Abstract
The present study aimed to investigate the potential mechanisms used during signal transduction by M3 muscarinic acetylcholine receptor (CHRM3) in prostate cancer. The microarray datasets of GSE3325, including 5 clinically localized primary prostate cancers and 4 benign prostate tissues, were downloaded from the Gene Expression Omnibus database. The differentially-expressed genes (DEGs) in primary prostate cancer tissues compared with benign controls were screened using the Limma package. Gene Ontology and pathway enrichment analyses were performed using the Database for Annotation Visualization and Integrated Discovery. Next, a protein-protein interaction (PPI) network was constructed. Additionally, microRNAs (miRNAs) associated with DEGs were predicted and miRNA-target DEG analysis was performed using a Web-based Gene Set Analysis Toolkit. Finally, the PPI network and the miRNA-target DEG network were integrated using Cytoscape. In total, 224 DEGs were screened in the prostate cancer tissues, including 113 upregulated and 111 downregulated genes. CHRM3 and epidermal growth factor (EGF) were enriched in the regulation of the actin cytoskeleton. EGF and v-myc avian myelocytomatosis viral oncogene homolog (Myc) were enriched in the mitogen-activated protein kinase (MAPK) signaling pathway. EGF with the highest degree of connectivity was the hub node in the PPI network, and miR-34b could interact with Myc directly in the miRNA-target DEG network. EGF and Myc may exhibit significant roles in the progression of prostate cancer via regulation of the actin cytoskeleton and the MAPK signaling pathway. CHRM3 may activate these two pathways in prostate cancer progression. Thus, these two key factors and pathways may be crucial mechanisms during signal transduction by CHRM3 in prostate cancer.
Keywords: prostate cancer, M3 muscarinic acetylcholine receptor, differentially-expressed genes, pathway enrichment analysis, protein-protein interaction network
Introduction
Prostate cancer is one of the leading causes of cancer-associated mortality in males, accounting for >240,000 new cancer cases and 28,000 fatalities annually in males in the United States (1,2). Although effective surgical and radiation treatments exist for clinically localized prostate cancer, the majority of patients with metastatic prostate cancer eventually succumb to the disease (3). Therefore, in order to develop more effective outcomes for the diagnosis and treatment of prostate cancer, articulation of the genetic underpinning and novel therapeutic targets are critically required.
Recently, the presence and function of muscarinic acetylcholine receptors (mAChRs) in the human prostate have aroused wide concern (4,5). mAChR and its ligands have been found to play key roles in regulating cellular proliferation and cancer progression (6). mAChRs are preferentially localized to the glandular epithelium of the prostate and promote the paracrine/autocrine actions within the prostate gland, which are critical for cancer cell survival, proliferation and migration (4,6,7). mAChRs consist of five distinct subtypes (M1-M5), and the M3 mAChR (also known as CHRM3) has been found to be a key member involved in prostate cancer. The stimulation of CHRM3 is strongly associated with the tumor growth of prostate carcinomas (4). Additionally, CHRM3 can effectively mediate the contractions of the mouse prostate elicited by acetylcholine (8). A great deal of attention has been focused on the roles of CHRM3 in prostate cancer, however, the signal transduction by CHRM3 in the pathophysiology of prostate cancer is not well understood. Therefore, investigation of the signal transduction by CHRM3 will herald a prominent expansion in our understanding of the molecular mechanisms of prostate cancer.
The microarray data of GSE3325 has been used to reveal critical genomic regions in the prostate tumor microenvironment for investigating novel biomarkers (9) or to reveal signatures of the metastatic progression of prostate cancer (10). In contrast to previous findings, the present study downloaded the GSE3325 microarray data and utilized comprehensive bioinformatics methods to identify the differentially-expressed genes (DEGs) associated with prostate cancer. Additionally, functional enrichment analysis of the DEGs was performed, and a protein-protein interaction (PPI) network and microRNA (miRNA)-target DEG network was constructed. The study aimed to elucidate the potential mechanisms used during signal transduction by CHRM3 in prostate cancer.
