Abstract
The vimentin gene is a hallmark of epithelial-to-mesenchymal transition and has been observed to be overexpressed in various types of tumor cell line and tissue. Previous studies have reported correlations between vimentin DNA methylation levels and subsequent vimentin expression levels in solid tumors, including breast and colorectal cancer; however, to the best of our knowledge, such a correlation has not been reported for gastric cancer (GC) using Lauren classification. Therefore, the present study aimed to quantify DNA methylation levels of the vimentin gene using quantitative (q) methylation-specific polymerase chain reaction (PCR) in intestinal-type GC cell lines (MKN-28, AGS and MKN-1), diffuse-type GC cell lines (SGC-7901, SNU-5 and KATO III), the GES-1 immortalized human non-neoplastic gastric epithelial cell line, as well as in tumor and paratumor normal tissue samples. Furthermore, the present study analyzed the messenger RNA expression of the vimentin gene in these cell lines and tissues by reverse transcription-qPCR. A comparison of the clinicopathological features was conducted between patients, grouped according to the Lauren classification. The present study identified that the vimentin promoter region was hypermethylated in all GC cell lines and tumor tissue samples when compared with immortalized normal gastric epithelial cells and paratumor normal tissues. In addition, vimentin promoter methylation levels were observed to be higher in intestinal-type cell lines when compared with those of diffuse-type lines and tissues. Correspondingly, vimentin expression levels were lower in intestinal-type gastric cell lines compared with those of diffuse-type cell lines and tissues, and were lowest in the non-neoplastic gastric cell line and paratumor normal tissues. Patients with diffuse-type GC were on average younger (P=0.023), and exhibited higher tumor (P=0.020), node (P=0.032) and TNM classification of malignant tumor stage (P=0.039) than those with intestinal-type GC. Following treatment of AGS cells (which demonstrated the highest methylation level of the vimentin gene) with 5-aza-2′-deoxycytidine, vimentin expression was restored significantly. Thus, the present study revealed that vimentin promoter methylation levels are inversely correlated with vimentin expression levels in GC (according to Lauren classification). High levels of methylation in the vimentin gene promoter region may be involved in carcinogenesis and the development of GC, and may provide a novel molecular classification for GC.
Keywords: DNA methylation, gastric cancer, vimentin, epigenetic, Lauren classification
Introduction
Gastric cancer (GC) is a frequently occurring type of malignancy and the second most common cause of cancer-associated mortality worldwide (1). Gastric tumorigenesis is a complex, multi-step process involving alterations of numerous genes (2). Aberrant promoter methylation is an important mechanism for silencing certain tumor suppressor and tumor-associated genes, and is significant during the pathogenesis and progression of certain types of human cancer (3,4), including GC (5,6). Data suggests that DNA methylation may be a useful biomarker for cancer risk evaluation (3,5), early diagnosis (5), prognosis (4,5) and evaluation of sensitivity to chemotherapy (7).
The vimentin gene encodes an intermediate filament protein reported to be involved in cytoskeletal architecture (8–10), the immune response and stabilization of collagen messenger RNAs (mRNAs) (11). Given its multiple functions, vimentin is considered to be significant in the epithelial-to-mesenchymal transition (EMT), including upregulation of EMT-associated genes, adaptive responses to wound healing and pathological responses during cell invasion and metastasis (12). Lauren classification has been globally adopted and separates gastric carcinoma into intestinal and diffuse subtypes according to the morphological features of the tumor (13).
GC is generally classified into two histological types, intestinal and diffuse, and each type develops through a distinct carcinogenic pathway (14). Phenotypically, the two types exhibit distinct macroscopic appearances, reflecting the differences in their microscopic growth patterns and molecular signaling pathways (15). In intestinal-type carcinoma, the macroscopic margins approximately correspond with microscopic spread, while the poor differentiation of diffuse-type carcinoma facilitates submucosal extension beyond the macroscopic borders (15). This difference in tumor extension is of clinical significance in the selection of a suitable treatment strategy.
