Abstract
The high frequency of a unique neonatal preleukaemic syndrome, Transient Abnormal Myelopoiesis (TAM), and subsequent acute myeloid leukaemia in early childhood in patients with trisomy 21 (Down syndrome) points to a specific role for trisomy 21 in transforming foetal haematopoietic cells. N-terminal truncating mutations in the key haematopoietic transcription factor GATA1 are acquired during foetal life in virtually every case. These mutations are not leukaemogenic in the absence of trisomy 21. In mouse models, deregulated expression of chromosome 21-encoded genes is implicated in leukaemic transformation, but does not recapitulate the effects of trisomy 21 in a human context. Recent work using primary human foetal liver and bone marrow cells, human embryonic stem cells and iPS cells cells shows that prior to acquistion of GATA1 mutations, trisomy 21 itself alters human foetal haematopoietic stem cell and progenitor cell biology causing multiple abnormalities in myelopoiesis and B-lymphopoiesis. The molecular basis by which trisomy 21 exerts these effects is likely to be extremely complex, to be tissue- and lineage-specific and to be dependent on ontogeny-related characteristics of the foetal microenvironment.
Keywords: trisomy 21, foetal liver, Transient Abnormal Myelopoiesis, Acute Megakaryoblastic Leukaemia, Neonatal leukaemia, Down syndrome
Introduction
It has long been recognised that children with constitutional trisomy 21 (Down syndrome; DS) have a markedly increased risk of acute leukaemia [1]. Remarkably, this susceptibility to haematopoietic malignancies manifests as an increased risk both of acute megakaryocyte (MK)-erythroid leukaemia (known as ML-DS) by 150-fold and of acute B-lineage lymphoblastic leukaemia (B-ALL) by 33-fold compared to children without DS [1, 2]. The unique features of DS-associated leukaemias not only indicate the crucial role played by trisomy 21 in their pathogenesis, but also represent potentially tractable models of multistep leukaemogenesis [3-5]. Furthermore, it is now clear that the myeloid, and possibly the lymphoblastic, leukaemias originate in foetal life.
Foetal origin of trisomy 21-associated leukaemias
ML-DS is characterised by a clinical presentation virtually confined to the first 5 years of childhood [1, 4]; and an antecedent, clonally-linked preleukaemic condition (termed Transient Abnormal Myelopoiesis, TAM) in most cases. TAM, a clonal myeloproliferative syndrome unique to DS, presents in foetal life or a few days after birth [3, 6-9]. Typically, the syndrome is characterised by the presence of high numbers of circulating blasts, together with diffuse hepatic infiltration with abnormal megakaryocytes and megakaryoblasts [6-9]. We, and others, have shown that virtually all cases of TAM and ML-DS have acquired N-terminal truncating mutations in the erythroid-megakaryocyte transcription factor gene GATA1 [10-15]. These mutations, which are not leukaemogenic in the absence of trisomy 21 [16], are present at, or before, birth and disappear when TAM (or ML-DS) enters remission [10]. While some cases of TAM progress to ML-DS, most cases spontaneously resolve within 3 months of birth (Figure 1)[6-8]. Thus, clinical, biological and molecular data indicate that TAM is a foetal haematopoietic disorder.