Materials and methods
Affymetrix microarray data
The microarray data of GSE3325 were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/), based on the platform of GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array; Affymetrix Inc., Santa Clara, CA, USA). The gene expression files were deposited by Varambally et al (10). A total of 19 samples were applied to the development of the array data, including 6 benign, 7 clinically localized primary and 6 metastatic prostate cancer tissues. Each category contained 2 pooled samples. In order to better investigate the molecular pathogenesis of prostate cancer, pooled samples were discarded, and the expression profiles analyzed in this study were derived from 9 samples, including 5 clinically localized primary prostate cancer and 4 benign prostate tissues.
Data preprocessing and DEG screening
The raw data were first preprocessed by the robust multiarray average algorithm (11) with application of Affy package (12) in R language. The gene expression matrix of the samples was then acquired.
The DEGs in the primary prostate cancer tissues compared with the benign controls were screened by Limma package (13,14) in R language. The P-value was adjusted using Student's t-test (13) in Limma. Fold-change (FC) of the gene expression was also observed for differential-expression test. The DEGs with adjusted P-values <0.05 and |log2 FC| >1 were considered to be significant.
Gene Ontology (GO) and pathway enrichment analysis
The GO database (15) is a large-scale collection of gene annotation terms. The Kyoto Encyclopedia of Genes and Genomes (KEGG) (16) is a pathway-related database for classification of correlating gene sets into their respective pathways. The Database for Annotation Visualization and Integrated Discovery (DAVID) (17) is an online tool used for systematically associating the functional terms with large gene or protein lists.
In order to analyze the function of DEGs, GO annotation associated with biological process (BP) and KEGG pathway enrichment analysis of DEGs by DAVID online tool were performed. P<0.05 and gene counts >2 were set as the threshold.
PPI network construction
The Search Tool for the Retrieval of Interacting Genes (STRING) (18) is an online database that provides comprehensive information on predicted and experimental interactions of proteins in a given cell. The interactions of protein pairs are displayed with a combined score. In the present study, the DEGs were mapped into the STRING database to construct a PPI network with a combined score of protein pairs of >0.4 as the cut-off value. In addition, the connectivity degree of each node in the PPI network was calculated and the hub node was then identified (19).
Prediction of miRNAs associated with DEGs
The Web-based Gene Set Analysis Toolkit (WebGestalt) (20) is a web-based integrated system for analyzing large-scale gene sets in various functional categories, such as transcription factor and miRNA targets. In order to screen the potential miRNAs associated with DEGs, the DEG lists were uploaded to the WebGestalt system, and a miRNA-target DEG analysis was performed by WebGestalt. Enriched gene counts of ≥2 and rawP<0.05 were defined as the cut-off value.
Then miRNA-target DEG network composed of miRNAs and their target DEGs was visualized in the application of Cytoscape (21) software.
Integration of PPI network and miRNA-target DEG network
In the present study, the miRNA-target DEGs and PPI networks were further integrated, and the PPI pair-miRNA network composed of miRNAs and their target protein pairs was established with Cytoscape (21) software.
Results
DEG screening
Using the cut-off value of adjusted P-values <0.05 and |log2 FC| >1, a total of 224 DEGs in the primary prostate cancer tissues compared with the benign controls were screened, including 113 upregulated and 111 downregulated genes.
GO and pathway enrichment analysis
GO and pathway analyses were performed for the upregulated and downregulated genes, respectively. The overrepresented GO-BP terms of the upregulated DEGs were mainly associated with amine transport, amino acid transport and the enzyme-linked receptor protein signaling pathway (Table I). The downregulated DEGs were mainly involved in the regulation of cell proliferation, the response to organic substances and the negative regulation of the nitrogen compound metabolic process (Table II).
Table I.