Although upregulation of levels of vimentin expression during EMT have been well characterized in GC, to the best of our knowledge, no study has reported epigenetic regulation of the vimentin gene in GC using Lauren classification. In the present study, vimentin promoter DNA methylation, and vimentin expression in gastric cell lines and human tissue samples, grouped according to Lauren classification, were examined to determine whether a correlation between the two existed.
Materials and methods
GC cell lines and human tissue samples
Six gastric cancer cell lines (MKN-28, AGS, MKN-1, SGC-7901, SNU-5 and KATO III) and one normal gastric epithelial cell line (GES-1) (American Type Culture Collection, Manassas, VA, USA) were used in the present study. The MKN-28 cells were checked for contamination. All cell lines were cultured and maintained in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS) (Gibco Life Technologies, Carlsbad, CA, USA) and 1% penicillin/streptomycin (100 U/ml penicillin and 100 µg/ml streptomycin; Invitrogen Life Technologies, Carlsbad, CA, USA) and incubated in 5% CO2 at 37°C.
Tumor and paratumor normal tissue samples of 64 patients with GC, who underwent radical gastrectomy at the Affiliated Hospital of Qingdao University (Qingdao, China), were collected between January 2008 and October 2011, and stored in liquid nitrogen. Written informed consent was obtained from all patients and the procedure of the present study was approved by the institutional review board of the Affiliated Hospital of Qingdao University. All samples were confirmed by histology and 36 diffuse-type and 28 intestinal-type samples, as well as their paired paratumor normal tissues, were selected for analysis. Clinicopathological features of patients enrolled in the present study are presented in Table I.
Table I.
Clinicopathological features of patients exhibiting gastric cancer according to Lauren classification.
| Lauren classification | ||||
|---|---|---|---|---|
| Clinicopathological features | Cases, n | Diffuse-type (n=36) | Intestinal-type (n=28) | P-value |
| Age, years | – | 54.2±9.1a | 69.7±9.6a | 0.023 |
| Gender | 0.072 | |||
| Male | 41 | 15 | 18 | |
| Female | 23 | 21 | 10 | |
| Maximal tumor size, mm | – | 56.3±23.7a | 51.4±22.6a | 0.264 |
| T stage (AJCC) | 0.020 | |||
| T1 | 6 | 1 | 5 | |
| T2 | 10 | 4 | 6 | |
| T3 | 39 | 28 | 11 | |
| T4 | 9 | 7 | 2 | |
| N stage (AJCC) | 0.032 | |||
| N0 | 18 | 6 | 12 | |
| N1 | 9 | 4 | 5 | |
| N2 | 16 | 11 | 5 | |
| N3 | 21 | 16 | 5 | |
| TNM stage (AJCC) | 0.039 | |||
| I | 15 | 4 | 11 | |
| II | 22 | 13 | 9 | |
| III | 27 | 18 | 9 | |
Data are expressed as the mean ± standard deviation. AJCC, American Joint Committee on Cancer; TNM, TNM classification of malignant tumors; T, tumor; N, (lymph) nodes; M, metastasis.
DNA and RNA isolation from cell lines and tissue samples
Genomic DNA extraction was conducted with the DNAeasy® Blood & Tissue kit (Qiagen GmbH, Hilden, Germany). RNAiso Plus Reagent (Takara Bio, Inc., Otsu, Japan) was used to extract total RNA, which was utilized in the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) prior to DNA isolation. For RT-qPCR, RNA was reverse transcribed using PrimeScript RT reagent (Takara Bio, Inc.) and the reaction product was treated with RNAse-free DNase I. Details of the RT-qPCR reaction are described below.
