Abstract
Background
The uncultivated species Dasypyrum breviaristatum carries novel diseases resistance and agronomically important genes of potential use for wheat improvement. The development of new wheat-D. breviaristatum derivatives lines with disease resistance provides an opportunity for the identification and localization of resistance genes on specific Dasypyrum chromosomes. The comparison of wheat-D. breviaristatum derivatives to the wheat-D. villosum derivatives enables to reveal the genomic divergence between D. breviaristatum and D. villosum.
Results
The mitotic metaphase of the wheat- D. breviaristatum partial amphiploid TDH-2 and durum wheat -D. villosum amphiploid TDV-1 were studied using multicolor fluorescent in situ hybridization (FISH). We found that the distribution of FISH signals of telomeric, subtelomeric and centromeric regions on the D. breviaristatum chromosomes was different from those of D. villosum chromosomes by the probes of Oligo-pSc119.2, Oligo-pTa535, Oligo-(GAA)7 and Oligo-pHv62-1. A wheat line D2139, selected from a cross between wheat lines MY11 and TDH-2, was characterized by FISH and PCR-based molecular markers. FISH analysis demonstrated that D2139 contained 44 chromosomes including a pair of D. breviaristatum chromosomes which had originated from the partial amphiploid TDH-2. Molecular markers confirmed that the introduced D. breviaristatum chromosomes belonged to homoeologous group 7, indicating that D2139 was a 7Vb disomic addition line. The D2139 displayed high resistance to wheat stripe rust races at adult stage plant, which may be inherited from, D. breviaristatum chromosome 7Vb.
Conclusion
The study present here revealed that the large divergence between D. breviaristatum and D. villosum with respected to the organization of different repetitive sequences. The identified wheat- D. breviaristatum chromosome addition line D2139 will be used to produce agronomically desirable germplasm for wheat breeding.
Keywords: Dasypyrum breviaristatum, Fluorescence in situ hybridization, Molecular marker, Wheat
Background
The genus Dasypyrum (or Haynaldia) consists of only two diploid species, the annual Dasypyrum villosum and perennial D. breviaristatum [1]. The genomes of diploid D. villosum and D. breviaristatum were assigned the symbols V and Vb, respectively [2, 3]. Based on the sequences comparison of nr5S DNA multigene family, Baum et al. [4] suggested that the genome constitution of 4x D. breviaristatum should be considered as an allotetraploid VVVbVb. Dasypyrum species possess agronomically important genes such as disease resistance, high protein quality and drought tolerance, all of which represent valuable resources for global wheat breeding [3]. The species D. villosum has been extensively hybridized with wheat for at least 6 decades, and several disease resistance genes have been successfully transferred to wheat [5–7]. Above all, over 20 elite cultivars carrying the wheat- D. villosum chromosome T6AL · 6VS translocation with powdery mildew resistance gene Pm21 have been released into agricultural production in China [8, 9]. Given the widespread success of this introgression from D. villosum, researches have been conducted with a similar aim to transfer useful genes from D. breviaristatum into wheat. Subsequently, the wheat- D. breviaristatum partial amphiploid [10] and wheat- D. breviaristatum introgression lines with multiply disease resistances have developed [11, 12].
Molecular and cytogenetic methods have been previously employed to assess the level of chromosomal divergence of these two Dasypyrum species [13, 14]. A large number of interspecific and intraspecific chromosome variations and significant genomic diversification were observed among different Dasypyrum accessions, probably due to the out-crossing characteristic of these two Dasypyrum species [2]. Each pair of Dasypyrum chromosomes were transferred into a wheat background after intergeneric hybridizations. The wheat- Dasypyrum chromosome addition lines with different Dasypyrum species or accession origins allow comparison of the different Dasypyrum genomes in wheat backgrounds. In the present study, fluorescent in situ hybridization (FISH) was carried out to characterize differences between D. breviaristatum and D. villosum chromosomes by comparing karyotypes between the wheat- D. breviaristatum partial amphiploid TDH-2 and Triticum turgidum - D. villosum amphiploid TDV-1, The FISH and molecular markers were applied to identify new wheat- D. breviaristatum addition line with stripe rust resistance, which will be a useful germplasm for wheat genetics and breeding.