GO-BP terms and KEGG pathways enriched by upregulated genes.
| A, GO-BP terms | |||
|---|---|---|---|
| Term | Description | Count | P-value |
| GO:0015837 | Amine transport | 5 | 5.95×10−3 |
| GO:0006865 | Amino acid transport | 4 | 1.78×10−2 |
| GO:0007167 | Enzyme-linked receptor protein signaling pathway | 7 | 1.85×10−2 |
| GO:0051674 | Localization of cell | 6 | 4.03×10−2 |
| GO:0048870 | Cell motility | 6 | 4.03×10−2 |
| GO:0035239 | Tube morphogenesis | 4 | 4.32×10−2 |
| GO:0001501 | Skeletal system development | 6 | 4.62×10−2 |
| GO:0035295 | Tube development | 5 | 4.63×10−2 |
| B, Enriched KEGG pathways | |||
| Term | Description | Count | P-value |
| hsa04012 | ErbB signaling pathway | 3 | 8.21×10−2 |
| hsa05216 | Thyroid cancer | 2 | 1.48×10−1 |
| hsa00512 | O-glycan biosynthesis | 2 | 1.53×10−1 |
| hsa04514 | CAMs | 3 | 1.64×10−1 |
| hsa00071 | Fatty acid metabolism | 2 | 1.99×10−1 |
| hsa05219 | Bladder cancer | 2 | 2.08×10−1 |
| hsa05213 | Endometrial cancer | 2 | 2.51×10−1 |
| hsa05221 | Acute myeloid leukemia | 2 | 2.75×10−1 |
| hsa03320 | PPAR signaling pathway | 2 | 3.19×10−1 |
| hsa04810 | Regulation of actin cytoskeleton | 3 | 3.33×10−1 |
| hsa04350 | TGF-β signaling pathway | 2 | 3.84×10−1 |
| hsa04060 | Cytokine-cytokine receptor interaction | 3 | 4.27×10−1 |
| hsa05200 | Pathways in cancer | 3 | 5.48×10−1 |
| hsa04310 | Wnt signaling pathway | 2 | 5.71×10−1 |
| hsa04144 | Endocytosis | 2 | 6.45×10−1 |
| hsa04510 | Focal adhesion | 2 | 6.78×10−1 |
| hsa04080 | Neuroactive ligand-receptor interaction | 2 | 7.66×10−1 |
| hsa04010 | MAPK signaling pathway | 2 | 7.80×10−1 |
| hsa04740 | Olfactory transduction | 2 | 8.86×10−1 |
Term, identification number of GO term or KEGG pathway; Description, name of GO term or KEGG pathway; Count, number of genes enriched in GO term or KEGG pathway; GO, Gene Oncology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CAMs, cell adhesion molecules; MAPK, mitogen-activated protein kinase; TGF-β, transforming growth factor-β; PPAR, peroxisome proliferator-activated receptor.
Table II.
Top 10 GO-BP terms and KEGG pathways enriched by downregulated genes.
| A, GO-BP terms | |||
|---|---|---|---|
| Term | Description | Count | P-value |
| GO:0042127 | Regulation of cell proliferation | 11 | 9.36×10−3 |
| GO:0010033 | Response to organic substance | 9 | 3.99×10−2 |
| GO:0051172 | Negative regulation of nitrogen compound metabolic process | 8 | 2.16×10−2 |
| GO:0031327 | Negative regulation of cellular biosynthetic process | 8 | 3.13×10−2 |
| GO:0009890 | Negative regulation of biosynthetic process | 8 | 3.45×10−2 |
| GO:0044092 | Negative regulation of molecular function | 7 | 9.21×10−3 |
| GO:0008015 | Blood circulation | 6 | 3.29×10−3 |
| GO:0003013 | Circulatory system process | 6 | 3.29×10−3 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 6 | 1.44×10−2 |
| GO:0045892 | Negative regulation of transcription, DNA-dependent | 6 | 4.31×10−2 |
| B, The enriched KEGG pathways | |||
| Term | Description | Count | P-value |
| hsa00982 | Drug metabolism | 3 | 3.14×10−2 |
| hsa04360 | Axon guidance | 3 | 1.14×10−1 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 2 | 2.39×10−1 |
| hsa04512 | ECM-receptor interaction | 2 | 3.19×10−1 |
| hsa04350 | TGF-β signaling pathway | 2 | 3.28×10−1 |
| hsa04916 | Melanogenesis | 2 | 3.64×10−1 |
| hsa04630 | Jak-STAT signaling pathway | 2 | 5.10×10−1 |
| hsa04020 | Calcium signaling pathway | 2 | 5.56×10−1 |
| hsa04510 | Focal adhesion | 2 | 6.05×10−1 |
| hsa05200 | Pathways in cancer | 2 | 7.85×10−1 |
Term, identification number of GO term or KEGG pathway; Description, name of GO term or KEGG pathway; Count, number of genes enriched in GO term or KEGG pathway. BP, biological process; GO, Gene Oncology; KEGG, Kyoto Encyclopedia of Genes and Genomes; TGF-β, transforming growth factor-β; ECM, extracellular matrix; Jak-STAT, Janus kinase-signal transducer and activator of transcription.