Sodium bisulfate modification and quantitative methylation-specific PCR (qMSP)
A total of 1 µg genomic DNA was subjected to bisulfate treatment using an Epitect Bisulfate kit (Qiagen GmbH). The bisulfate-treated DNA was amplified by qMSP using a LightCycler480 (Roche Diagnostics Ltd., Rotkreuz, Switzerland). Briefly, thermocycling was conducted in a final volume of 25 µl, containing 1.0 µl treated DNA sample, 100 nM of each primer and 12.5 µl SYBR Premix Ex Taq II (Takara Bio, Inc.). The PCR primer sequences for vimentin (GeneChem, Shanghai, China) were as follows: Methylated sense, 5′-TCGTTTCGAGGTTTTCGCGTTAGAGAC-3′ and vimentin methylated antisense, 5′-CGACTAAAACTCGACCGACTCGCGA-3′. PCR amplification consisted of 40 cycles (95°C for 5 sec and 55°C for 30 sec) following an initial denaturation step (95°C for 10 sec). Genomic DNA, methylated in vitro by CpG methyltransferase (Sss I; New England BioLabs, Inc., Ipswich, MA, USA), served as a positive control and a water blank served as a negative control. GAPDH served as an internal control for normalization. The percentage of vimentin promoter methylation in each sample was estimated using the following formula:
RT-qPCR gene expression analysis in gastric cell lines and tissue samples
Gene expression in gastric cell lines and tissue samples was quantified by RT-qPCR using a 7500 Real-Time PCR Platform (Applied Biosystems, Foster City, CA, USA). Vimentin and GAPDH expression levels were assessed using pre-designed TaqMAN probes VIM-Hs00185584_m1 and GAPDH-Hs02758991_g1, respectively (Applied Biosystems Life Technologies). Complementary DNA samples from three independent biological experiments were examined by RT-qPCR (50°C for 2 min, denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C annealing for 1 min). For each experiment, the samples were analyzed in duplicate. Normalized vimentin expression was determined using the comparative Ct (ΔΔCt) method using Relative Quantification Study software (7300 Sequence Detection system, version 1.4; Applied Biosystems Life Technologies).
5-Aza-2′-deoxycytidine (5-Aza-dC) treatment
To further examine whether vimentin expression is regulated by DNA promoter methylation, the AGS cell line, which demonstrated the highest level of vimentin gene methylation among the six cell lines, was selected for further study. The AGS cell line was cultured in RPMI-1640 medium supplemented with 10% FBS. Cells in the logarithmic proliferative phase were seeded into a 96-well plate (Corning, New York, NY, USA) at a density of 5,000 cells/well and cultured in an incubator (Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 saturated humidity for 24 h. Cells were subsequently treated with 1 µM 5-Aza-dC (Sigma-Aldrich, St. Louis, CA, USA) for 72 h, with replenishment of fresh medium containing 5-Aza-dC every 24 h. Cells in the treatment and control groups were then harvested. Demethylation of the vimentin promoter regions was detected using MSP, and vimentin mRNA expression levels were measured using RT-qPCR. MSP and RT-qPCR were performed as described previously in the current study.
Statistical analysis
Statistical analyses were performed using SPSS (version 17.0; SPSS, Inc., Chicago, IL, USA). Vimentin DNA methylation was compared between diffuse-type and intestinal-type cell lines or between tumor and paratumor normal tissue samples by Student's t-test, and correlation of vimentin promoter methylation with vimentin expression was assessed by Pearson's correlation analysis. The χ2 test or Student's t-test were utilized to compare the clinicopathological data. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinicopathological features of patients with GC are associated with Lauren classification
Clinicopathological data were assessed for differences between diffuse- and intestinal-type GC cells. There were 36 (56.3%) patients with diffuse-type and 28 (43.7%) patients with intestinal-type GC. As shown in Table I, a significant difference was observed between diffuse- and intestinal-type GC for age (P=0.023), tumor (T) stage (P=0.020), node (N) stage (P=0.020) and Tumor-Node-Metastasis (TNM) stage (P=0.039). There was no significant difference observed for gender (P=0.072) or maximal tumor size (P=0.264).
