Abstract
Metastasis constantly occurs in the majority of cases of primary breast cancer at late stage or following surgical treatment. Survivin, a member of the inhibitor of apoptosis protein family, has long been recognized as a promising anticancer target, but its antitumor effects remain largely unexplored. In order to elucidate the role of survivin in breast cancer metastasis, short interfering RNA (siRNA) was used in the present study to specifically downregulate survivin expression in the murine breast cancer cell line 4T1. The results demonstrated that blocking the expression of survivin by siRNA inhibited the proliferation, migration and invasion abilities of murine breast cancer cells in vitro. Vascular endothelial growth factor (VEGF)-C is a lymphatic endothelial cell-stimulating factor that may lead to the formation of lymphatic vessels in lymph nodes. In the present study, the inhibition of survivin by siRNA was able to reduce the overexpression of VEGF-C in 4T1 cells. Furthermore, intratumoral injections of the survivin-siRNA significantly inhibited the growth of orthotopically transplanted 4T1 tumors in vivo. In addition, the number of pulmonary metastases and the microlymphatic vessel density were significantly reduced in vivo, following transfection with survivin-siRNA. The results of the present study suggested that the Akt/hypoxia-inducible factor-1α signaling pathway participates in the survivin-mediated downregulation of VEGF-C expression observed in breast cancer cells treated with survivin-siRNA. Therefore, the use of siRNA specifically targeting survivin may be a potential anticancer method in the future.
Keywords: breast cancer, short interfering RNA, survivin, VEGF-C, migration, tumor inhibition
Introduction
Breast cancer is the most frequently diagnosed type of cancer, and the leading cause of cancer-associated mortality among females, accounting for 23% of the total cases of cancer and 14% of the cancer-associated mortalities (1). Metastasis, characterized by its complex multistep processes and mechanism, is the most common aspect and malignant phenotype of breast cancer, and is responsible for ~90% of the lethality associated with this type of cancer (2). Despite early detection and opportune treatments, metastasis remains hardly inevitable, and predicts a poor prognosis in patients with breast cancer (3,4). The current treatments for breast cancer, including surgery, chemotherapy and radiation therapy, are not efficient in targeting metastasis. Therefore, the development of novel strategies for the treatment of breast cancer is required, particularly those targeting metastasis.
Metastasis via the lymphatic system is the primary step in the progression of breast cancer. The majority of patients with breast cancer present with lymph node metastasis when initially diagnosed or following surgery, which usually results in a poor prognosis (5,6). Vascular endothelial growth factor (VEGF)-C, a member of the VEGF family, has been defined as a lymphangiogenic growth factor, which participates in tumor lymphangiogenesis and spreading of tumor cells to lymph nodes in breast cancer (7–9). It has been previously observed that the overexpression of VEGF-C in tumor tissues of patients with breast cancer is negatively correlated with the survival rate and prognosis of these patients (10,11). Previous studies have demonstrated that knocking down the expression of VEGF-C with a functional blocking antibody or with siRNA prevents lymphogenesis and enhances chemosensitivity in human breast cancer cells (12). Therefore, VEGF-C can be considered a representative marker to estimate lymphogenesis and lymphatic metastasis in breast cancer. Previous studies have reported that the presence of lymphatic metastases and the expression levels of VEGF-C are associated with the levels of survivin in breast cancer (13).
Survivin, the smallest but most efficient member of the inhibitory apoptotic protein family, is involved in cell proliferation and inhibition of apoptosis (14,15). It has been previously reported that the downregulation of the expression or function of survivin is able to inhibit tumor growth and increase spontaneous apoptosis in different tumor models (16,17). Furthermore, previous studies have implicated survivin in cell motility, which may suggest a role for this protein in promoting tumor metastasis (18). In addition, survivin was observed to be positively correlated with tumor metastasis in laryngeal carcinoma, small adenocarcinoma of the lung and breast cancer (19–21). The overexpression of survivin in patients with cancer usually indicates a poor prognosis and high risk of mortality of these patients, since survivin is commonly expressed in neoplasms and embryonic tissue, but rarely detected in normal differentiated adult tissues (22). Therefore, survivin may be a promising target for the diagnosis and therapy of cancer.
Survivin and VEGF-C are highly expressed in breast cancer tissues, and a positive correlation exists between the expression levels of these proteins in cancer (23). However, to the best of our knowledge, the complex association between survivin and lymphatic metastasis remains poorly characterized. In the present study, a murine breast cancer cell line termed 4T1, which exhibits metastatic characteristics similar to those of human breast cancer, was employed to investigate whether knocking down survivin by siRNA provided antitumor effects in these cells, and to study the association between survivin and lymphatic metastasis in mouse breast cancer.
Materials and methods
Cell culture
Murine mammary carcinoma 4T1 cells were acquired from the Shanghai Laboratory Animal Center of the Chinese Academy of Sciences (Shanghai, China). The cells were maintained in high glucose Dulbeccos modified Eagle medium (DMEM) (Hyclone, GE Healthcare Life Sciences, Logan, UT, USA), containing 10% fetal bovine serum (Gibco™; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA), 1 mM mixed non-essential amino acids, 2 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin, and incubated at 37°C in a humidified atmosphere with 5% CO2. Cells in exponential growth phase were used throughout the experiments.