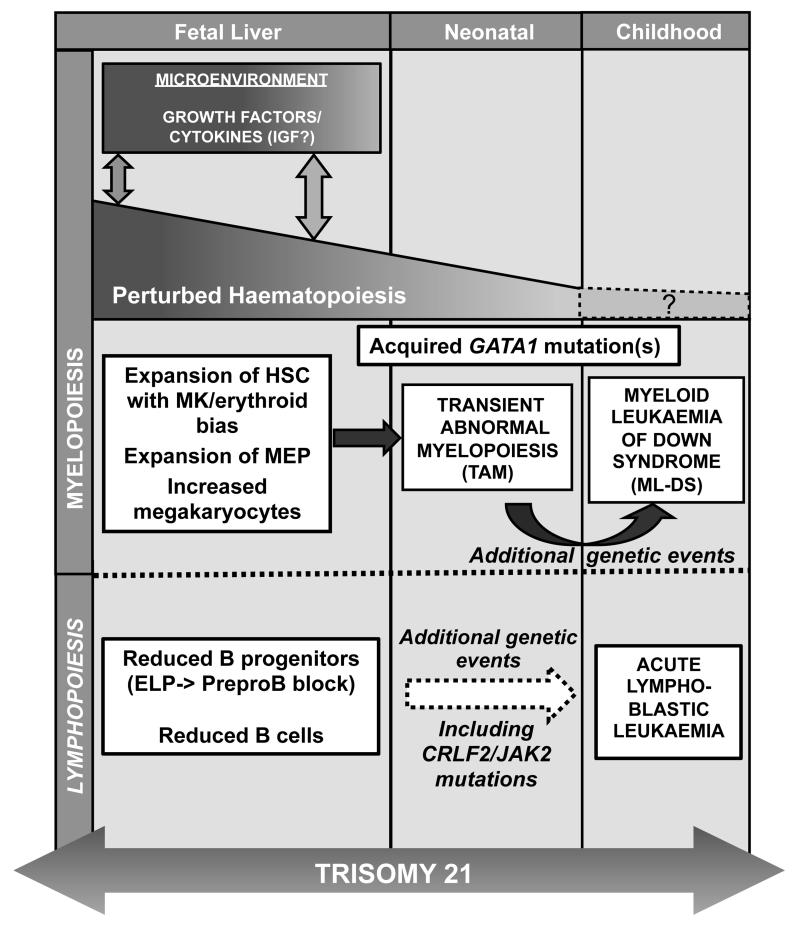
Figure 1. Impact of trisomy 21 on foetal and post-natal hematopoiesis.
Schematic representation of the effect of trisomy 21 (T21) on foetal, neonatal and childhood haematopoiesis. Foetal liver cells trisomic for chromosome 21 demonstrate perturbed haematopoiesis with an expansion of the haematopoietic stem cell compartment (HSC), megakaryocyte (MK)-erythroid progenitors (MEP) and megakaryocytes together with reduced B lymphopoiesis. Interaction of haematopoietic cells with the T21 foetal liver microenvironment may play an important role in initiating or maintaining abnormal foetal haematopoiesis, providing a susceptible HSC/progenitor pool upon which subsequent acquisition of N-terminal truncating GATA1 mutations would have a selective advantage. Expansion of the foetal liver mutant GATA1 HSC/progenitor population results in Transient Abnormal Myelopoiesis (TAM) in late foetal or early neonatal life. Although most cases of TAM resolve spontaneously, up to 30% of cases develop Down syndrome-associated acute myeloid leukaemia (ML-DS) before the age of 5 years as a result of additional genetic/ epigenetic events. Reduced foetal B lymphopoiesis may also underlie the increased susceptibility of children with Down Syndrome (T21) to acute lymphoblastic leukaemia (ALL).
Trisomy 21 and human foetal haematopoiesis
Trisomy 21 may impact on haematopoietic cell biology in multiple complex ways [17]. Trisomic genes, individually or collectively, may be directly involved through gene dosage; their effects may be haematopoietic cell autonomous or via other cell types, and the effects may be exerted indirectly via disomic genes. To address this, we have investigated the cellular consequences of trisomy 21 in primary human foetal haematopoietic cells, prior to the acquisition of GATA1 mutations.
Perturbation of second trimester haematopoiesis by trisomy 21
We [18], and others [19] found a specific and marked expansion of the megakaryocyte-erythroid progenitor (MEP) compartment in second trimester DS foetal liver in the absence of detectable GATA1 mutations. To investigate whether the abnormalities in the myeloid progenitor compartment of trisomy 21 foetal liver were confined to the MEP compartment, or extended to the HSC or multipotential progenitor (MPP) level, we have recently gone on to perform detailed immunophenotypic and functional analyses of the HSC/MPP, committed myeloid and B-lymphoid compartments of trisomy 21 foetal liver and compared these with normal foetal liver [20]. We demonstrated for the first time, that in human foetal liver, trisomy 21 itself increases immunophenotypic HSC frequency, clonogenicity, MK-erythroid output and biases erythroid-megakaryocyte primed gene expression with associated MEP expansion. In addition, immunohistochemical studies of trisomy 21 foetal liver sections showed that megakaryocytes were both increased [20] and morphologically abnormal. Furthermore, we found severe impairment of B-lymphoid development with ~10-fold reduction in pre-pro B-cells and B-cell potential of HSC, in tandem with reduced HSC lymphoid gene expression priming suggesting that multiple genes are likely to be involved at distinct foetal stages of haematopoiesis (see below).