Vimentin promoter methylation and vimentin gene expression vary according to Lauren classification
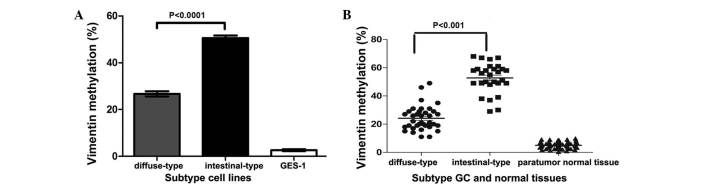
In order to investigate epigenetic silencing mechanisms of the vimentin gene in GC, qMSP technology was applied to evaluate the DNA methylation status of the vimentin promoter. GES-1 immortalized human non-neoplastic gastric epithelial cells, intestinal-type GC cell lines (MKN-28, AGS, MKN-1) and diffuse-type GC cell lines (SGC-7901, SNU-5, KATO III), as well as tumor and paratumor normal tissue samples were analyzed. All GC cell lines and tissues that were assessed demonstrated promoter hypermethylation (>10%), and methylation levels were significantly higher in intestinal-type compared with diffuse-type (P<0.0001; Fig. 1A and B). Conversely, GES-1 and paratumor normal tissues exhibited hypomethylation at the vimentin promoter (P<0.0001; Fig. 1A and B).
Figure 1.
DNA methylation of the vimentin promoter in gastric cell lines and tissues. (A) DNA methylation of the vimentin promoter was analyzed using methylation specific-quantitative polymerase chain reaction technology in diffuse-type GC cell lines (SGC-7901, SNU-5, KATO III), intestinal-type GC cell lines (MKN-28, AGS, MKN-1) and immortalized human non-neoplastic gastric epithelial cells (GES-1), as well as in (B) tumor and paratumor normal tissues. GC, gastric cancer.
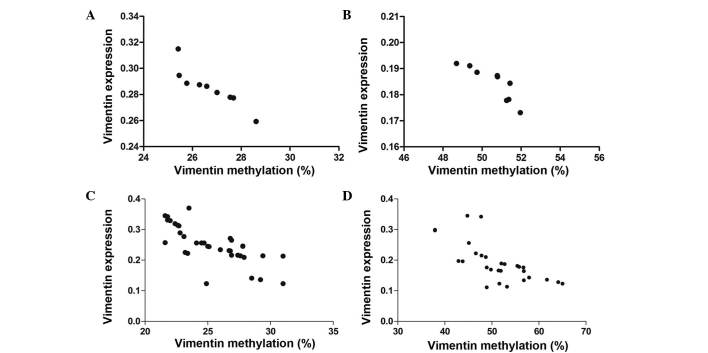
Subsequently, RT-qPCR was conducted to investigate the correlation between the vimentin promoter methylation and expression levels. Vimentin expression levels were strongly detected in diffuse-type cell lines and tissues, weakly detected in intestinal-type cell lines and tissues, and markedly reduced in GES-1 and paratumor normal tissues. Through comparison of vimentin promoter methylation and vimentin mRNA expression levels, an inverse correlation was identified (Table II) in the GC cell lines (Fig. 2A and B) and tissues (Fig. 2C and D). This may suggest that expression is negatively regulated by vimentin promoter DNA methylation.
Table II.
Spearman's correlation analysis for vimentin methylation and expression.
| Vimentin | ||||
|---|---|---|---|---|
| Source | Methylation, % | Expression | Pearson's r | P-value |
| Cell line | ||||
| Diffuse-type | 26.71±1.04 | 0.285±0.014 | −0.901 | <0.001 |
| Intestinal-type | 50.60±1.03 | 0.184±0.006 | −0.869 | <0.001 |
| GC tissue | ||||
| Diffuse-type | 19.48±5.26 | 0.342±0.0521 | −0.821 | <0.001 |
| Intestinal-type | 61.02±9.19 | 0.158±0.055 | −0.879 | <0.001 |
Data are expressed as the mean ± standard deviation. GC, gastric cancer.