siRNA design and cell transfection
The sequences of the siRNA oligonucleotides were designed as follows: Survivin, 5′-GGAAUUGGAAGGCUGGGAATT-3 (sense) and 5′-UUCCCAGCCUUCCAAUUCCTT-3 (antisense); and VEGF-C, 5′-GCAAGACGUUGUUUGAAAUTT-3 (sense) and 5′-AUUUCAAACAACGUCUUGCTT-3 (antisense). In order to enhance the stability of the siRNAs, the sense strands were methylated, while the antisense strands were not modified. Scrambled siRNA (siRNAscr), 5′-UUCUCCGAACGUGUCACGUTT-3 (sense) and 5′-ACGUGACACGUUCGGAGAATT-3 (antisense), was used as negative control, since the selected sequences did not share any homology with any of the known mRNA databases. The 3′-end of the siRNAscr was labeled with fluorescein isothiocyanate to evaluate the efficiency of the transfection. All the siRNA sequences were chemically synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
The siRNAs were transfected into cultured cells at 50% confluence, which had been seeded on dishes 24 h earlier. For transfection, Invitrogen™ Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.) was used, according to the manufacturers protocol. Three groups of cells were established, as follows: i) An untransfected (UT) group, serving as blank control; ii) a group transfected with 100 nmol/l siRNAscr; and iii) a group transfected with 100 nmol/l survivin-siRNA. The cells in the UT group were treated with Opti-MEM (Thermo Fisher Scientific, Inc.), while the cells in the other groups were treated with Lipofectamine 2000-Opti-MEM. At 5–6 h post-transfection, the medium was replaced with 10% serum-supplemented DMEM, and the cells were incubated for additional 24–96 h. Next, the cells were harvested by centrifugation, rinsed with phosphate-buffered saline, and subjected to total RNA or protein extraction.
RNA extraction and quantitative polymerase chain reaction (qPCR)
Total RNA was isolated from the cells using Invitrogen™ TRIzol reagent (Thermo Fisher Scientific, Inc.), and cDNA was prepared with First Strand cDNA Synthesis Kit (Roche Diagnostics, Basel, Switzerland), according to the manufacturers instructions. Next, the cDNA amplified by qPCR, using the following primers (Shanghai Sangon Biotech Co., Ltd., Shanghai, China): Survivin, forward 5′-ATCGCCACCTTCAAGAACTG-3 and reverse 5′-GGCCAAATCAGGCTCGTTCT-3; VEGF-C, forward 5′-AAACCTCAGCTGTCTGGTCC-3 and reverse 5′-GAACCATGTGGATTACTGCG-3; and β-actin, forward 5′-GCGGACTGTTACTGAGCTGCGTAACG-3 and reverse 5′-GAAGCAATGCTGTCACCTTCCC-3′, which was used as reference gene. The reaction conditions were as follows: 95°C for 30 sec, followed by 30 cycles of annealing at 58 or 57°C (in the case of survivin or VEGF-C and β-actin, respectively) for 30 sec, and extension at 72°C for 30 sec. qPCR amplification was performed on a Corbett Rotor-Gene 3000 (Qiagen, Inc., Valencia, CA, USA), using FastStart Universal SYBR Green Master (Rox) (Roche Diagnostics). Each RNA sample was analyzed in triplicate, and the data were quantified according to the 2-ΔΔCq method.
Western blotting
The western blot assay was performed as previously described (24). Briefly, total protein content was extracted from the cells with RIPA lysis buffer (Beyotime Institute of Biotechnology, Nantong, China), which contained phosphatase inhibitors (in addition to protease inhibitors) when detecting phosphorylated proteins. The protein concentration in the cell lysates was determined with Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL, USA). Protein samples were separated by SDS-PAGE using 12% polyacrylamide gels (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and then transferred to polyvinylidene difluoride membranes (EMD Millipore, Bedford, MA, USA). The membranes were blocked for 1 h with Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% non-fat milk at room temperature. Subsequently, primary antibodies were added to the membrane and incubated overnight at 4°C. Rabbit polyclonal antibodies against survivin (cat. no. sc-10811), VEGF-C (cat. no. sc-25783), hypoxia-inducible factor (HIF)-1α (cat. no. sc-10790) and β-actin (cat. no. sc-7210; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), were diluted 1:300, while the rabbit monoclonal antibodies against Akt (cat. no. 9272) and phosphorylated (p)-Akt (Thr308; cat. no. 9275) (Cell Signaling Technology, Inc., Danvers, MA, USA) were diluted 1:1,000. Following 3 washes of 5 min each with TBST, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no. ZDR-5306; Zhongshan Jinqiao Biotechnology, Co., Ltd., Beijing, China) for 1 h at room temperature, before subjection to 3 washes of 5 min each with TBST. Proteins were visualized using the enhanced chemiluminescence method, and protein expression was quantified with a Gel EDAS 293 analysis system (Cold Spring USA Corp., Cherry Hill, NJ, USA).
Cell proliferation assay
Cell viability was examined by MTT assay (Roche Diagnostics). For the experiment, 4T1 cells were seeded in a 96-well plate (Corning Life Sciences, New York, NY, USA) at a density of 5×103 cells/well, and transfected 24 h later with 100 nM survivin-siRNA or siRNAscr. At 24, 48 and 72 h post-transfection, MTT (5 mg/ml) was added to the cells, which were incubated at 37°C for additional 4 h. Next, the supernatant was removed, and 150 µl dimethyl sulfoxide (Sigma-Aldrich) was added to each well, followed by 10-min agitation. The absorbance of each well at 490 nm was measured using an ELISA plate reader (Thermo Multiskan MK3; Thermo Fisher Scientific, Shanghai, China). Quintuplicate wells were used for each experimental condition, and all the experiments were repeated at least three times.