Results
Comparative FISH karyotype of TDH-2 and TDV-1
The mitotic metaphase chromosomes of the wheat- D. breviaristatum partial amphiploid TDH-2, were hybridized with probes Oligo-pSc119.2, Oligo-pTa535, Oligo-(GAA)7 by sequential multicolor-FISH (Fig. 1). As shown in Fig. 1a, the FISH hybridization signals of the probes Oligo-pSc119.2 and Oligo-pTa535 can easily identify the 28 wheat chromosomes from 1A-7A and 1B-7B based on the standard FISH karyotype of wheat chromosomes using the same probes described by Tang et al. [15]. Yang et al. [10] reported that the partial amphiploid TDH-2 was produced by the elimination of some chromosomes from the wheat Chinese Spring (CS)- D. breviaristatum decaploid amphiploid. It is likely that the A and B chromosomes of TDH-2 originated from CS. By comparing the FISH patterns of Oligo-pSc119.2 and Oligo-pTa535 probes of TDH-2 to those of CS by Tang et al. [15], we found additional signals corresponding to probe Oligo-pSc119.2 on the terminal regions of 1BS and 2BL in TDH-2 (Fig. 1b). After comparing the (GAA)n signal distribution on TDH-2 chromosomes with those of CS reported by Danilova et al. [16], we observed that the (GAA)n signals on chromosome 7A of CS were absent in TDH-2. The results suggest that at least three wheat chromosomes have undergone structural change(s) which may be related to the presence of D. breviaristatum chromosomes. Each of the seven pairs of D. breviaristatum chromosomes were also distinguished using probes Oligo-pSc119.2, Oligo-pTa535 in TDH-2 (Fig. 1b). These chromosomes were temporarily named Vb1-Vb7 (Fig. 1b).
Fig. 1.
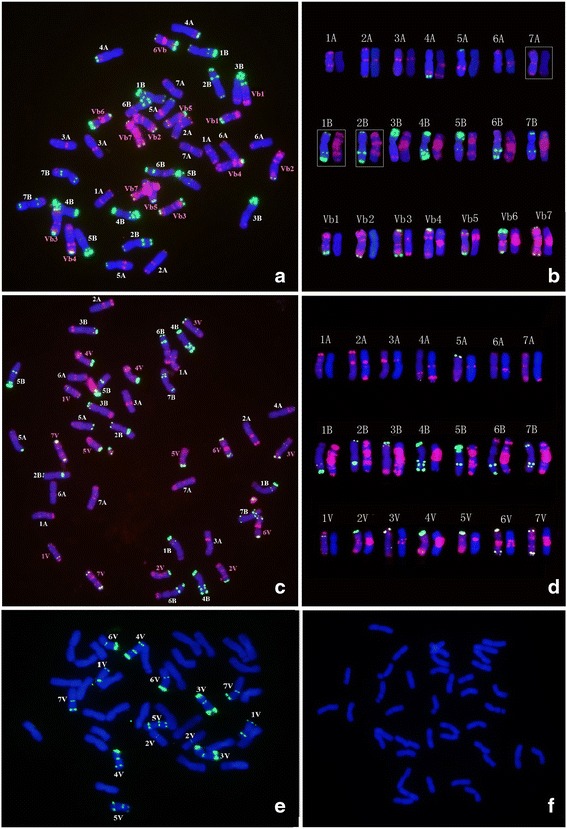
FISH and karyotypes of TDH-2 (a, b, f) and TDV-1(c, d, e). The mitotic metaphase chromosomes (a, c) after hybridization with probes Oligo-pTa535 (red) and Oligo-pSc119.2 (green), or (b, d) hybridized with Oligo-(GAA)7 (red) The boxes shows the modified chromosomes compared to its parents. Figures e and f are hybridized by probe of pHv62-3 (green)
FISH using probes Oligo-pSc119.2, Oligo-pTa535, Oligo-(GAA)7 were also carried out on the chromosomes of the Triticum turgidum cv. Jorc-69- D. villosum amphiploid TDV-1 (Fig. 1c and d). We found that the signals of probe Oligo-pSc119.2 were mainly located on terminal sites, while the hybridization signals of Oligo-pTa535 were distributed along the chromosome arms of D. villosum. The probe Oligo- (GAA)7 hybridized to 2V-7V of D. villosum chromosomes at their centromeric or sub-terminal regions. Moreover, we produced a high tandem repeat sequences probe Oligo-pHv62-1 as reported by Li et al. [17]. FISH revealed that Oligo-pHv62-1 present in terminal or sub-terminal heterochromatic C-banding regions of D. villosum chromosomes in TDV-1, but was absent in D. breviaristatum chromosomes of TDH-2 (Fig. 1e and f). The comparative FISH karyotypes of the D. breviaristatum and D. villosum chromosomes allows easily to distinguish each individual Dasypyrum chromosome in wheat background.