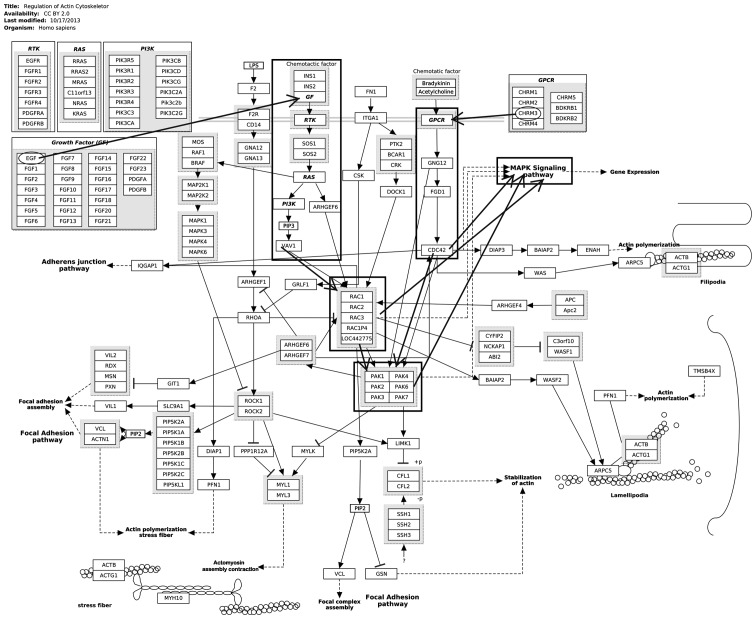
The upregulated DEGs were significantly enriched in the ErbB signaling pathway, thyroid cancer and O-glycan biosynthesis (Table I), while downregulated DEGs were significantly enriched in drug metabolism, axon guidance and metabolism of xenobiotics by cytochrome P450 (Table II). Notably, CHRM3 and epidermal growth factor (EGF) were significantly enriched in regulation of the actin cytoskeleton (Fig. 1). EGF and v-myc avian myelocytomatosis viral oncogene homolog (Myc) were enriched in the mitogen-activated protein kinase (MAPK) signaling pathway.
Figure 1.
Regulation of actin cytoskeleton pathway.
PPI network analysis
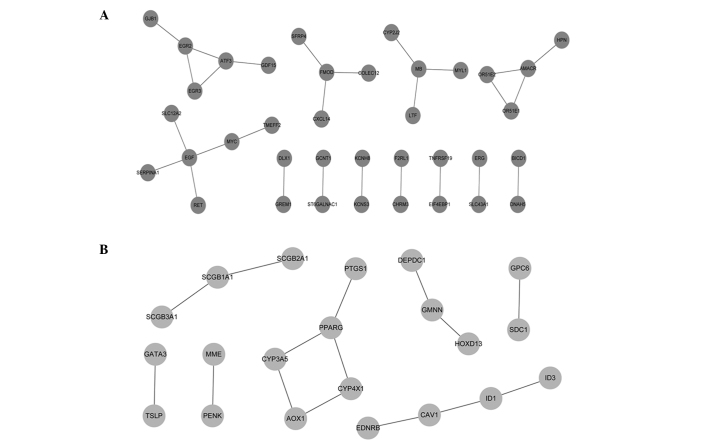
Based on the STRING database, a total of 27 upregulated PPIs (Fig. 2A) and 15 downregulated PPIs (Fig. 2B) with a combined score of >0.4 were obtained. The upregulated nodes with a connectivity degree of >2 were EGF (4), myoglobin (MB) (3), α-methylacyl-CoA racemase (3), early growth response 2 (3), fibromodulin (3) and activating transcription factor 3 (3). The downregulated node with a connectivity degree of >2 was peroxisome proliferator-activated receptor γ.