Cell invasion and migration assay
Matrigel invasion assay was performed using transwell chambers (Corning Life Sciences). For the assay, Matrigel (1 mg/ml; BD Biosciences, Franklin Lakes, NJ, USA) was diluted in serum-free DMEM, and 70 µl of the diluted Matrigel were then coated in 8-µm pore size filters, followed by incubation for 5–6 h at room temperature. At 24 h post-transfection, 100 µl 4T1 cells in serum-free medium (5×105 cells/ml) were placed in the upper chamber, while the lower chamber was filled with 600 µl NIH 3T3-conditioned medium (prepared in our laboratory). Following 24 h incubation, the non-invasive cells on the upper chamber were removed with cotton swabs, and the migrated cells were stained with 0.1% crystal violet (#C6158; Sigma-Aldrich) for 10 min at room temperature, prior to be examined and imaged by light microscopy at ×200 magnification (Olympus BX60; Olympus Corporation, Tokyo, Japan). Quantification of cellular migration was conducted by counting the number of stained cells in five randomly selected fields. The migration assay was performed following the same method used for the aforementioned invasion assay, with the exception of the exclusion of the Matrigel matrix and the use of polycarbonate membranes.
Tumor growth in mice
Female BALB/c mice of 4 weeks of age were acquired from the Laboratory Animal Center of Shandong University (Jinan, China), and acclimated to the laboratory conditions (temperature, 24±2°C; humidity, 50%; and day/night cycle, 12 h day/night). Experiments were conducted in the Animal Laboratory of the Medical College of Qingdao University (Qingdao, China). The animals received humane care, and experiments were conducted according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals (25) and with approval of the Animal Care and Use Committee of the Medical College of Qingdao University. The tumor generation assay in vivo was performed as previously described, with minor modifications (26). Briefly, 5×104 4T1 cells suspended in 50 µl DMEM were injected into the inguinal mammary fat pads of mice of 5 weeks of age (10 mice/group), following acclimation. Animal body weight and tumor size were measured and recorded regularly. Tumor size was measured every two days in two perpendicular dimensions (a=length and b=width) with a vernier caliper, and the size recorded was calculated as volume (mm3), according to the formula (a × b2)/2. When the tumors reached 40–50 mm3 in size, mice were randomly divided into 3 groups: i) The UT group, which was intratumorally injected with 5% glucose every two days; ii) the siRNAscr group, which was treated with siRNAscr (1 mg/kg body weight), in vivo-jetPEI (Polyplus-transfection SA, Illkirch, France) and 5% glucose every two days; and iii) the survivin-siRNA group, which was treated with survivin-siRNA (1 mg/kg body weight), in vivo-jetPEI and 5% glucose every two days, in a total volume of 200 µl. Following 7 injections, the mice were sacrificed by cervical dislocation, and their tumors were removed and weighed.
Immunohistochemical staining
For immunostaining, the tumor specimens were fixed in 4% formaldehyde, embedded in paraffin and sectioned at 4 µm. Next, a rabbit anti-mouse antibody against podoplanin (cat. no. sc-134483) was used as the primary antibody (Santa Cruz Biotechnology, Inc.), and for detection, a biotinylated anti-rabbit antibody (cat. no. cw-0103, Cwbio, Beijing, China) was used as the secondary antibody. The sections were dewaxed, rehydrated and subjected to antigen retrieval, prior to be subjected to microlymphatic vessel density (MLVD) evaluation, according to the procedure previously described (27). To quantify the MLVD, each slide was scanned at low power magnification (×100), and two ‘hot spot’ areas with a relatively high number of newly formed vessels were identified, which were subsequently scanned at high power magnification (×400). Five random fields of each ‘hot spot’ area were analyzed.
Lung metastases
The lungs were fixed and blanched in ethanol for the quantification of metastases. The lungs were harvested and fixed for 24 h in Bouins solution, a mixture of saturated picric acid, formaldehyde and acetic acid, at a ratio of 75:25:5, respectively. Next, the color was restored by soaking the tissues in 70% ethanol.
Statistical analysis
SPSS software version 16.0 was used to for the statistical analysis of the data. Data are presented as the mean ± SD. Statistical analysis was performed using one-way analysis of variance or Students t-test. P<0.05 was considered to indicate a statistical significant difference.
Results
Survivin-siRNA inhibited survivin expression in 4T1 cells
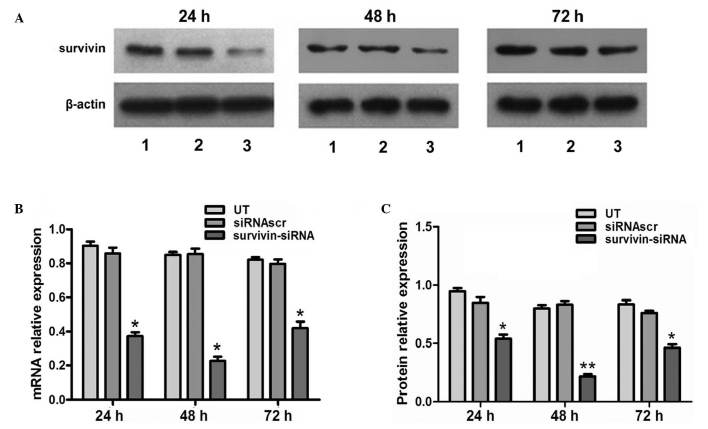
siRNA was transfected into 4T1 cells with high efficiency. In the survivin-siRNA-transfected cells, the mRNA levels of survivin were significantly reduced at 24, 48 and 72 h post-transfection, compared with the UT and siRNAscr groups (Fig. 1B). In order to confirm the qPCR results, and better characterize the knocking down of survivin at the protein level, western blot analysis was performed. The results indicated that the protein expression levels of survivin were significantly reduced at 24, 48 and 72 h post-transfection in the survivin-siRNA group, compared with the UT and siRNAscr groups (Fig. 1A and C).
Figure 1.