The same pattern of foetal liver haematological abnormalities was seen in every trisomy 21 foetal sample over the gestation range we investigated (14-22 weeks) [20]. This confirmed our earlier observations of consistent foetal abnormalities despite the absence of GATA1 mutations which was independently confirmed in two labs [18,19] and is found also in neonates with DS (submitted). These data support the contention that an extra copy of Hsa21 in foetal liver HSC is sufficient to perturb their growth and differentiation. This in turn would lead to increased foetal liver MEP and, following acquisition of GATA1 mutation(s), to a selective expansion of a leukaemic erythro-megakaryocytic blast cell population manifesting as the clinical condition TAM in late foetal, or early neonatal life (Figure 1).
Also in support of the notion that trisomy 21 itself directly perturbs haematopoiesis, and particularly megakaryocyte/erythroid lineage development, was recently reported by MacLean et al, using human trisomy 21 human embryonic stem (hES) cells and induced pluripotent stem (iPS) cells. They found that trisomy 21 hES and iPS cells, when they were differentiated under foetal liver-like conditions, displayed increased erythroid and megakaryocyte colony forming potential compared to isogenic disomic clones [21]. By contrast, in similar experiments using an iPS cells model of trisomy 21 yolk sac hematopoiesis, trisomy 21 selectively enhanced erythropoiesis while megakaryocyte production was normal and myelopoiesis was reduced [22] suggesting that the effects of trisomy 21 are likely to be developmental stage specific.
Impact of trisomy 21 on perinatal haematopoiesis
The profound abnormalities of second trimester haematopoiesis in Down syndrome raise the question of whether these changes are confined to the second trimester when haematopoiesis is maximal in foetal liver or whether trisomy 21 also perturbs peri- and post-natal haematopoiesis. To address this we have recently performed a systematic analysis of blood counts and blood cell morphology in newborn infants with DS. DS neonates almost all had quantitative and morphological haematological abnormalities independent of their GATA1 mutation status (submitted). The most common erythrocyte abnormality was macrocytosis, a consistent finding in individuals with Down Syndrome [23] and murine models of Down syndrome [24,25], suggesting a direct link to altered expression of chromosome 21 genes on postnatal haematopoiesis. Dysplastic platelets and leucocytes similar to other myelodysplastic conditions [26,27] were also common in neonates with Down syndrome supporting the contention that trisomy 21 itself causes trilineage perturbation of neonatal, as well as foetal, haematopoiesis.
Molecular basis for perturbation of haematopoiesis by trisomy 21
Although there are ~300 protein-encoding and RNA genes on human chromosome 21 (Hsa21)[28], the functional correlation between levels of expression of Hsa21 genes and phenotype, is unknown [17]. Several approaches are being used to try to identify genes linked to increased leukaemia susceptibility. Studies in rare patients with partial trisomy suggest that leukaemia risk is confined to an 8.5Mb region on chromosome 21 (Hsa21) although conclusions are limited by the very small number of cases of leukaemia identified in these patients [29]. Others have investigated gene expression by hES cells or iPS cells from primary trisomy 21 samples or used a series of elegant transgenic mouse models including the Tc1 mouse, which contains a copy of most of human Hsa21 [25], and mice containing additional copies of all or part of mouse chromosomal regions syntenic with Hsa21 (see below)[24, 30, 31].
Human studies
Several Hsa21 genes, such as RUNX1, BACH1, ETS2, ERG, DYRK1A, and GABPA encode proteins with known functions in haematopoietic cells [17,32-37]. RUNX1, for example, has been reported to interact with GATA1 during megakaryocyte differentiation [32] and plays an important role in the pathogenesis of non-DS AML [38]. However, RUNX1 trisomy is not required for the abnormal megakaryopoiesis in mouse models [24] and gene expression studies do not show significantly increased RUNX1 expression in foetal liver [18-20], trisomy 21-derived ES or iPS cells cells [21,22] or in ML-DS compared to non-DS AMKL [39].