Figure 2.
Correlations between vimentin methylation and relative expression levels in GC cell lines and tissues. Reverse transcription-quantitative polymerase chain reaction was used to analyze relative vimentin messenger RNA expression levels. A negative correlation was identified between vimentin DNA methylation and expression levels in (A) diffuse-type and (B) intestinal-type GC cell lines. (C and D) Comparable correlations were observed in diffuse- and intestinal-type of GC tissue. GC, gastric cancer.
Vimentin expression levels are lower in intestinal- than diffuse-type GCs
To evaluate the expression of vimentin in the various Lauren histological types, vimentin expression levels in intestinal-type cell lines and GC tissues were compared with those in diffuse-type cell lines and GC tissues. The results revealed significantly lower vimentin expression levels in intestinal-type cells and GC tissues when compared with diffuse-type cells and GC tissues (P<0.001; Fig. 3).
Figure 3.
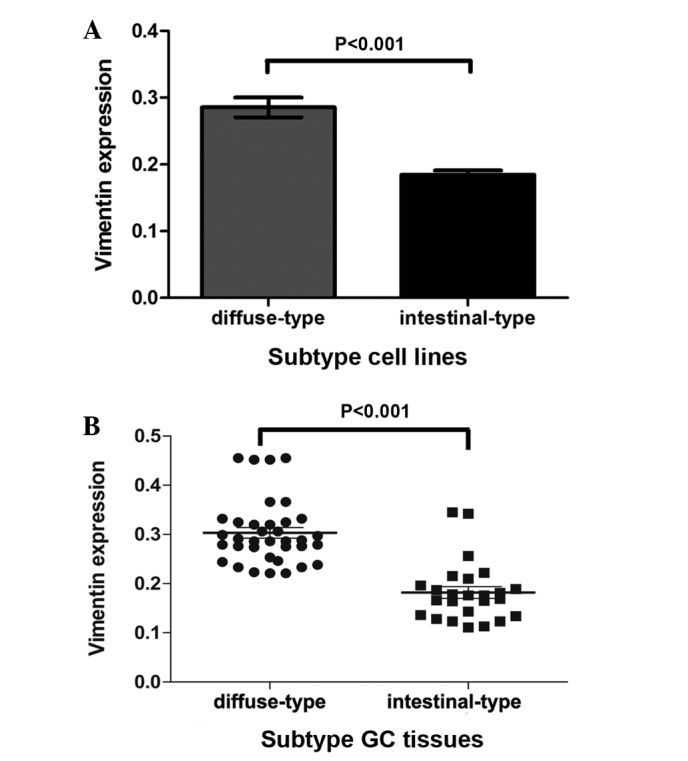
Diffuse-type GC exhibits higher vimentin expression levels compared with intestinal-type GC. (A) GC cell lines and (B) GC tissues. Statistical analysis was performed using Student's t test. GC, gastric cancer.
5-Aza-dC treatment restores vimentin expression
Among the six cell lines examined, AGS cells demonstrated the highest levels of vimentin gene methylation and thus, this cell line was selected for further study. To further examine whether vimentin expression was regulated by vimentin promoter DNA methylation, AGS cells were treated with a DNA demethylating agent (5-Aza-dC) for 72 h. Following treatment, promoter DNA methylation was significantly reduced, which was accompanied by reactivation of vimentin expression (P<0.01; Table III). Although significantly increased, vimentin expression was still in the range for GC.
Table III.
Varying expression levels of vimentin messenger RNA following 5-Aza-dC treatment in AGS cells (n=3).
| Group | Vimentin methylation, % | P-value | Vimentin expression | P-value |
|---|---|---|---|---|
| Control | 51.53±0.31 | 0.002 | 0.1764±0.0023 | 0.014 |
| 5-Aza-dC | 31.66±0.17 | 0.2251±0.0019 |
Data are expressed as the mean ± standard deviation. 5-Aza-dC, 5-Aza-2′-deoxycytidine.