FISH of wheat- D. breviaristatum addition line D2139
Sequential multi-color ND-FISH by probes Oligo-pSc119.2, Oligo-pTa535, Oligo-(GAA)7 was conducted to characterize the mitotic metaphase cells of D2139 (Fig. 2). The chromosome number of D2139 is 2n = 44, including all the 42 wheat chromosomes and two alien chromosomes added in the wheat background. The probes Oligo-pSc119.2, Oligo-pTa535, showed a pair of chromosomes with faint Oligo-pSc119.2 hybridization signals at the telomeric region of long arm, and strong hybridization signals of Oligo-pTa535 along the long and short arm in D2139 (Fig. 2). The FISH hybridization pattern of the chromosomes was identical to D. breviaristatum chromosomes Vb7 (Fig. 1). Therefore, we concluded that the line D2139 was a chromosome Vb7 addition line. Comparing the FISH patterns of D2139 parents MY11 [15] and TDH-2 (Fig. 1), it appeared that the D2139 line inherited the A and B- genome chromosomes from MY11 and/or TDH-2. Based on the FISH patterns, D2139 inherited chromosomes 5A, 7A, 1B, and 7B which were identical to the TDH-2 parent, while 6B, 2B appeared to be from MY11. Since there is no D-genome in the partial amphiploid TDH-2, D2139 would have inherited D-chromosomes from MY11. As shown in Fig. 2c, chromosomes 1D and 3D revealed clear differences in the distribution of Oligo-pSc119.2 signals compared to previously published FISH patterns of D-genome chromosomes of MY11 [15]. The terminal region of 1DL showed strong Oligo-pSc119.2 signals, while the Oligo-pSc119.2 signals were absent from the 3DS terminal region. The observation implies that the transmission of the D. breviaristatum chromosomes may be associated with structural changes in wheat chromosomes.
Fig. 2.
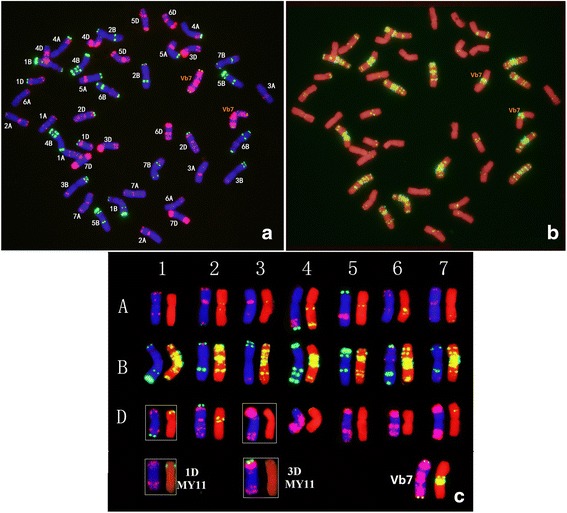
Sequential FISH karyotypes of D2139. Left figure (a) was stained with DAPI (blue), Oligo-pTa535 (red) and Oligo-pSc119.2 (green), while the right figure (b) chromosomes were stained by PI (red) and Oligo-GAA (green). The box shows the modified chromosomes compared to the parent line MY11 (c)
Molecular markers analysis
PLUG primers were designed from rice genomic DNA sequences specific for the syntenic regions, in the expectation that they would presumably amplify fragments from the corresponding linkage group(s) of wheat genomes [18]. Our previous studies showed that the PLUG markers were useful for producing alien chromosome-specific markers [19, 20]. A total of 21 PLUG markers from wheat homoeologous group 7 were tested on D2139 compared to its parents MY11 and TDH-2 (Table 1). Based on the amplification of nulli-tetrasomic lines of Chinese Spring, the PLUG markers give rise to the 7A, 7B and 7D specific bands, respectively (Fig. 3). A total of 13 pairs of primers generated the identical bands from D. breviaristatum VbVb and TDH-2 to those from the disomic addition line D2139. These results suggested that the D. breviaristatum chromosome in D2139 belonged to homoeologous group 7. Five markers out of the 13 had previously been mapped onto the short arms and eight mapped onto the long arms of group 7 (Table 1). After combining these PCR results with the FISH patterns, we concluded that the D2139 was a 7Vb addition line, and the Vb7 was chromosome 7Vb. The wheat CS - D. villosum 7V addition line TA7683 was also used to test the PLUG markers. We found that seven of 13 primer pairs showed polymorphic amplification differences between the D. breviaristatum 7Vb and D. villosum 7V chromosomes (Table 1). The results suggested that sequence divergence may have occurred among Dasypyrum species during their evolution.
Table 1.