Chou et al found increased expression of only 4 Hsa21 genes using gene expression arrays DYRK1a, BACH1, GABPA and SON, The increased levels of these genes was modest but close to those predicted as a result of an additional copy of 21. This illustrates one of the difficulties in analysing expression data from aneuploid cells where the modest differences in expression level may be difficult to differentiate from interindividual differences [40]. Indeed, MacLean et al, investigating gene expression in iPS cells differentiated under foetal liver-like conditions observed no consistent change in transcript levels between differentiated isogenic disomic and trisomic iPS cells cells [21]. There are also 5 Hsa21 microRNAs (miRs) on Hsa21, four of which are known to be expressed in megakaryocyte lineage cells [41]. However, no differences in expression of any of these miRs were seen between trisomic and disomic iPS cells cells [21]. Interestingly, in DS-ML and TAM miR-125b has been reported to be overexpressed in comparison with normal megakaryocytes [42].
Mouse models of trisomy 21
Mouse models of DS have provided important insight into the function of a number of genes on chromosome 21 but none of the models fully recapitulates the human disease. The Tc1 mouse, which contains a copy of ~80% of Hsa21 genes, developed macrocytic anaemia, splenomegaly and increased megakaryopoiesis but the changes were notable mainly in older, rather than foetal/neonatal mice, none of the mice developed leukaemia or a true myeloproliferative disorder and no specific genes were identified as responsible for the haematological abnormalities [25]. More recently multiple structural rearrangements/deletions have been identified in Tc1 mice which may help to better refine the contribution of individual genes since it is now clear that 50 of the Hsa21 genes in this model, including RUNX1 for example, are disomic [43].
Using mice trisomic for a variable number of genes on mouse chromosome 16 syntenic with Hsa21, two groups have also reported macrocytic anaemia and abnormal megakaryopoiesis [30, 31]. One of these models (Ts65Dn), which contains ~104 Hsa21 orthologues, develops a myeloproliferative disorder in adult rather than neonatal life [31]. This has been linked to overexpression of ERG, which is necessary for the myeloproliferative disorder in Ts65Dn mice [44] and is able to promote megakaryopoiesis and induce megakaryoblastic leukaemia in the absence of a trisomy 21 background [35]. However, there is no evidence to date that ERG is overexpressed in human trisomy 21 foetal liver cells, ES/iPS cells cells or in ML-DS [20-22, 38]. To identify other genes responsible for the haematopoietic abnormalities in Ts65Dn mice, John Crispino's lab used a refined mouse model (Ts1Rhr) which is only trisomic for 33 Hsa21 orthologues. Although neither TAM nor ML-DS develop spontaneously in these mice, by crossing Ts1Rhr with GATA1s knock in mice and over-express a transforming MPL allele (MPLW515L), they recently produced the first mouse model of trisomy 21-dependent ML-DS [45]. Using this model, both shRNA and gene expression profiling and functional studies identified DYRK1A (and possibly CHAF1B, HLCS and ERG) as mediators of abnormal megakaryopoiesis and, for DYRK1A, as a megakaryoblastic tumour-promoting gene in the setting of partial trisomy 21 and GATA1s [45]. Further studies using this model or the newly characterised Tc1 mice, in tandem with studies in primary human cells at different stages of leukaemic transformation should provide interesting data over the next few years.
The role of the foetal haematopoietic microenvironment in perturbation of haematopoiesis by trisomy 21
Several lines of indirect evidence suggest that foetal liver may provide the specialised microenvironment necessary for driving and/or maintaining abnormal haematopoiesis in DS. First, as mentioned above, pathological and molecular studies clearly show that TAM (and by inference ML-DS) arises in the foetal liver [7, 9, 10, 46, 47]. TAM is predominantly a foetal liver disease with little bone marrow involvement [9] and spontaneous resolution of the majority of cases in the first 6 months of life strongly suggests that the foetal microenvironment is important for the maintenance of mutant GATA1 clones [6-8]. Second, our previous work suggested that, in contrast to the marked abnormalities in foetal liver myeloid progenitors, there were no significant differences in foetal bone marrow myeloid progenitors between foetuses with and without trisomy 21 [18]. More recently we have characterised foetal bone marrow haematopoiesis in more detail and identified trisomy 21-associated haematopoietic abnormalities which are distinctly different from those in foetal liver (unpublished data). Third, differential production and responsiveness of foetal tissues, including haematopoietic cells, to insulin-like growth factors (IGFs) is one of the few consistently reported differences between adult and foetal haematopoiesis [48-51]. While osteoblast-derived IGF1 is important for survival and expansion of adult HSC [48], foetal HSC expansion is supported by IGF2 produced by unique foetal liver stromal cells [49, 50]. In addition, foetal, but not adult, murine megakaryocyte progenitors are dependent for their survival and proliferation on the IGF signalling pathway [51]. Since ML-DS and TAM cells also both depend upon increased IGF signalling pathway, it is plausible that developmentally-regulated IGF signalling, mediated by foetal liver-derived IGFs, is responsible for the HSC MK-erythroid bias and MEP expansion in trisomy 21 foetal liver.