Figure 2.
Protein-protein interaction networks of differentially-expressed genes. (A) The dark gray nodes indicate upregulated genes and (B) the light gray nodes indicate downregulated genes. The lines indicate interactions between these genes.
Prediction of miRNAs associated with DEGs
The miRNAs associated with DEGs were predicted by WebGestalt. A total of 11 upregulated miRNAs, including miR-23A, miR-23B, miR-219 and miR-143, and 4 downregulated miRNAs, including miR-193a, miR-193b, miR-124a and miR-302c, were selected (Table III). Notably, miR-34b could interact with Myc directly.
Table III.
Predicted miRNAs associated with DEGs.
| A, Upregulated | ||
|---|---|---|
| miRNA | Count | P-value |
| hsa_GACAATC, miR-219 | 5 | 1.10×10−3 |
| hsa_TCATCTC, miR-143 | 5 | 1.20×10−3 |
| hsa_AATGTGA, miR-23a, miR-23b | 8 | 2.00×10−3 |
| hsa_ACTGCCT, miR-34b | 5 | 6.20×10−3 |
| hsa_AAGCAAT, miR-137 | 5 | 6.30×10−3 |
| hsa_GAGCCAG, miR-149 | 4 | 7.60×10−3 |
| hsa_TCTGATA, miR-361 | 3 | 1.34×10−2 |
| hsa_CATGTAA, miR-496 | 4 | 1.41×10−2 |
| hsa_GCAAGAC, miR-431 | 2 | 2.33×10−2 |
| hsa_TACTTGA, miR-26a, miR-26b | 5 | 2.33×10−2 |
| hsa_CAGTGTT, miR-141, miR-200a | 5 | 2.61×10−2 |
| B, Downregulated | ||
| miRNA | Count | P-value |
| hsa_GGCCAGT, miR-193a, miR-193b | 3 | 1.11×10−2 |
| hsa_TGCCTTA, miR-124a | 7 | 2.22×10−2 |
| hsa_ATGTTAA, miR-302c | 4 | 3.31×10−2 |
P-values are calculated from hypergeometric tests (rawP) and adjusted by the multiple test adjustment. Count, number of genes that interact with miRNAs; miRNAs, microRNAs; DEGs, differentially-expressed genes.
Integration of PPI and miRNA-target DEG networks
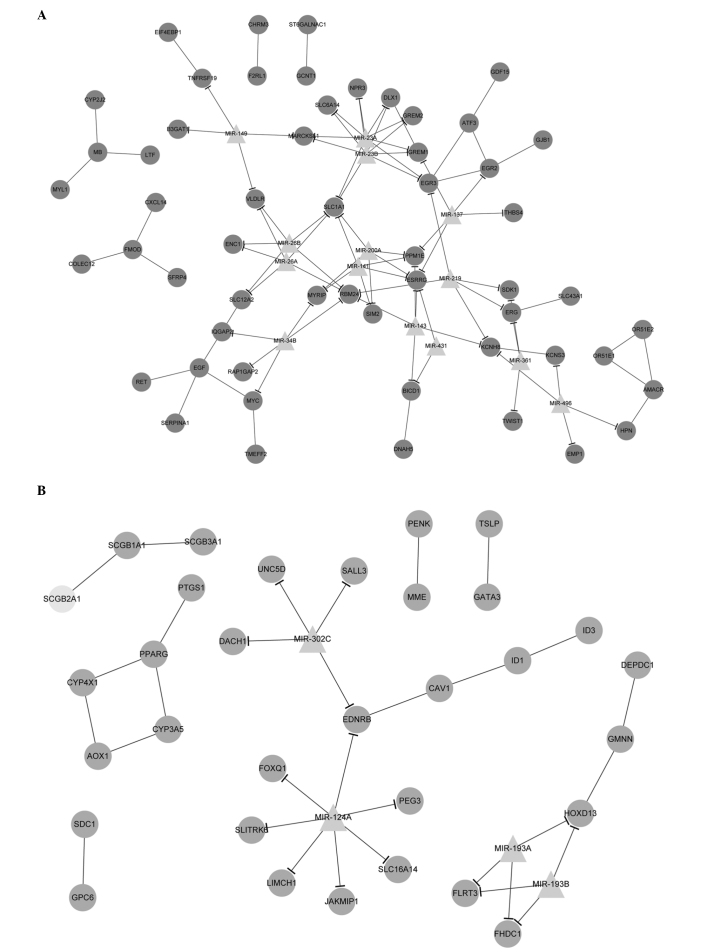
With Cytoscape software, the PPI and miRNA-target DEG networks were successfully integrated together. The upregulated and downregulated PPI pair-miRNA networks with significant protein pairs and their corresponding miRNAs were also established (Fig. 3).