Discussion
Vimentin is a type III intermediate filament protein involved in cell attachment, migration and signaling (9,16). Vimentin regulates integrins, which are heterodimeric transmembrane cell adhesion receptors, and stimulates development and turnover of adhesive structures, particularly in endothelial cells (17,18). Vimentin also affects the activity of certain cell membrane-associated signaling pathways, including mitogen-activated protein kinase (19,20). Overexpression of vimentin in cancer cells strongly correlates with levels of invasiveness and poor prognosis (21). Increased vimentin expression levels have been observed in a number of types of epithelial cancer, including prostate, gastrointestinal, central nervous system, breast, malignant melanoma and lung (21). Consistent with previous findings, the present study demonstrated vimentin overexpression in GC cell lines and clinical GC tissues.
Over the past decade, knowledge of the importance of epigenetic events in the control of normal cellular processes, and aberrant events leading to tumor development and progression has increased (3–7,22). DNA methylation is a major epigenetic mechanism that has been intensively investigated in the context of gene regulation and abnormal gene silencing in cancer cells (3–6). DNA methylation-associated biomarkers are under investigation in various types of human cancer (3–5,7). Studies have previously investigated epigenetic alterations in the vimentin gene, with data demonstrating differential vimentin DNA methylation levels in solid tumors, including colorectal, cervical, bladder and pancreatic cancer (23–26). In patients with GC, frequent methylation of vimentin DNA in serum (27) and tissues (28,29) has been detected. The frequency of vimentin DNA methylation in GC cell lines and tissues observed in the present study is consistent with the findings of the above-mentioned studies. Additionally, the results of the present study demonstrated an inverse correlation between vimentin DNA methylation and vimentin expression in GC cell lines and tissues grouped according to Lauren classification. Furthermore, restoration of vimentin expression in AGS cells was achieved through DNA demethylation with 5-Aza-dC. Data from the present study indicates that the inverse correlation between vimentin DNA methylation level and vimentin expression level may be cancer-specific, as it was not observed in GES-1 cells (a non-neoplastic gastric epithelial cell line) or paratumor normal tissues. GES-1 cells and paratumor normal tissue demonstrated reduced levels of vimentin expression, while the vimentin promoter was observed to be hypomethylated. These results suggest the existence of an alternative mechanism for the suppression of vimentin expression in normal cells, which requires further investigation.
The difference in tumor extension between intestinal and diffuse-type carcinoma is of clinical significance in the selection of a suitable treatment strategy (14,15). In the present study, the younger age (P=0.023), higher T (P=0.020), N (P=0.032) and TNM stage (P=0.039) of patients with diffuse-type GC indicated that diffuse-type was more invasive than intestinal-type GC, which may contribute to the poorer prognosis of patients with diffuse-type carcinoma (30). An improved understanding of the underlying pathogenesis and molecular events of the two histological subtypes may facilitate the development of novel diagnostic, therapeutic and preventive strategies for GC. The present study revealed differential hypermethylation at the vimentin gene promoter and an inverse correlation between vimentin DNA methylation and transcriptional expression levels among cancer cell lines and tissues, which were derived from the two types of GC. Intestinal-type GC exhibited higher promoter methylation levels and lower vimentin expression levels than those of diffuse-type. Considering the disparity in vimentin DNA methylation between the two types of GC and the marked correlation between vimentin expression, invasiveness and poor prognosis, vimentin DNA methylation may be involved in a mechanism that induces the distinct morphology and behavior of the two GC types by regulating vimentin mRNA expression. It has been reported that vimentin DNA methylation in colorectal cancer (CRC) tissues is significantly more frequent than that in normal and benign tissues and, therefore, vimentin promoter methylation may be involved in the carcinogenesis of CRC (31). The results of the present study demonstrated hypermethylation of the vimentin promoter in GC, which is comparable with CRC on a cytological and histological level. This suggests that the high level of methylation in the vimentin gene promoter region may be involved in carcinogenesis and the development of GC, and may serve as a novel molecular marker for GC.