Summary and conclusions
Recent data from primary human foetal liver, as well as ES cells and iPS cells, show that trisomy 21 itself alters human foetal HSC and progenitor biology causing multiple defects in megakaryocyte/erythroid and B lymphoid lineage development. These data provide clues to mechanisms by which trisomy 21, or aneuploidy in general, may perturb haematopoietic cell growth and differentiation and a model with which to investigate these. The molecular basis of these effects is likely to be extremely complex, to be both tissue- and lineage-specific and to be dependent on the foetal liver, and possibly bone marrow, microenvironment.
Trisomy 21 is likely to impact on hematopoietic cell biology in multiple complex ways. Several genes on chromosome 21 (Hsa21), such as RUNX1, ERG and DYRK1A, encode proteins or microRNAs, such as miR-125b, with relevant functions in hematopoietic cells. However, while trisomic genes, individually or collectively, may be directly involved through gene dosage either in a hematopoietic cell-autonomous fashion or via other cell types, the effects may also be exerted indirectly via disomic genes. To address this, several investigators have studied mouse models of DS [4]. Although these models implicate deregulated expression of Hsa21-encoded genes as tumour-promoting, most evidence suggests that the mouse may not be a suitable model [4]. Critically, none of the models spontaneously develop TAM and/or ML-DS. Furthermore, the hematopoietic phenotype of germline N-terminal GATA1 mutations in disomic humans [5] is markedly different to mouse.
Conclusion
In conclusion, recent data from primary human FL [10], as well as foetal BM, ES cells and iPS cells [8-9], indicate that T21 itself alters human foetal HSC and progenitor biology causing multiple defects in lympho-myelopoiesis. These data provide clues to possible mechanisms through which T21, or aneuploidy in general, may perturb hematopoietic cell growth and differentiation and a model with which to investigate these. However, the molecular basis through which T21 exerts these effects is likely to be extremely complex, to be both tissue- and lineage-specific and to be dependent on the FL, and possibly foetal BM, microenvironment, analogous to the role of the specialised tumour microenvironment in enabling and sustaining neoplastic cancer cells.
References
- [1].Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 2.Zipursky A. Susceptibility to leukemia and resistance to solid tumors in Down syndrome. Pediatr Res. 2000;47:704. doi: 10.1203/00006450-200006000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Roy A, Roberts I, Norton A, Vyas P. Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: a multistep model of leukaemogenesis. Br J Haematol. 2009;147:3–12. doi: 10.1111/j.1365-2141.2009.07789.x. [DOI] [PubMed] [Google Scholar]
- 4.Lange BJ, Kobrinsky N, Barnard DR, et al. Distinctive demography, biology, and outcome of acute myeloid leukemia and myelodysplastic syndrome in children with Down syndrome: Children's Cancer Group Studies 2861 and 2891. Blood. 1998;91:608–615. [PubMed] [Google Scholar]
- 5.Whitlock JA, Sather HN, Gaynon P, et al. Clinical characteristics and outcome of children with Down syndrome and acute lymphoblastic leukemia: a Children's Cancer Group study. Blood. 2005;106:4043–4049. doi: 10.1182/blood-2003-10-3446. [DOI] [PubMed] [Google Scholar]
- 6.Massey GV, Zipursky A, Chang MN, et al. A prospective study of the natural history of transient leukemia (TL) in neonates with Down syndrome (DS): Children's Oncology Group (COG) study POG-9481. Blood. 2006;107:4606–13. doi: 10.1182/blood-2005-06-2448. [DOI] [PubMed] [Google Scholar]
- 7.Klusmann JH, Creutzig U, Zimmermann M, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood. 2008;111:2991–2998. doi: 10.1182/blood-2007-10-118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muramatsu H, Kato K, Watanabe N, et al. Risk factors for early death in neonates with Down syndrome and transient leukaemia. Br J Haematol. 2008;142:610–615. doi: 10.1111/j.1365-2141.2008.07231.x. [DOI] [PubMed] [Google Scholar]