The primers used to localize the Vb7 specific amplification in D2139
| No. | Primers name | Primer sequences (5’–3’) | Wheat bin map | Restriction enzyme | Length of Vb7 bands |
|---|---|---|---|---|---|
| 1 | TNAC1776 | F: ATCATCCTGCTGCTACTGTGC | 7AS2-0.73–0.83 | – | |
| R: CCTTCTCAGCTTAGCGATGTG | 4AL2-0.75–0.80 | ||||
| 7DS4-0.73–1.00 | |||||
| 2 | TNAC1782 | F: TCACTGAACAGCCTAGACATGG | 7AS2-0.73–0.83 | HaeIII | 690 bp |
| R: ATTCGCAGACCGCATCTATC | 7BS2-0.27–1.00 | ||||
| 7DS4-0.73–1.00 | |||||
| 3 | TNAC1803 | F: TGCGACCAGTCTCTTTGAAAT | C-7AL1-0.39 | HaeIII | 800 bp* |
| R: GTCGGAGCCTGGATCTCTAGT | 7BL2-0.38–0.63 | ||||
| 7DL5-0.30–0.61 | |||||
| 4 | TNAC1806 | F: ATTCCTCGTGAATTGCTGGAT | 7AS8-0.45–0.59 | TaqI | 350 bp* |
| R: TCTGCAGTTAGGGACTTGAAA | 7BS2-0.27–1.00 | ||||
| 7DS2-0.61–0.73 | |||||
| 5 | TNAC1811 | F: CTGCTCAACGAGTTCATCGAC | 7AL1-0.39–0.63 | TaqI | 740 bp |
| R: TTGGAGTGGACGTTGCATT | 7BL2-0.38–0.63 | ||||
| 7DL5-0.30–0.61 | |||||
| 6 | TNAC1812 | F: ACTTCGCTTGGTCTCCTCAAT | 7AL5-0.63–0.71 | TaqI | 860 bp |
| R: GAGAAGTGTGCCAATTCCAAA | 7BL7-0.63–0.78 | ||||
| 7DL5-0.30–0.61 | |||||
| 7 | TNAC1815 | F: AGCAGACATCAGCAAGTTTGAG | 7AL1-0.39–0.63 | TaqI | 600 bp* |
| R: ACTGACAAGCCCATGATTGAC | 7BL2-0.38–0.63 | ||||
| 7DL5-0.30–0.61 | |||||
| 8 | TNAC1822 | F: CCCTCCGTCCGTGCAAAT | 7AL5-0.63–0.71 | TaqI | 730 bp* |
| R: GGCTGATGATGGAGACGTG | 7BL2-0.38–0.63 | ||||
| 7DL2-0.61–0.82 | |||||
| 9 | TNAC1867 | F: GCCTTTCCTTTGGTAGTCTGG | C-7AL1-0.39 | – | 840 bp |
| R: CGATCCAAATGATCCTGAAGA | 7BL2-0.38–0.63 | ||||
| 7DL1-0.14–0.30 | |||||
| 10 | TNAC1903 | F: TCGCTTCTTCTGCTTGTTCTT | C-7AL1–0.39 | TaqI | 920 bp* |
| R: CTGCTACTAGGCCACCCAAA | C-7BL2–0.38 | ||||
| 7DL1-0.14–0.30 | |||||
| 11 | TNAC1926 | F: CGTCAGCTACAGCGACATCTA | C-7AS8–0.45 | TaqI | 700 bp* |
| R: AACTTGAGCAGCGTGGTGTT | 7BS2-0.27–1.00 | ||||
| 7DS3-0.15–0.36 | |||||
| 12 | TNAC1943 | F: GCTGCTATGGTCCACGAATTA | 7AS5-0.59–0.73 | HaeIII | 600 bp* |
| R: AGAGTATCGTATCCGGGCAAT | 7BS2-0.27–1.00 | ||||
| 7DS4-0.73–1.00 | |||||
| 13 | TNAC1957 | F: TCAACATTTGCAGGATTGTCA | 7AL21-0.74–0.86 | – | 730 bp |
| R: TTTCACAGGAACCTCTGCATC | 7BL10-0.78–0.84 | ||||
| 7DL2-0.61–0.82 |
The primers and the location in wheat bin map were referred to Ishikawa et al. [18]. The star indicated the Vb7 specific bands were polymorphic to 7V band
Fig. 3.