Figure 3.
Integration of protein-protein interaction and miRNA-target differentially-expressed gene networks. (A) The dark gray circular nodes indicate upregulated genes and (B) the light gray circular nodes indicate downregulated genes. Gray triangular nodes indicate microRNAs. The lines indicate interactions between these nodes.
Discussion
In the present study, the bioinformatics approach was used to investigate the potential mechanisms used during signal transduction by CHRM3 in prostate cancer. The results showed that CHRM3 and EGF were significantly enriched in regulation of actin cytoskeleton. EGF and Myc were enriched in the MAPK signaling pathway. Notably, as shown in Fig. 1, the activation of the regulation of the actin cytoskeleton may induce the activation of MAPK signaling pathway to a certain extent, thus promoting the progression of prostate cancer. Therefore, these DEGs and the aforementioned two pathways may be potential mechanisms during signal transduction by CHRM3 in prostate progression.
EGF, with the highest degree of connectivity, was the hub protein in the PPI network. Growth factors (GFs), including EGF, and their transmembrane receptor tyrosine kinases (RTKs) play important roles in the growth, proliferation, migration and differentiation of various human tumor cells (22,23). The EGF receptor (EGFR) family of RTKs is formed by four members: EGFR/ErbB1, human epidermal growth factor (HER)2/ErbB2, HER3/ErbB3 and HER4/ErbB4 (24). Increased expression and/or amplification of EGFR and HER2 have been recorded in a range of human cancer types (24). Additionally, stimulation with EGF results in the activation of the lipoprotein kinase, phosphatidylinositol 3-kinase (PI3K), which phosphorylates phosphatidylinositol 4,5-bisphosphate, generating the second messenger phosphatidylinositol (3,4,5)-trisphosphate (25). PI3K-Akt signaling pathway activation promotes prostate cancer cell metastasis and invasion (26,27). In the present study, the PI3K-Akt signaling pathway activated by EGF was a part of the regulation of the actin cytoskeleton. Therefore, the present results are in line with previous findings and suggest that EGF may play roles in the metastasis and invasion of prostate cancer via regulation of the actin cytoskeleton.
Furthermore, EGFR and HER2 have been identified to be essential pathway elements in the signaling from G protein-coupled receptors (GPCRs) (24). CHRM3 belongs to the GPCRs and is coupled to MAPK via EGFR (28). GPCRs may interact with Rho guanosine triphosphatases, including RhoA and Cdc42, which play roles in regulating cell motility (29). Moreover, GPCRs can induce EGFR transactivation, thus generating signals defining the required biological response (24). The roles of EGFR are as aforementioned. In addition, previous findings have strongly linked the excessive activation of the GPCR and RTK pathways to prostate cancer metastasis (26). Therefore, as shown in Fig. 1, CHRM3 may activate the regulation of the actin cytoskeleton via EGFR or GPCRs for the promotion of the metastasis of prostate cancer.