In conclusion, the present study demonstrated that vimentin promoter methylation was inversely correlated with vimentin expression, and that the levels of vimentin expression were significantly different in the Lauren histological types of human GC. Vimentin gene hypermethylation may be associated with the occurrence and progression of GC, and detection of methylation of the vimentin gene may serve as a diagnostic marker for GC classification, which may provide guidance for the treatment and evaluation of GC.
Acknowledgements
The present study was supported by Shandong Excellent Young Scientist Research Award Fund Project (grant nos. 2006BSB14114 and BS2010YY013), Shandong Natural Science Foundation (grant nos. ZR2015HM085 and 2009HW024) and the National Natural Science Foundation of China (grant no. 81472338).
References
- 1.globocan.iarc.fr/Pages/fact_sheets_population.aspx. [Jul 14;2014 ];World Health Organization: Estimated cancer incidence, mortality, and prevalence worldwide in 2012. Accessed. [Google Scholar]
- 2.David S, Meltzer SJ. Stomach - genetic and epigenetic alterations of preneoplastic and neoplastic lesions. Cancer Biomark. 2010;9:493–507. doi: 10.3233/CBM-2011-0169. [DOI] [PubMed] [Google Scholar]
- 3.Peng D, Belkhiri A, Hu T, Chaturvedi R, Asim M, Wilson KT, Zaika A, El-Rifai W. Glutathione peroxidase 7 protects against oxidative DNA damage in oesophageal cells. Gut. 2012;61:1250–1260. doi: 10.1136/gutjnl-2011-301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge MH, Chen C, Xu JJ, Ling ZQ. Critical regions and spreading of runt-related transcription factor-3 C-phosphate-G (CpG) island methylation in human salivary gland adenoid cystic carcinoma. Hum Pathol. 2011;42:1862–1872. doi: 10.1016/j.humpath.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Lu XX, Yu JL, Ying LS, Han J, Wang S, Yu QM, Wang XB, Fang XH, Ling ZQ. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer. 2012;118:5507–5517. doi: 10.1002/cncr.27604. [DOI] [PubMed] [Google Scholar]
- 6.Yu QM, Wang XB, Luo J, Wang S, Fang XH, Yu JL, Ling ZQ. CDH1 methylation in preoperative peritoneal washes is an independent prognostic factor for gastric cancer. J Surg Oncol. 2012;106:765–771. doi: 10.1002/jso.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlet J, Schnekenburger M, Brown KW, Diederich M. DNA demethylation increases sensitivity of neuroblastoma cells to chemotherapeutic drugs. Biochem Pharmacol. 2012;83:858–865. doi: 10.1016/j.bcp.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kokkinos MI, Wafai R, Wong MK, Newgreen DF, Thompson EW, Waltham M. Vimentin and epithelial-mesenchymal transition in human breast cancer - observations in vitro and in vivo. Cells Tissues Organs. 2007;185:191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 10.Katz E, Dubois-Marshall S, Sims AH, Gautier P, Caldwell H, Meehan RR, Harrison DJ. An in vitro model that recapitulates the epithelial to mesenchymal transition (EMT) in human breast cancer. PLoS One. 2011;6:e17083. doi: 10.1371/annotation/3aa3ef2b-b38e-4f37-ad5d-c1448d722b26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mor-Vaknin N, Punturieri A, Sitwala K, Markovitz DM. Vimentin is secreted by activated macrophages. Nat Cell Biol. 2003;5:59–63. doi: 10.1038/ncb898. [DOI] [PubMed] [Google Scholar]