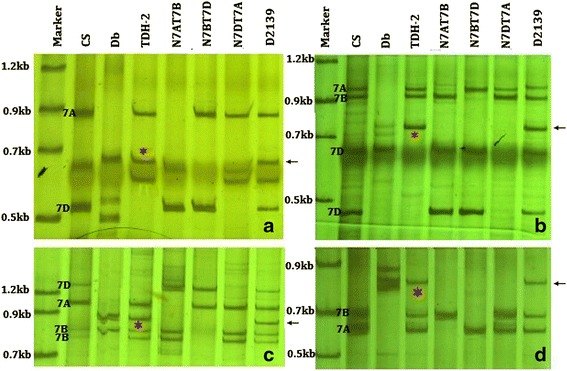
PCR using PLUG primers TNAC1782/HaeIII (a), TNAC1811/TaqI (b), TNAC1812/TaqI (c) and TNAC1867 (d). The stars and arrows indicate the D. breviaristatum specific bands in TDH-2 and D2139, respectively
Agronomic traits and rust resistance analysis
The spike phenotype and stripe rust resistance of lines D2139, TDH-2, CS and MY11 were observed. As shown in Fig. 4a, the spike length of D2139 was 11–12 cm, which was longer than the wheat parents (9–10 cm), while the D2139 had 22–22 spikelets per spike, closely resembling MY11. When inoculated with P. striiformis f. sp. tritici (PST) races CYR32 and CYR33 at adult plant stage, the TDH-2 and D2139 lines were highly resistance to the isolates, whereas wheat MY11 and CS were highly susceptible (Fig. 4b). These results indicated that the stripe rust resistance in D2139 was from the TDH-2 parent, and originates from D. breviaristatum.
Fig. 4.
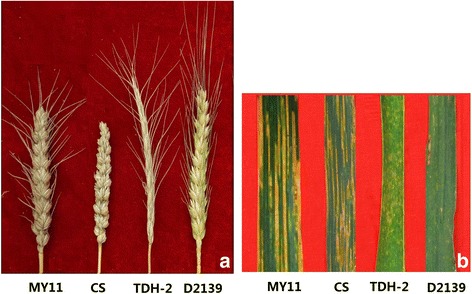
The spikes morphology (a) and leaf response to stripe rust (b) of the lines
Discussion
With respect to the genomic relationship between two Dasypyrum species, D. breviaristatum and D. villosum, cytogenetic and molecular evidence has revealed the huge genetic divergence between the two species. Friebe et al. [21] established the C-banded patterns of D. villosum, and then Linde-Laursen and Frederiksen [22] observed extensive C-band karyotype differences between the two genomes. De Pace et al. [23] isolated a repeat sequence that was mapped further distally on D. villosum chromosomes using FISH compared to wheat chromosomes. Galasso et al. [13] found that the differentiated GISH patterns reflected the large genomic divergence between the D. villosum and D. breviaristatum chromosomes. Liu et al. [14] used FISH probed by a ribosomal DNA sequence and proposed a hypothesis that the diploid D. villosum and tetraploid D. breviaristatum evolved in parallel from an ancestral species. Recently, we used the pDbC2 probe to hybridize to tetraploid D. breviaristatum and diploid D. villosum chromosomes, and found more Ty3-gypsy retrotransposon copy numbers in centromeric regions of D. villosum than those in D. breviaristatum [24]. In the present study, we compared D. breviaristatum and diploid D. villosum chromosomes through the distribution of FISH signals of Oligo-pSc119.2, Oligo-pTa535, Oligo-(GAA)7 by sequential multicolor-FISH (Fig. 1) and the molecular markers (Table 1) by PCR. The results suggested that strong evolutionary divergence involving copy number of repeated sequences and nucleotide sequence rearrangement may have occurred among Dasypyrum species during species evolution. Moreover, the Oligo-pHv62-1 can hybridize D. villosum chromosomes in TDV-1 (Fig. 1e). It confirmed that the high tandem repeat sequences present largely in telomeric heterochromatin regions of D. villosum as reported by Li et al. [17]. However, FISH revealed that Oligo-pHv62-1 was absent in D. breviaristatum chromosomes of TDH-2 (Fig. 1f). This significant amplification of different types of repetitive sequences between the D. villosum and D. breviaristatum chromosomes may be related to adaptation of the plant species to their environments [3]. The cytogenetic and molecular markers which are species-specific can be used to identify and characterize the introgression of D. villosum and D. breviaristatum chromosome segments into a wheat background.