Notably, the highly-conserved MAPK signaling pathway can be activated by EGFR (22,24,30). Previous studies have also suggested that the activation of the MAPK cascade can be mediated by GPCRs via several distinct pathways (31). Moreover, the MAPK signaling pathway may be activated by means of a PI3K-dependent feedback loop in human cancer (32). The PI3K/Akt/mTOR and MAPK signaling pathways are often observed in prostate tumors (33). Therefore, CHRM3 may be further involved in the MAPK signaling pathway via the roles of EGFR or the activation of the PI3K-Akt signaling pathway. Additionally, MAPK signaling pathway activation is necessary for inhibitor of DNA binding 1-induced serum-independent prostate cancer cell growth (34). Targeting MAPK signaling pathway activated by AKT/mTOR and MEK/ERK can inhibit the progression of prostate cancer in humans (35). Taken together, these results and the present study results suggest that CHRM3 may play important roles in prostate cancer progression via activating the MAPK signaling pathway.
In addition to EGF, Myc was found to be significantly enriched in the MAPK signaling pathway in the present study. Myc is a basic helix-loop-helix leucine zipper transcriptional factor that functions in cell proliferation, differentiation and death (36). An increased Myc gene copy number is found in human prostate cancer, and Pim-1 kinase can cooperate with Myc in tumorigenesis (37). c-Myc may link v-ets avian erythroblastosis virus E26 oncogene homolog to a major oncogenic pathway in prostate cancer (38). In addition, Myc could interact with miR-34b directly in the present study. miR-34 mediates androgen receptor-dependent p53-induced apoptosis in prostate cancer (39). The study by Corney et al confirmed that miR-34b is a target of p53 and plays important roles in the control of cell proliferation (40). Furthermore, the MAPK/heterogeneous nuclear ribonucleoprotein K pathway controls the oncogenic potential of breakpoint cluster region/Abelson murine leukemia viral oncogene homolog 1 (BCR/ABL) oncoprotein via the regulation of Myc mRNA translation (41). Therefore, Myc may play an important role in prostate cancer via the MAPK signaling pathway, and the present results are consistent with the findings that the MAPK signaling pathway may be the key mechanism used during signal transduction by CHRM3 in prostate cancer.
In conclusion, EGF and Myc may play significant roles in the progression of prostate cancer via regulation of the actin cytoskeleton and the MAPK signaling pathway. CHRM3 may activate these two pathways via EGFR or GPCRs in prostate cancer progression. Thus, regulation of the actin cytoskeleton, the MAPK signaling pathway and the two key factors of EGF and Myc may be crucial mechanisms during signal transduction by CHRM3 in prostate cancer. The present findings shed new light on the molecular mechanism of prostate cancer and have implications for future research. However, a relatively small sample size and no experimental validation are limitations to the present study, and further genetic studies are required to confirm these observations.
Acknowledgements
The present study was supported by the Doctoral Fund of the Ministry of Education of China (grant no. 20120131120070).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 4.Rayford W, Noble MJ, Austenfeld MA, Weigel J, Mebust WK, Shah GV. Muscarinic cholinergic receptors promote growth of human prostate cancer cells. Prostate. 1997;30:160–166. doi: 10.1002/(SICI)1097-0045(19970215)30:3<160::AID-PROS3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 5.Witte LP, Chapple CR, de la Rosette JJ, Michel MC. Cholinergic innervation and muscarinic receptors in the human prostate. Eur Urol. 2008;54:326–334. doi: 10.1016/j.eururo.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Shah N, Khurana S, Cheng K, Raufman J-P. Muscarinic receptors and ligands in cancer. Am J Physiol Cell Physiol. 2009;296:C221–C232. doi: 10.1152/ajpcell.00514.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura S, Pennefather J, Mitchelson F. Cholinergic innervation and function in the prostate gland. Pharmacol Ther. 2002;94:93–112. doi: 10.1016/S0163-7258(02)00174-2. [DOI] [PubMed] [Google Scholar]
- 8.White CW, Short JL, Haynes JM, Matsui M, Ventura S. Contractions of the mouse prostate elicited by acetylcholine are mediated by M(3) muscarinic receptors. J Pharmacol Exp Ther. 2011;339:870–877. doi: 10.1124/jpet.111.186841. [DOI] [PubMed] [Google Scholar]