- 9.Gamis AS, Alonzo TA, Ryan EA, et al. Natural history of transient myeloproliferative disorder clinically diagnosed in Down syndrome neonates: a report from the Children's Oncology Group Study A2971. Blood. 2011;118:6752–6759. doi: 10.1182/blood-2011-04-350017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed M, Sternberg A, Hall G, et al. Natural history of GATA1 mutations in Down syndrome. Blood. 2004;103:2480–2489. doi: 10.1182/blood-2003-10-3383. [DOI] [PubMed] [Google Scholar]
- 11.Wechsler J, Greene M, McDevitt MA, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 12.Rainis L, Bercovich D, Strehl S, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood. 2003;102:981–986. doi: 10.1182/blood-2002-11-3599. [DOI] [PubMed] [Google Scholar]
- 13.Hitzler JK, Cheung J, Li Y, Scherer SW, Zipursky A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood. 2003;10:4301–4304. doi: 10.1182/blood-2003-01-0013. [DOI] [PubMed] [Google Scholar]
- 14.Xu G, Nagano M, Kanezaki R, et al. Frequent mutations in the GATA-1 gene in the transient myeloproliferative disorder of Down syndrome. Blood. 2003;102:2960–2968. doi: 10.1182/blood-2003-02-0390. [DOI] [PubMed] [Google Scholar]
- 15.Groet J, McElwaine S, Spinelli M, et al. Acquired mutations in GATA1 in neonates with Down's syndrome with transient myeloid disorder. Lancet. 2003;361:1617–1620. doi: 10.1016/S0140-6736(03)13266-7. [DOI] [PubMed] [Google Scholar]
- 16.Hollanda LM, Lima CS, Cunha AF, et al. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet. 2006;38:807–812. doi: 10.1038/ng1825. [DOI] [PubMed] [Google Scholar]
- 17.Roper RJ, Reeves RH. Understanding the basis for Down syndrome phenotypes. PLoS Genet. 2006;2:e50. doi: 10.1371/journal.pgen.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tunstall-Pedoe O, Roy A, Karadimitris A, et al. Abnormalities in the myeloid progenitor compartment in Down syndrome foetal liver precede acquisition of GATA1 mutations. Blood. 2008;112:4507–4511. doi: 10.1182/blood-2008-04-152967. [DOI] [PubMed] [Google Scholar]
- 19.Chou ST, Opalinska JB, Yao Y, et al. Trisomy 21 enhances human foetal erythro-megakaryocytic development. Blood. 2008;112:4503–4506. doi: 10.1182/blood-2008-05-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy A, Cowan G, Mead AJ, et al. Perturbation of foetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc Natl Acad Sci U S A. 2012;109:17579–17584. doi: 10.1073/pnas.1211405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maclean GA, Menne TF, Guo G, et al. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proc Natl Acad Sci U S A. 2012;109:17567–17572. doi: 10.1073/pnas.1215468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou ST, Byrska-Bishop M, Tober JM, et al. Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:17573–17578. doi: 10.1073/pnas.1211175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starc TJ. Erythrocyte macrocytosis in infants and children with Down syndrome. J Pediatr. 1992;121:578–581. doi: 10.1016/s0022-3476(05)81149-7. [DOI] [PubMed] [Google Scholar]
- 24.Kirsammer G, Jilani S, Liu H, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alford KA, Slender A, Vanes L, et al. Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood. 2010;115:2928–2937. doi: 10.1182/blood-2009-06-227629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 27.Rajnoldi A. Cantu, Fenu S, Kerndrup G, et al. Evaluation of dysplastic features in myelodysplastic syndromes: experience from the morphology group of the European Working Group of MDS in Childhood (EWOG-MDS) Ann Hematol. 2005;84:429–433. doi: 10.1007/s00277-005-1034-4. [DOI] [PubMed] [Google Scholar]
- 28. http://www.ensembl.org/Homo_sapiens/Location/Chromosome?r=21:1-48129895.