Rapid genomic and epigenomic changes have been commonly found in some newly synthesized wheat-alien amphiploids [25, 26]. Alterations of alien chromosomal structure in wheat background have also been described especially wheat-rye chromosome addition, substitution and translocation lines [27, 28]. However, the variations in the karyotype of wheat chromosomes have been less reported. Recently, Fu et al. [29] reported that pSc119.2 FISH signals could be observed at the telomeric regions of 3DS arms which was not observed in the current material, and structural variation and abnormal mitotic behavior of the 3D chromosome were detected in the selfed progeny of wheat “MY11”-rye 6R monosomic addition line. Furthermore, Fu et al. [30] reported the occurrence of 14 chromosomal rearrangements in wheat -rye hybrids. Our studies found that chromosomes 1B, 2B and 7A of the wheat- D. breviaristatum partial amphiploid TDH-2 (Fig. 1), and chromosomes 1D and 3D of the 7Vb addition lines D2139 (Fig. 2) showed apparent structural changes revealed by FISH patterns compared to the parental lines. Patokar et al. [31] characterized several novel wheat-Thinopyrum bessarabicum recombinant lines carrying intercalary translocations and did not report any observable wheat chromosomal rearrangements using FISH. It is likely that the distant species of genera Secale and Dasypyrum may induce such structural changes while present in a wheat background, while chromosomes derived from Thinopyrum species may not have the same effect due to their close relationship to wheat [32]. Thus, we suggest that the introgression of chromosomes from closely related species may not lead to the significant structural changes of wheat chromosomes, although the introduction of closely related Aegilops chromosomes causes massive deletions of wheat chromosomes [33], which were mainly useful for physical mapping of genes. There is the other possibility that the recipient wheat genotype may also increase the chromosomal rearrangement with visible changes of representative repeats. Taking advantage of fast multicolor FISH methods [15], we recently identified some chromosomal changes in high yielding elite cultivars originating from wheat distant hybridization. The association between the visibly rearranged wheat chromosomes and the yield or disease resistances are being verified for breeding purpose.
D. villosum chromosomes are known to contain genetic variability of value for incorporation into wheat. At least three sets of D. villosum chromosomes addition lines in wheat background have been developed [34–36]. Novel genes including disease resistance and quality-related characters have been found in different wheat- D. villosum derived lines [7]. With the aim to transfer novel genes from D. breviaristatum to wheat, we identified the two D. breviaristatum chromosomes addition lines Y93-1-A6-4 and Y93-1-6-6, which showed novel resistance to powdery mildew isolates and stem rust Ug99 (pathotype TTKSK) [11]. Molecular marker and GISH analysis revealed that those introduced D. breviaristatum chromosomes were rearranged chromosomes involved groups 2, 6 and 7. Recently, Li et al. [12] reported a wheat - D. breviaristatum substitution line D11-5 possessing a pair of 2Vb chromosomes which had replaced wheat 2D. Based on the FISH analysis, we found that the 2Vb chromosome in line D11-5 was identical to chromosome Vb2 of TDH-2 (Fig. 1). We thus suggest that chromosome Vb2 can be provisionally assigned to linkage group 2, subject to confirmation using other genetic markers.
In the present study, we identified line D2139 which contained a pair of D. breviaristatum chromosomes confirmed herein to be “7Vb”. This disomic substitution line D2139 may be potentially useful germplasm for agronomic traits including enhance spike length and the stripe rust resistance from the D. breviaristatum 7Vb into the wheat genome using marker-assisted chromosome engineering [37]. The divergence between the individual D. villosum 7V and D. breviaristatum 7Vb chromosomes was revealed by FISH and molecular markers in wheat background, which will provide the basis for future detailed comparative genomics analysis. Guo et al. [38] compared chromosomes 7el1, 7el2, 7E(e), and 7Ei derived from different Thinopyrum species by molecular and cytological methods. In a similar manner, we intend to create hybrid populations between wheat- Dasypyrum 7 V and 7Vb addition lines for further and direct localization of genes on these alien chromosomes.
Conclusions
In summary, the different FISH patterns between D. breviaristatum and D. villosum chromosomes were observed clearly by using different repetitive sequences as probes, which allows to identify the individual Dasypyrum chromosomes in wheat background. The changes of FISH patterns of wheat chromosomes were induced by the induction of D. breviaristatum to wheat. The D. breviaristatum specific molecular markers can be used to assign the homologous group of D. breviaristatum to wheat. We identified wheat- D. breviaristatum chromosome 7Vb addition line with novel stripe rust resistances will be potential useful for wheat breeding. The molecular and cytogenetic markers will assist to trace the D. breviaristatum chromatin in wheat background.