Effect of survivin-siRNA on the mRNA and protein levels of survivin in 4T1 cells. (A) The protein levels of survivin were detected by western blot analysis at 24, 48 and 72 h post-transfection. Lanes 1, 2 and 3 correspond to the UT, siRNAscr and survivin-siRNA groups, respectively. (B) Survivin-siRNA suppressed the mRNA expression of survivin in 4T1 cells at 24 h post-transfection, and its effects lasted for 72 h. (C) The treatment with survivin-siRNA reduced the protein levels of survivin in 4T1 cells, as revealed by western blot analysis. Data are presented as the ratio of survivin to β-actin.*P<0.05 and **P<0.01 vs. UT. The experiments were performed in triplicate. siRNA, short interfering RNA; UT, untransfected; siRNAscr, scrambled siRNA.
Antiproliferative effect of survivin-siRNA on 4T1 cells
The effects of survivin-siRNA on the proliferation ability of 4T1 cells were assessed by MTT assay. Inhibition of cell proliferation was noticeable in 4T1 cells transfected with 100 nM survivin-siRNA at 48 h post-transfection. The proliferation ability of these cells was observed to be significantly suppressed in a time-dependent manner, whereas no inhibitory effect was observed in the UT or siRNAscr groups (Fig. 2).
Figure 2.
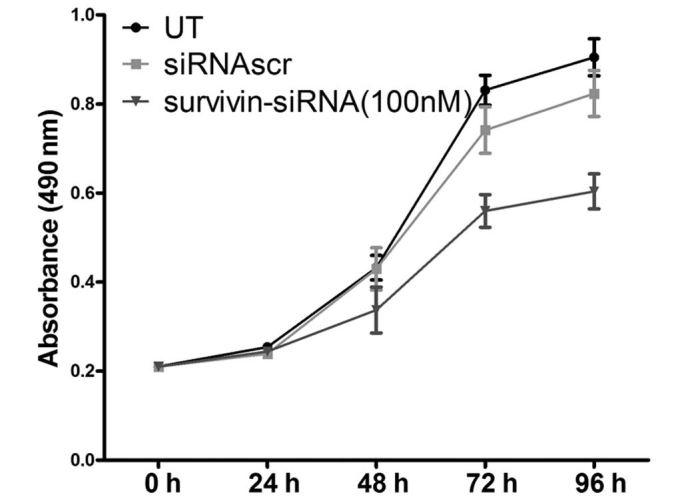
The knockdown of survivin inhibited the proliferation of 4T1 cells in vitro. The results indicated that the transfection with survivin-siRNA, but not with siRNAscr, inhibited the proliferation of 4T1 cells, compared with the UT group. The experiments were performed in triplicate. siRNA, short interfering RNA; siRNAscr, scrambled siRNA; UT, untransfected.
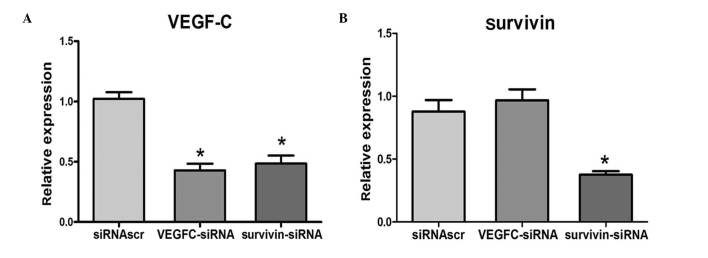
Positive correlation between the expression levels of VEGF-C and survivin in 4T1 cells
Following the transfection of 4T1 cells with survivin-siRNA, VEGF-C-siRNA or siRNAscr, the mRNA levels of survivin and VEGF-C were examined by qPCR. The results indicated that when survivin was downregulated in 4T1 cells, VEGF-C was also downregulated. However, the expression levels of survivin were not reduced when VEGF-C was downregulated. qPCR analysis revealed that the mRNA levels of VEGF-C were positively correlated with the mRNA levels of survivin (Fig. 3).
Figure 3.
Expression of VEGF-C and survivin at mRNA level in 4T1 cells treated with different siRNAs. mRNA expression levels of (A) VEGF-C and (B) survivin, according to the results of quantitative polymerase chain reaction analysis. *P<0.05 vs. UT. VEGF-C, vascular endothelial growth factor-C; siRNA, short interfering RNA; siRNAscr, scrambled siRNA; UT, untransfected.
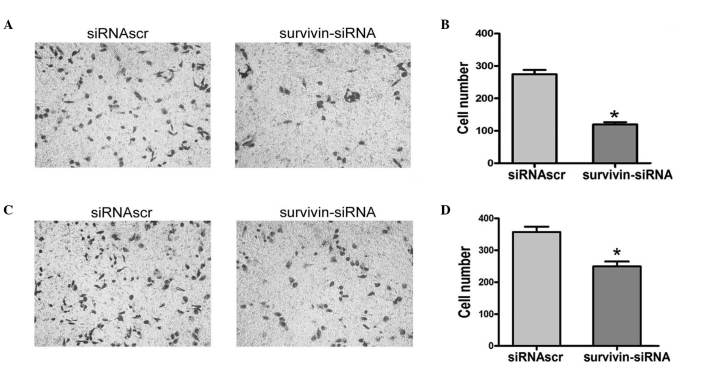
Survivin-siRNA inhibited the migration and invasion abilities of 4T1 cells in vitro
To assess the role of endogenous survivin in the invasion and migration abilities of 4T1 cells, the expression of survivin was downregulated using siRNA. Following incubation for 24 h, the invasion ability of the survivin-siRNA-transfected cells was inhibited, compared with the siRNAscr-transfected cells, indicating that knocking down survivin in 4T1 cells resulted in retarded cell invasion (Fig. 4A and B). Furthermore, a significant difference was observed between the number of migrated cells in the survivin-siRNA group, compared with the siRNAscr group (Fig. 4C and D). These results indicated that the expression of survivin in 4T1 cells was associated with the capacity of migration and invasion of these cells.