- 29.Korbel JO, Tirosh-Wagner T, Urban AE, et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci U S A. 2009;106:12031–12036. doi: 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmichael CL, Majewski IJ, Alexander WS, et al. Hematopoietic defects in the Ts1Cje mouse model of Down syndrome. Blood. 2009;113:1929–1937. doi: 10.1182/blood-2008-06-161422. [DOI] [PubMed] [Google Scholar]
- 31.Kirsammer G, Jilani S, Liu H, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elagib KE, Racke FK, Mogass M, et al. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 33.Xu G, Kanezaki R, Toki T, et al. Physical association of the patient-specific GATA1 mutants with RUNX1 in acute megakaryoblastic leukemia accompanying Down syndrome. Leukemia. 2006;20:1002–1008. doi: 10.1038/sj.leu.2404223. [DOI] [PubMed] [Google Scholar]
- 34.Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood. 2009;113:3337–3347. doi: 10.1182/blood-2008-08-174813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salek-Ardakani S, Smooha G, de Boer J, et al. ERG is an oncogene. Cancer Res. 2009;69:4665–4673. doi: 10.1158/0008-5472.CAN-09-0075. [DOI] [PubMed] [Google Scholar]
- 36.Toki T, Katsuoka F, Kanezaki R, et al. Transgenic expression of BACH1 transcription factor results in megakaryocytic impairment. Blood. 2005;105:3100–3108. doi: 10.1182/blood-2004-07-2826. [DOI] [PubMed] [Google Scholar]
- 37.Yu S, Cui K, Jothi R, et al. GABP controls a critical transcription regulatory module that is essential for maintenace and differentiation of hematopietic stem/progenitor cells. Blood. 2011;117:2166–2178. doi: 10.1182/blood-2010-09-306563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dicker S. Schnittger F., Kern W. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 39.Bourquin JP, Subramanian A, Langebrake C, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci U S A. 2006;103:3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birchler JA. Reflections on studies of gene expression in aneuploids. Biochem J. 2010;426:119–123. doi: 10.1042/BJ20091617. [DOI] [PubMed] [Google Scholar]
- 41.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klusmann JH, Li Z, Bohmer K, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24:478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gribble SM, Wiseman FK, Clayton S S, et al. Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS One. 2013;8:e60482. doi: 10.1371/journal.pone.0060482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng AP, Hyland CD, Metcalf D D, et al. Trisomy of ERG is required for myeloproliferation in a mouse model of Down syndrome. Blood. 2010;115:3966–3969. doi: 10.1182/blood-2009-09-242107. [DOI] [PubMed] [Google Scholar]
- 45.Malinge S, Bliss-Moreau S,M, Kirsammer G, et al. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J Clin Invest. 2012;122:948–962. doi: 10.1172/JCI60455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taub JW, Mundschau G, Ge Y, et al. Prenatal origin of GATA1 mutations may be an initiating step in the development of megakaryocytic leukemia in Down syndrome. Blood. 2004;104:1588–1589. doi: 10.1182/blood-2004-04-1563. [DOI] [PubMed] [Google Scholar]
- 47.Heald B, Hilden JM, Zbuk K, et al. Severe TMD/AMKL with GATA1 mutation in a stillborn fetus with Down syndrome. Nat Clin Pract Oncol. 2007;4:433–438. doi: 10.1038/ncponc0876. [DOI] [PubMed] [Google Scholar]
- 48.Garrett RW, Emerson SG. The role of parathyroid hormone and insulin-like growth factors in hematopoietic niches: physiology and pharmacology. Mol Cell Endocrinol. 2008;288:6–10. doi: 10.1016/j.mce.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Zhang CC, Lodish HF. Insulin-like growth factor 2 expressed in a novel foetal liver cell population is a growth factor for hematopoietic stem cells. Blood. 2004;103:2513–2521. doi: 10.1182/blood-2003-08-2955. [DOI] [PubMed] [Google Scholar]
- 50.Chou S, Lodish HF. Foetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc Natl Acad Sci U S A. 2010;107:7799–7804. doi: 10.1073/pnas.1003586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klusmann JH, Godinho FJ, Heitmann K, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in foetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010;24:1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]