Methods
Plant materials
D. breviaristatum accession PI 546317 was obtained from the National Small Grains Collection at Aberdeen, Idaho, USA. The wheat– D. breviaristatum partial amphiploid TDH-2 (genome AABBVbVb) was as described by Yang et al. [10]. The accession of Dasypyrum villosum TA10220 and the Chinese Spring- D. villosum chromosome 7 V addition line TA7683 [39] were obtained from Dr. Bernd Friebe of Wheat Genetic and Genomic Resources Center at Kansas State University, Manhattan, KS, USA. The T. turgidum cv. Jorc-69- D. villosum amphiploid TDV-1 (genome AABBVV) was developed and provided by Prof. Hua-Ren Jiang at Sichuan Agricultural University, China [40]. Line D2139 was obtained from a BC1F5 generation of the crosses between wheat cultivar ‘Mianyang 11’ (MY11) and TDH-2.
Fluorescence in situ hybridization (FISH)
Seedling root tips were collected and then treated with nitrous oxide followed by enzyme digestion, using the procedure of Han et al. [41]. The synthesized oligo-nucleotide probes Oligo-pSc119.2, Oligo-pTa535, Oligo-(GAA)7 were used for identifying the wheat chromosomes according to the description of Tang et al. [15]. A new probe, Oligo-pHv62-1 (5’ CGAAGGATTG AAAAAAGGAA CAATTTCGCA CTTACAGCTC AAAAATATA TGGGACA 3’) was synthesized and labeled at 5’ 6-carboxyfluorescein (FAM) based on high tandem repeat sequences pHv62 in D. villosum as reported by Li et al. [17]. The protocol of non-denaturing FISH by the synthesized probes was described by Fu et al. [30]. Photomicrographs of FISH chromosomes were taken with an Olympus BX-51 microscope equipped with a DP-70 CCD camera.
Molecular marker analysis
DNA was extracted from young leaves of D. breviaristatum, TDH-2, TDV-1, lines D11-5 and CS [42]. PCR-based Landmark Unique Gene (PLUG) primers were designed according to Ishikawa et al. [18]. Polymerase chain reaction (PCR) was performed in an Icycler thermalcycler (Bio-RAD Laboratories, Emeryville, CA) in a 25 μl reaction, containing 10 mmol Tris–HCl (pH 8.3), 2.5 mmol MgCl2, 200 μmol of each dNTP, 100 ng template DNA, 0.2 U Taq polymerase (Takara, Japan) and 400 nmol of each primer. The cycling parameters were 94 °C for 3 min for denaturation; followed by 35 cycles at 94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min; and a final extension at 72 °C for 10 min. The amplified products were separated by 8 % PAGE gel as described by Hu et al. [43].
Disease resistance screening
Wheat- D. breviaristatum derivative lines were evaluated for adult-plant resistance to Pst strains CYR32 and CYR33 during the 2013 and 2015 cropping seasons as described by Li et al. [12].
Acknowledgement
We particularly thank Dr. I. Dundas at University of Adelaide, Australia, for reviewing the manuscript. We thank the National Natural Science Foundation of China (No. 31171542), and Sichuan wheat breeding community for the financial support.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GL and ZY designed the study. GL, DG, HZ, JL conducted the experiments. HW, SL and JM helped to conduct experiment and data analysis. GL and ZY participated in paper writing. All authors read and approved the final manuscript.
Contributor Information
Guangrong Li, Email: ligr28@uestc.edu.cn.
Dan Gao, Email: gaodangaodan2010@163.com.
Hongjun Zhang, Email: hongjunzh01@sina.com.
Jianbo Li, Email: lijianbo199061@163.com.
Hongjin Wang, Email: 201521090326@std.uestc.edu.cn.
Shixiao La, Email: 201521090303@std.uestc.edu.cn.
jiwei Ma, Email: 201521090304@std.uestc.edu.cn.
Zujun Yang, Email: yangzujun@uestc.edu.cn.