Figure 4.
Invasion and migration assays were performed using the transwell chamber assay. (A) Invasion assay for the siRNAscr and survivin-siRNA groups. (B) The average number of cells penetrating the Matrigel matrix and pores was significantly reduced in the survivin-siRNA group, compared with the siRNAscr group (274.20 and 119.40 cells, respectively, vs. cells). (C) Migration assay for the siRNAscr and survivin-siRNA groups. (D) The transfection of survivin-siRNA led to a significant reduction in the average number of cells penetrating the pores, compared with the siRNAscr group (357.00 and 249.60 cells, respectively, vs. cells). *P<0.05 vs. siRNAscr group. siRNA, short interfering RNA; siRNAscr, scrambled siRNA; UT, untransfected.
Effects of survivin-siRNA on the VEGF-C and Akt/HIF-1α signaling pathways
The phosphoinositide 3-kinase (PI3K)/Akt signaling pathway is known to participate in the malignant proliferation, angiogenesis, metastasis and resistance to chemotherapy of tumor cells. In various types of cancer, the expression of HIF-1α is positively correlated with lymphatic metastasis. Therefore, to investigate how survivin regulates VEGF-C, the expression levels of p-Akt and HIF-1α were measured in the survivin-siRNA and control groups. The results indicated that the expression levels of VEGF-C and HIF-1α and the phosphorylation levels of Akt were reduced in 4T1 cells following transfection of survivin-siRNA (Fig. 5).
Figure 5.
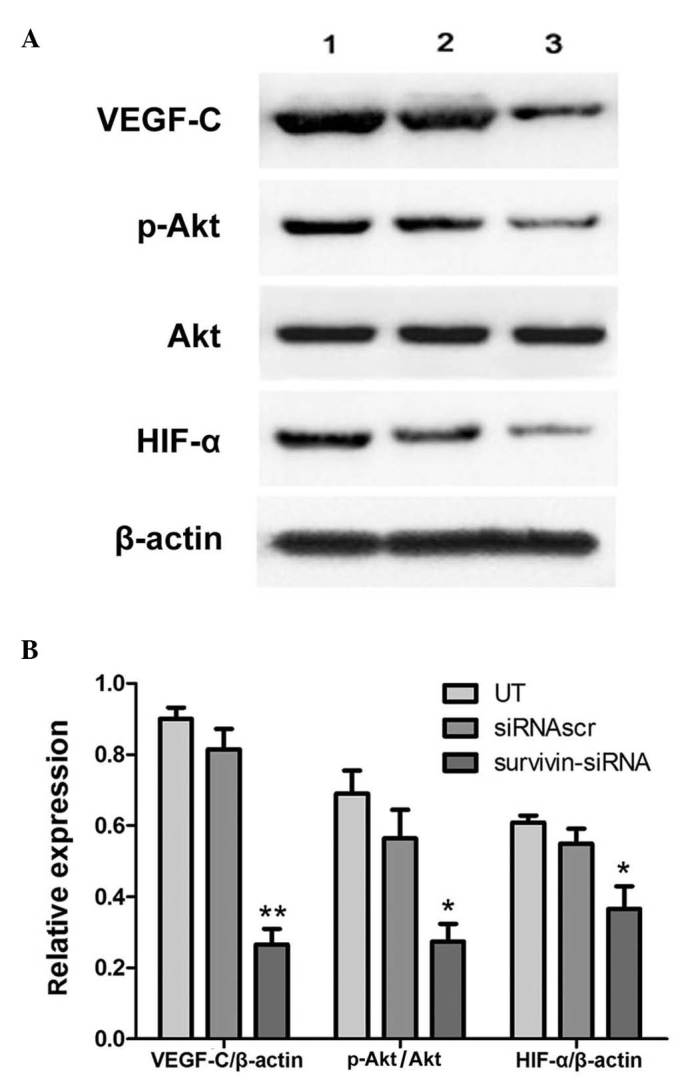
Effects of survivin-siRNA on the VEGF-C and Akt/HIF-1α signaling pathways. (A) Transfection with survivin-siRNA led to reduced protein expression levels of VEGF-C, HIF-1α and p-Akt in 4T1 cells at 48 h post-transfection. (B) Quantification of the results from panel A. The protein ratios were determined by the relative intensities of the protein bands. Each bar represents the mean ± SD of three independent experiments. *P<0.05 and **P<0.01 vs. UT group. siRNA, short interfering RNA; VEGF-C, vascular endothelial growth factor-C; HIF-1α, hypoxia-inducible factor-1α; p, phosphorylated; UT, untransfected; siRNAscr, scrambled siRNA.
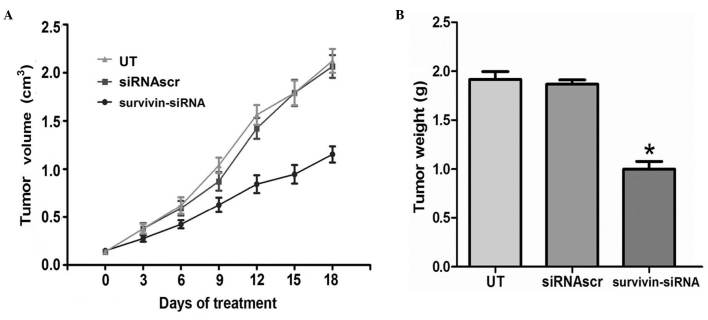
Survivin-siRNA inhibited tumor proliferation in vivo
In the present study it was observed that survivin-siRNA exhibited significant antitumor abilities in vitro. Therefore, in order to explore the applicability of the survivin-siRNA system in vivo, a murine metastatic breast tumor model mimicking stage IV of human breast cancer was established. Survivin-siRNA was able to reduce tumor volume and weight in this xenograft model (Fig. 6). These results indicated that survivin-siRNA may exert its antitumor effects on this model of breast cancer by inhibiting tumor proliferation.