- 12.Ivaska J. Vimentin: Central hub in EMT induction? Small GTPases. 2011;2:51–53. doi: 10.4161/sgtp.2.1.15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vauhkonen M, Vauhkonen H, Sipponen P. Pathology and molecular biology of gastric cancer. Best Pract Res Clin Gastroenterol. 2006;20:651–674. doi: 10.1016/j.bpg.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauren P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 16.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 17.Gonzales M, Weksler B, Tsuruta D, Goldman RD, Yoon KJ, Hopkinson SB, Flitney FW, Jones JC. Structure and function of a vimentin-associated matrix adhesion in endothelial cells. Mol Biol Cell. 2001;12:85–100. doi: 10.1091/mbc.12.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homan SM, Mercurio AM, LaFlamme SE. Endothelial cells assemble two distinct alpha6beta4-containing vimentin-associated structures: Roles for ligand binding and the beta4 cytoplasmic tail. J Cell Sci. 1998;111:2717–2728. doi: 10.1242/jcs.111.18.2717. [DOI] [PubMed] [Google Scholar]
- 19.Perlson E, Michaelevski I, Kowalsman N, Ben-Yaakov K, Shaked M, Seger R, Eisenstein M, Fainziber M. Vimentin binding to phosphorylated Erk sterically hinders enzymatic dephosphorylation of the kinase. J Mol Biol. 2006;364:938–944. doi: 10.1016/j.jmb.2006.09.056. [DOI] [PubMed] [Google Scholar]
- 20.Janosch P, Kieser A, Eulitz M, et al. The Raf-1 kinase associates with vimentin kinases and regulates the structure of vimentin filaments. FASEB J. 2000;14:2008–2021. doi: 10.1096/fj.99-0883com. [DOI] [PubMed] [Google Scholar]
- 21.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, Eknaes M, Skotheim RI, Rodrigues A, Magalhães JS, Oliveira J, Lothe RA, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16:5842–5851. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]
- 23.Kisiel JB, Yab TC, Taylor WR, et al. Stool DNA testing for the detection of pancreatic cancer: Assessment of methylation marker candidates. Cancer. 2012;118:2623–2631. doi: 10.1002/cncr.26558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X, Chen S. Epigenetic regulation of cytochrome P450 enzymes and clinical implication. Curr Drug Metab. 2015;16:86–96. doi: 10.2174/138920021602150713114159. [DOI] [PubMed] [Google Scholar]
- 25.Jung S, Yi L, Kim J, Jeong D, Oh T, Kim CH, Kim CJ, Shin J, An S, Lee MS. The role of vimentin as a methylation biomarker for early diagnosis of cervical cancer. Mol Cells. 2011;31:405–411. doi: 10.1007/s10059-011-0229-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 27.Shirahata A, Sakuraba K, Kitamura Y, Yokomizo K, Gotou T, Saitou M, Kidawa G, Menoto H, Sanada Y, Hibi K. Detection of vimentin methylation in the serum of patients with gastric cancer. Anticancer Res. 2012;32:791–794. [PubMed] [Google Scholar]
- 28.Kitamura YH, Shirahata A, Sakata M, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, Hibi K. Frequent methylation of Vimentin in well-differentiated gastric carcinoma. Anticancer Res. 2009;29:2227–2229. [PubMed] [Google Scholar]
- 29.Moinova H, Leidner RS, Ravi L, Lutterbaugh J, Barnholtz-Sloan JS, Chen Y, Chak A, Markowitz SD, Willis JE. Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol Biomarkers Prev. 2012;21:594–600. doi: 10.1158/1055-9965.EPI-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu MZ, Cai MY, Zhang DS, Wang ZQ, Wang DS, Li YH, Xu RH. Clinicopathological characteristics and prognostic analysis of Lauren classification in gastric adenocarcinoma in China. J Transl Med. 2013;11:58. doi: 10.1186/1479-5876-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YW, Kong FM, Zhou JP, Dong M. Aberrant promoter methylation of the vimentin gene may contribute to colorectal carcinogenesis: A meta-analysis. Tumour Biol. 2014;35:6783–6790. doi: 10.1007/s13277-014-1905-1. [DOI] [PubMed] [Google Scholar]