:221–230. [PubMed] [Google Scholar]
- 3.Nieder C, Astner ST, Mehta MP, Grosu AL, Molls M. Improvement, clinical course, and quality of life after palliative radiotherapy for recurrent glioblastoma. Am J Clin Oncol. 2008;31:300–305. doi: 10.1097/COC.0b013e31815e3fdc. [DOI] [PubMed] [Google Scholar]
- 4.Shang C, Lu YM, Meng LR. MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2. Med Sci Monit. 2012;18:BR149–BR155. doi: 10.12659/MSM.882617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Guo Y, Shang C, Song Y, Wu B. MiR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80:1298–1302. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Hu J, Cheng Y, Li Y, Jin Z, Pan Y, Liu G, Fu S, Zhang Y, Feng K, Feng Y. MicroRNA-128 plays a critical role in human non-small cell lung cancer tumourigenesis, angiogenesis and lymphangiogenesis by directly targeting vascular endothelial growth factor-C. Eur J Cancer. 2014;50:2336–2350. doi: 10.1016/j.ejca.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Lin L, Chen X, Peng X, Zhou J, Kung HF, Lin MC, Jiang S. microRNA-128 promotes cell-cell adhesion in U87 glioma cells via regulation of EphB2. Oncol Rep. 2013;30:1239–1248. doi: 10.3892/or.2013.2596. [DOI] [PubMed] [Google Scholar]
- 8.Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian X, Yin Y, Zhao P, Wang YY, Wang XF, et al. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One. 2012;7:e32709. doi: 10.1371/journal.pone.0032709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adlakha YK, Saini N. MicroRNA-128 downregulates Bax and induces apoptosis in human embryonic kidney cells. Cell Mol Life Sci. 2011;68:1415–1428. doi: 10.1007/s00018-010-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Fu W, Wo L, Shu X, Liu F, Li C. MiR-128 and its target genes in tumorigenesis and metastasis. Exp Cell Res. 2013;319:3059–3064. doi: 10.1016/j.yexcr.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Shang C, Zhang H, Guo Y, Hong Y, Liu Y, Xue Y. MiR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol Biol Rep. 2014;41:2521–2527. doi: 10.1007/s11033-014-3110-0. [DOI] [PubMed] [Google Scholar]
- 12.Kumar M, Singh M, Singh SB. Optimization of conditions for expression of recombinant interferon-γ in E. coli. Mol Biol Rep. 2014;41:6537–6543. doi: 10.1007/s11033-014-3537-3. [DOI] [PubMed] [Google Scholar]
- 13.Dahlrot RH. The prognostic value of clinical factors and cancer stem cell-related markers in gliomas. Dan Med J. 2014;61:B4944. [PubMed] [Google Scholar]
- 14.Lim YC, Roberts TL, Day BW, Stringer BW, Kozlov S, Fazry S, Bruce ZC, Ensbey KS, Walker DG, Boyd AW, Lavin MF. Increased sensitivity to ionizing radiation by targeting the homologous recombination pathway in glioma initiating cells. Mol Oncol. 2014;8:1603–1615. doi: 10.1016/j.molonc.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukai J, Koizumi F, Nakao N. Enhanced anti-tumor effect of zoledronic acid combined with temozolomide against human malignant glioma cell expressing O6-methylguanine DNA methyltransferase. PLoS One. 2014;9:e104538. doi: 10.1371/journal.pone.0104538. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wang F, Zamora G, Sun CH, Trinidad A, Chun C, Kwon YJ, Berg K, Madsen SJ, Hirschberg H. Increased sensitivity of glioma cells to 5-fluorocytosine following photo-chemical internalization enhanced nonviral transfection of the cytosine deaminase suicide gene. J Neurooncol. 2014;118:29–37. doi: 10.1007/s11060-014-1410-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Wang XY, Li C, Xu SJ. Downregulation of microRNA-124 predicts poor prognosis in glioma patients. Neurol Sci. 2015;36:131–135. doi: 10.1007/s10072-014-1895-1. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Shi ZM, Jiang CF, Liu X, Chen QD, Qian X, Li DM, Ge X, Wang XF, Liu LZ, et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-inducedapoptosis in glioma. Oncotarget. 2014;5:5416–27. doi: 10.18632/oncotarget.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Zhao J, Zhang PY, Zhang Y, Sun SY, Yu SY, Xi QS. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit. 2012;18:BR299–BR308. doi: 10.12659/MSM.883262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonjedo M, Poch E, Mocholí E, Hernández-Sánchez M, Ivorra C, Franke TF, Guasch RM, Pérez-Roger I. The Rho family member RhoE interacts with Skp2 and is degraded at the proteasome during cell cycle progression. J Biol Chem. 2013;288:30872–30882. doi: 10.1074/jbc.M113.511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y, Hu C, Yang H, Cao L, Li Y, Deng P, Huang L. Rnd3 regulates lung cancer cell proliferation through notch signaling. PLoS One. 2014;9:e111897. doi: 10.1371/journal.pone.0111897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23:4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma W, Wong CC, Tung EK, Wong CM, Ng IO. RhoE is frequently down-regulated in hepatocellular carcinoma (HCC) and suppresses HCC invasion through antagonizing the Rho/Rho-kinase/myosin phosphatase target pathway. Hepatology. 2013;57:152–161. doi: 10.1002/hep.25987. [DOI] [PubMed] [Google Scholar]
- 24.Poch E, Miñambres R, Mocholí E, Ivorra C, Pérez-Aragó A, Guerri C, Pérez-Roger I, Guasch RM. RhoE interferes with Rb inactivation and regulates the proliferation and survival of the U87 human glioblastoma cell line. Exp Cell Res. 2007;313:719–731. doi: 10.1016/j.yexcr.2006.11.006. [DOI] [PubMed] [Google Scholar]