Figure 6.
Antitumor effects of survivin-siRNA on breast cancer in vivo. Stable transfection of cancer cells with survivin-siRNA resulted in reduced tumor (A) size and (B) weight. siRNA, short interfering RNA; siRNAscr, scrambled siRNA; UT, untransfected.
Survivin-siRNA inhibited lymphangiogenesis and lung metastasis in vivo
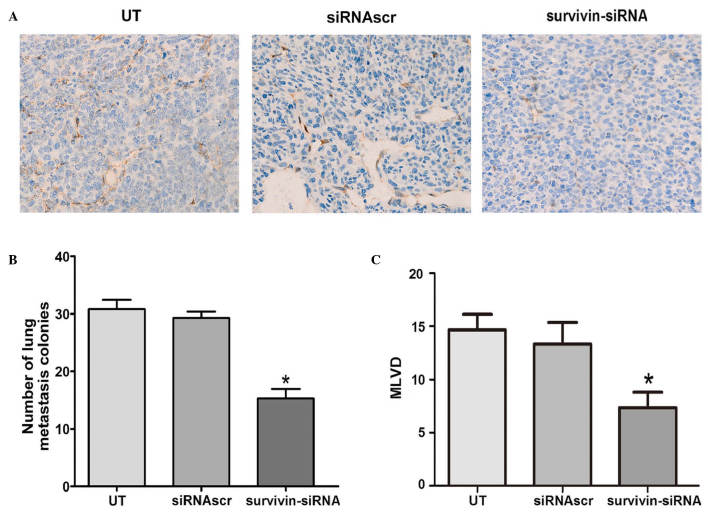
To determine whether the treatment with survivin-siRNA affected lymphangiogenesis in vivo, MLVD was detected by podoplanin staining in the animal tumors. According to the results obtained, the median numbers of MLVD were apparently reduced in the tumors of the survivin-siRNA-treated group, compared with the control groups (Fig. 7A and C). Furthermore, to investigate the effect of survivin-siRNA on metastasis, the lungs of the sacrificed mice were harvested, and their metastatic nodules were counted. The results are presented as the mean ± SD (n=10 tumors). Compared with the control mice, the survivin-siRNA-treated mice exhibited significantly fewer lung metastases (Fig. 7B). These results indicated that survivin participates in the lymphangiogenesis and metastasis of breast cancer tumors.
Figure 7.
Survivin-siRNA inhibited lymphangiogenesis and lung metastasis in vivo. (A) Representative images of the tumor sections examined by immunohistochemical staining for podoplanin, revealing the tumor vasculature (magnification, ×400). (B) Murine breast cancer 4T1 cells that were stably transfected with survivin-siRNA displayed significantly fewer pulmonary metastatic colonies than those cells in the UT and siRNAscr groups. (C) The expression levels of podoplanin and the mean vessel density within the tumors were reduced following stable injection of survivin-siRNA, compared with the UT and siRNAscr groups. Each bar represents the average vessel number for each group. All data are expressed as the mean ± SD. *P<0.05 vs UT. siRNA, short interfering RNA; siRNAscr, scrambled siRNA; UT, untransfected; MLVD, microlymphatic vessel density.
Discussion
Survivin is normally expressed in neoplasms and fetal tissues, but is rarely present in normal adult tissues. Survivin generally acts as an anti-apoptotic protein by inhibiting the activation of caspases (28). Previous studies have suggested that survivin participates in the lymphatic metastasis of breast cancer by regulating the expression of VEGF-C in tumor cells (13). Lymphatic metastasis is the process by which detached tumor cells may enter the lymph nodes located near the primary tumor via lymphatic vessels, and is considered one of the most important mechanisms of breast cancer systemic metastasis. VEGF-C is the most important lymphangiogenic growth factor in the progression of lymphogenesis and lymphatic metastasis (9). Downregulation of the expression or function of VEGF-C has been previously demonstrated to supress lymphogenesis and lymphatic metastasis in various tumor xenograft models (29,30).
siRNA technology is a recently developed molecular biological technique widely used in gene therapy and functional genetics studies (31,32). In the present study, chemically synthesized siRNAs specifically targeting mouse survivin inhibited the expression of survivin at the mRNA and protein level in the murine breast cancer cell line 4T1. The results of MTT assay indicated that the proliferation ability of 4T1 cells was significantly reduced in vitro following transfection with survivin-siRNA, compared with the UT and siRNAscr groups. Additionally, the migration and invasion assay revealed that the motility of 4T1 cells was significantly inhibited upon transfection with survivin-siRNA.
In the present study, the potential effects of survivin on cellular signaling pathways that regulate the expression of VEGF-C in tumor cells, were explored. The results indicated that the PI3K/Akt signal transduction pathway may be involved in the survivin-mediated regulation of the expression of VEGF-C in breast cancer. Previous studies have suggested that HIF-1α, a transcription factor involved in cellular adaptive responses to hypoxia, may be responsible for the overexpression of VEGF-C in cancer cells (33). Previous studies have demonstrated that the activity of HIF-1α can be blocked by the PI3K-inhibitor LY294002, indicating that the activation of HIF-1α may be regulated by the PI3K/Akt pathway. Akt is a serine/threonine protein kinase that functions as a critical regulator of cell adhesion, migration and apoptosis (34). The activation of Akt has been previously observed to contribute to tumorigenesis and tumor metastasis in various types of human cancer (35,36). McKenzie et al (18) reported that survivin may trigger the activation of Akt, thus enhancing the motility of melanoma cells. The activation of Akt is regulated by phosphatase and tensin homolog (PTEN), which is reported to be affected by survivin (18). Therefore, it can be hypothesized that survivin may suppress lymphatic metastasis in murine breast cancer by downregulating VEGF-C through the Akt/HIF-1α pathway.