- 9.Ashida S, Orloff MS, Bebek G, Zhang L, Zheng P, Peehl DM, Eng C. Integrated analysis reveals critical genomic regions in prostate tumor microenvironment associated with clinicopathologic phenotypes. Clin Cancer Res. 2012;18:1578–1587. doi: 10.1158/1078-0432.CCR-11-2535. [DOI] [PubMed] [Google Scholar]
- 10.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 12.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 13.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 14.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York, NY: Springer; 2005. pp. 397–420. [DOI] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium: Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2:e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server):W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 22.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–S8. doi: 10.1016/S0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 23.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 24.Prenzel N, Fischer OM, Streit S, Hart S, Ullrich A. The epidermal growth factor receptor family as a central element for cellular signal transduction and diversification. Endocr Relat Cancer. 2001;8:11–31. doi: 10.1677/erc.0.0080011. [DOI] [PubMed] [Google Scholar]
- 25.Bjorge JD, Chan T-O, Antczak M, Kung H-J, Fujita DJ. Activated type I phosphatidylinositol kinase is associated with the epidermal growth factor (EGF) receptor following EGF stimulation. Proc Natl Acad Sci USA. 1990;87:3816–3820. doi: 10.1073/pnas.87.10.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin J, Xie Y, Wang B, Hoshino M, Wolff DW, Zhao J, Scofield MA, Dowd FJ, Lin MF, Tu Y. Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene. 2009;28:1853–1863. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla S, Maclennan GT, Hartman DJ, Fu P, Resnick MI, Gupta S. Activation of PI3K-Akt signaling pathway promotes prostate cancer cell invasion. Int J Cancer. 2007;121:1424–1432. doi: 10.1002/ijc.22862. [DOI] [PubMed] [Google Scholar]
- 28.Slack BE. The m3 muscarinic acetylcholine receptor is coupled to mitogen-activated protein kinase via protein kinase C and epidermal growth factor receptor kinase. Biochem J. 2000;348:381–387. doi: 10.1042/0264-6021:3480381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie D, Guo Y, Yang D, Tang Y, Chen Y, Wang MT, Zacharek A, Qiao Y, Che M, Honn KV. Thromboxane A2 receptors in prostate carcinoma: Expression and its role in regulating cell motility via small GTPase Rho. Cancer Res. 2008;68:115–121. doi: 10.1158/0008-5472.CAN-07-1018. [DOI] [PubMed] [Google Scholar]
- 30.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta. 2007;1773:1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naor Z, Benard O, Seger R. Activation of MAPK cascades by G-protein-coupled receptors: The case of gonadotropin-releasing hormone receptor. Trends Endocrinol Metab. 2000;11:91–99. doi: 10.1016/S1043-2760(99)00232-5. [DOI] [PubMed] [Google Scholar]
- 32.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Kobayashi T, Floc'h N, Kinkade CW, Aytes A, Dankort D, Lefebvre C, Mitrofanova A, Cardiff RD, McMahon M, et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res. 2012;72:4765–4776. doi: 10.1158/0008-5472.CAN-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling M-T, Wang X, Ouyang X-S, Lee TK, Fan TY, Xu K, Tsao SW, Wong YC. Activation of MAPK signaling pathway is essential for Id-1 induced serum independent prostate cancer cell growth. Oncogene. 2002;21:8498–8505. doi: 10.1038/sj.onc.1206007. [DOI] [PubMed] [Google Scholar]
- 35.Kinkade CW, Castillo-Martin M, Puzio-Kuter A, Yan J, Foster TH, Gao H, Sun Y, Ouyang X, Gerald WL, Cordon-Cardo C, et al. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J Clin Invest. 2008;118:3051–3064. doi: 10.1172/JCI34764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 37.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/S1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 38.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whitman E, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rokhlin OW, Scheinker VS, Taghiyev AF, Bumcrot D, Glover RA, Cohen MB. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther. 2008;7:1288–1296. doi: 10.4161/cbt.7.8.6284. [DOI] [PubMed] [Google Scholar]
- 40.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 41.Notari M, Neviani P, Santhanam R, Blaser BW, Chang JS, Galietta A, Willis AE, Roy DC, Caligiuri MA, Marcucci G, et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107:2507–2516. doi: 10.1182/blood-2005-09-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]