Previous studies have demonstrated that binding of VEGF-C to VEGF receptor(R)-3 on the membrane of breast cancer and endothelial cells results in the phosphorylation and subsequent activation of the tyrosine kinase activity of VEGFR3, which consequently stimulates lymphogenesis and mitosis in these cancer cells (17,18). This ligand-induced phosphorylation of VEGFR-3 has been previously suggested to activate downstream signaling molecules such as Akt, by stimulating the kinase activity of the receptor (17,18). Therefore, the knockdown of survivin and the downregulation of VEGF-C may affect the phosphorylation of Akt. In the present study, the results of western blotting revealed that the knockdown of survivin in vitro led to the downregulation of the expression of VEGF-C, HIF-1α and p-Akt at 48 h post-transfection, while the downregulation of survivin was noticeable 24 h earlier. These findings indicated the existence of a time window between the effects exerted by survivin-siRNA on the expression of survivin and on the activation of VEGFR-3. Therefore, it can be hypothesized that survivin suppresses the Akt/HIF-1α signaling pathway, which results in the downregulation of the expression of VEGF-C.
Furthermore, to evaluate the effects of survivin-siRNA on breast cancer growth in vivo, the antitumor efficacy of survivin-siRNA was examined in a mouse metastatic breast cancer model. In this model, the intratumoral injection of survivin-siRNA significantly inhibited the tumor growth and pulmonary metastasis of the orthotopically implanted 4T1 cells. In addition, immunohistochemical staining, hematoxylin and eosin staining and qPCR assays were performed on the tumor tissues derived from the survivin-siRNA-injected mice. The data obtained from these analyses suggested that the injection of survivin-siRNA into the tumor exerted significant antitumor effects in vivo, similar to those exhibited in vitro. According to these results, it can be concluded that survivin participates in cell proliferation and metastasis of mouse breast cancer.
In conclusion, the results of the present study revealed that survivin-siRNA possessed potent in vitro and in vivo antitumor and anti-metastatic abilities, by inhibiting the proliferation and lymphogenesis of cancer cells. Therefore, knocking down survivin by siRNA may be considered a potential therapeutic approach for the treatment of breast cancer. In addition, the present study has demonstrated that the ability of survivin-siRNA to attenuate metastasis in murine breast cancer 4T1 cells may be due to the downregulation of VEGF-C, which is associated with the PTEN/Akt/HIF-1α pathway in these cells. However, the signals that affect metastasis function in complex regulatory networks rather than a single pathway, and these signals are differently activated in different types of tumors. Therefore, the mechanisms by which survivin regulates metastasis remain to be further investigated.
Acknowledgements
This study was supported by grants from Shandong Science and Technology Development Planning (no. 2014GGH218023).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. (Erratum in CA Cancer J Clin 61: 134, 2011) [DOI] [PubMed] [Google Scholar]
- 2.Ali SM, Harvey HA, Lipton A. Metastatic breast cancer: Overview of treatment. Clin Orthop Relat Res. 2003;415(Suppl):S132–S137. doi: 10.1097/01.blo.0000092981.12414.7b. [DOI] [PubMed] [Google Scholar]
- 3.Bidard FC, Fehm T, Ignatiadis M, Smerage JB, Alix-Panabières C, Janni W, Messina C, Paoletti C, Müller V, Hayes DF, et al. Clinical application of circulating tumor cells in breast cancer: Overview of the current interventional trials. Cancer Metastasis Rev. 2013;32:179–188. doi: 10.1007/s10555-012-9398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackey JR, Kerbel RS, Gelmon KA, et al. Controlling angiogenesis in breast cancer: A systematic review of anti-angiogenic trials. Cancer Treat Rev. 2012;38:673–688. doi: 10.1016/j.ctrv.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Ran S, Volk L, Hall K, Flister MJ. Lymphangiogenesis and lymphatic metastasis in breast cancer. Pathophysiology. 2010;17:229–251. doi: 10.1016/j.pathophys.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989;63:181–187. doi: 10.1002/1097-0142(19890101)63:1<181::AID-CNCR2820630129>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 7.Timoshenko AV, Rastogi S, Lala PK. Migration-promoting role of VEGF-C and VEGF-C binding receptors in human breast cancer cells. Br J Cancer. 2007;97:1090–1098. doi: 10.1038/sj.bjc.6603993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun P, Gao J, Liu YL, Wei LW, Wu LP, Liu ZY. RNA interference (RNAi)-mediated vascular endothelial growth factor-C (VEGF-C) reduction interferes with lymphangiogenesis and enhances epirubicin sensitivity of breast cancer cells. Mol Cell Biochem. 2008;308:161–168. doi: 10.1007/s11010-007-9624-1. [DOI] [PubMed] [Google Scholar]
- 9.Joukov V, Pajusola K, Kaipainen A, Chilov D, Lahtinen I, Kukk E, Saksela O, Kalkkinen N, Alitalo K. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO. 1996;15:290–298. [PMC free article] [PubMed] [Google Scholar]
- 10.Valtola R, Salven P, Heikkilä P, Taipale J, Joensuu H, Rehn M, Pihlajaniemi T, Weich H, de Waal R, Alitalo K. VEGFR-3 and its ligand VEGF-C are associated with angiogenesis in breast cancer. Am J Pathol. 1999;154:1381–1390. doi: 10.1016/S0002-9440(10)65392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinoshita J, Kitamura K, Kabashima A, Saeki H, Tanaka S, Sugimachi K. Clinical significance of vascular endothelial growth factor-C (VEGF-C) in breast cancer. Breast Cancer Res Treat. 2011;66:159–164. doi: 10.1023/A:1010692132669. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Varney ML, Backora MW, Cowan K, Solheim JC, Talmadge JE, Singh RK. Down-regulation of vascular endothelial cell growth factor-C expression using small interfering RNA vectors in mammary tumors inhibits tumor lymphangiogenesis and spontaneous metastasis and enhances survival. Cancer Res. 2005;65:9004–9011. doi: 10.1158/0008-5472.CAN-05-0885. [DOI] [PubMed] [Google Scholar]
- 13.Cai X, Ma S, Gu M, Zu C, Qu W, Zheng X. Survivin regulates the expression of VEGF-C in lymphatic metastasis of breast cancer. Diagn Pathol. 2012;7:52. doi: 10.1186/1746-1596-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M, Shiraki K. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- 16.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 17.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22:8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 18.McKenzie JA, Liu T, Goodson AG, Grossman D. Survivin enhances motility of melanoma cells by supporting Akt activation and {α}5 integrin upregulation. Cancer Res. 2010;70:7927–7937. doi: 10.1158/0008-5472.CAN-10-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marioni G, Bertolin A, Giacomelli L, Marchese-Ragona R, Savastano M, Calgaro N, Marino F, De Filippis C, Staffieri A. Expression of the apoptosis inhibitor protein Survivin in primary laryngeal carcinoma and cervical lymph node metastasis. Anticancer Res. 2006;26:3813–3817. [PubMed] [Google Scholar]
- 21.Kedinger V, Meulle A, Zounib O, et al. Sticky siRNAs targeting survivin and cyclin B1 exert an antitumoral effect on melanoma subcutaneous xenografts and lung metastases. BMC Cancer. 2013;13:338. doi: 10.1186/1471-2407-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikehara M, Oshita F, Kameda Y, Ito H, Ohgane N, Suzuki R, Saito H, Yamada K, Noda K, Mitsuda A. Expression of survivin correlated with vessel invasion is a marker of poor prognosis in small adenocarcinoma of the lung. Oncol Rep. 2002;9:835–838. [PubMed] [Google Scholar]
- 23.Li X, Dang X, Sun X. Expression of survivin and VEGF-C in breast cancer tissue and its relation to lymphatic metastasis. Eur J Gynaecol Oncol. 2012;33:178–182. [PubMed] [Google Scholar]
- 24.Xue M, Ge Y, Zhang J, Liu Y, Wang Q, Hou L, Zheng Z. Fucoidan inhibited 4T1 mouse breast cancer cell growth in vivo and in vitro via downregulation of Wnt/β-catenin signaling. Nutr Cancer. 2013;65:460–468. doi: 10.1080/01635581.2013.757628. [DOI] [PubMed] [Google Scholar]
- 25.Guide for the Care and Use of Laboratory Animals (Guide) (revised 1985) Bethesda: National Institutes of Health; 1989. National Institutes of Health, Department of Health and Human Services. NIH Publication No. 85–23. [Google Scholar]
- 26.Xue M, Ge Y, Zhang J, Wang Q, Hou L, Liu Y, Sun L, Li Q. Anticancer properties and mechanisms of fucoidan on mouse breast cancer in vitro and in vivo. PloS One. 2012;7:e43483. doi: 10.1371/journal.pone.0043483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong XP, Xiao TH, Dong H, Jiang N, Zhao XG. Endostar combined with cisplatin inhibits tumor growth and lymphatic metastasis of lewis lung carcinoma xenografts in mice. Asian Pac J Cancer Prev. 2013;14:3079–3083. doi: 10.7314/APJCP.2013.14.5.3079. [DOI] [PubMed] [Google Scholar]
- 28.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 29.Hirakawa S, Brown LF, Kodama S, Paavonen K, Alitalo K, Detmar M. VEGF-C-induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood. 2007;109:1010–1017. doi: 10.1182/blood-2006-05-021758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skobe M, Hawighorst T, Jackson DG, Prevo R, Janes L, Velasco P, Riccardi L, Alitalo K, Claffey K, Detmar M. Induction of tumor lymphangiogenesis by VEGF-C promotes breast cancer metastasis. Nat Med. 2001;7:192–198. doi: 10.1038/84643. [DOI] [PubMed] [Google Scholar]
- 31.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 32.Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;23:4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- 33.Brito LGO, Schiavon VF, Andrade JM, Tiezzi DG, Peria FM, Marana HR. Expression of Hypoxia-inducible factor 1-α and vascular endothelial growth factor-C in locally advanced breast cancer patients. Clinics. 2011;66:1313–1320. doi: 10.1590/S1807-59322011000800002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West KA, Castillo SS, Dennis PA. Activation of the PI3K/Akt pathway and chemotherapeutic resistance. Drug Resist Updat. 2002;5:234–248. doi: 10.1016/S1368-7646(02)00120-6. [DOI] [PubMed] [Google Scholar]
- 36.Goc A, Al-Husein B, Kochuparambil ST, Liu J, Heston WW, Somanath PR. PI3 kinase integrates Akt and MAP kinase signaling pathways in the regulation of prostate cancer. Int J Oncol. 2011;38:267–277. [PubMed] [Google Scholar]