References
- 1.Frederiksen S. Taxonomic studies in Dasypyrum (Poaceae) Nord J Bot. 1991;11:135–142. doi: 10.1111/j.1756-1051.1991.tb01813.x. [DOI] [Google Scholar]
- 2.Gradzielewska A. The genus Dasypyrum-part 1. The taxonomy and relationships within Dasypyrum and with Triticeae species. Euphytica. 2006;152:429–440. doi: 10.1007/s10681-006-9232-2. [DOI] [Google Scholar]
- 3.De Pace C, Vaccino P, Cionini PG, Pasquini M, Bizzarri M, Qualset CO. Dasypyrum. In: Kole C, editor. Wild Crop Relatives, Genomic and Breeding Resources, Cereals. Heidelberg: Springer; 2011. pp. 185–292. [Google Scholar]
- 4.Baum BR, Edwards T, Johnson DA. What does the nr5S DNA multigene family tell us about the genomic relationship between Dasypyrum breviaristatum and D. villosum (Triticeae: Poaceae)? Mol Genet Genomics. 2014;289:553–565. doi: 10.1007/s00438-014-0825-5. [DOI] [PubMed] [Google Scholar]
- 5.Chen PD, Qi LL, Zhang SZ, Liu DJ. Development and molecular cytogenetic analysis of wheat-Haynaldia 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet. 1995;91:1125–1128. doi: 10.1007/BF00223930. [DOI] [PubMed] [Google Scholar]
- 6.Yildirim A, Jones SS, Murray TD. Mapping a gene conferring resistance to Pseudocercosporella herpotrichoides on chromosome 4 V of Dasypyrum villosum in a wheat background. Genome. 1998;41:1–6. doi: 10.1139/gen-41-1-1. [DOI] [Google Scholar]
- 7.Qi LL, Pumphrey MO, Friebe B, Zhang P, Qian C, Bowden RL, et al. A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor Appl Genet. 2011;123:159–67. [DOI] [PubMed]
- 8.He ZH, Xia XC, Chen XM, Zhuang QS. Progress of wheat breeding in China and the future perspective. Acta Agron Sin. 2011;37:202–215. doi: 10.3724/SP.J.1006.2011.00202. [DOI] [Google Scholar]
- 9.Bie T, Zhao R, Zhu S, Chen S, Cen B, Zhang B, et al. Efficient marker-assisted screening of structural changes involving Haynaldia villosa chromosome 6V using a double-distal-marker strategy. Mol Breeding. 2015;35:34.
- 10.Yang ZJ, Li GR, Feng J, Jiang HR, Ren ZL. Molecular cytogenetic characterization and disease resistance observation of wheat - Dasypyrum breviaristatum partial amphiploid and its derivatives. Hereditas. 2005;142:80–85. doi: 10.1111/j.1601-5223.2005.01918.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Li G, Yan H, Zhou J, Hu L, Lei M, et al. Molecular and cytogenetic identification of new wheat- D. breviaristatum additions conferring resistance to stem rust and powdery mildew. Breeding Sci. 2011;61:366–72. [DOI] [PMC free article] [PubMed]
- 12.Li GR, Zhao JM, Li DH, Yang EN, Huang YF, Liu C, et al. A novel wheat–Dasypyrum breviaristatum substitution line with stripe rust resistance. Cytogenet Genome Res. 2014;143:280–7. [DOI] [PubMed]
- 13.Galasso I, Blanco A, Katsiotis A, Pignone D, Heslop-Harrison HS. Genomic organization and phylogenetic relationships in the genus Dasypyrum analysed by Southern and in situ hybridization of total genomic and cloned DNA probes. Chromosoma. 1997;106:53–61. doi: 10.1007/s004120050224. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Li GR, Sunish S, Jia JQ, Yang ZJ, Friebe B, et al. Genome relationships in the genus Dasypyrum: evidence from molecular phylogenetic analysis and in situ hybridization. Plant Syst Evol. 2010;288:149–56.
- 15.Tang ZX, Yang ZJ, Fu SL. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet. 2014;55:313–318. doi: 10.1007/s13353-014-0215-z. [DOI] [PubMed] [Google Scholar]
- 16.Danilova TV, Friebe B, Gill BS. Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma. 2012;121:597–611. doi: 10.1007/s00412-012-0384-7. [DOI] [PubMed] [Google Scholar]
- 17.Li WL, Chen PD, Qi LL, Liu DJ. Isolation, characterization and application of a species-specific repeated sequence from Haynaldia villosa. Theor Appl Genet. 1995;90:526–533. doi: 10.1007/BF00221999. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa G, Nakamura T, Ashida T, Saito M, Nasuda S, Endo T, et al. Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor Appl Genet. 2009;118:499–514. [DOI] [PubMed]
- 19.Jia J, Li G, Liu C, Zhou J, Yang Z. Sequence variations of PDHA1 gene in Triticeae species allow for identifying wheat-alien introgression lines. Frontiers Agric China. 2010;4:137–144. doi: 10.1007/s11703-010-0110-2. [DOI] [Google Scholar]
- 20.Hu LJ, Liu C, Zeng ZX, Li GR, Song XJ, Yang ZJ. Genomic rearrangement between wheat and Thinopyrum elongatum revealed by mapped functional molecular markers. Genes Genomics. 2012;34:67–75. doi: 10.1007/s13258-011-0153-7. [DOI] [Google Scholar]
- 21.Friebe B, Cermeno MC, Zeller FJ. C-banding polymorphism and the analysis of nucleolar activity in Dasypyrum villosum (L.) Candargy, its added chromosomes to hexaploid wheat and the amphiploid Triticum dicoccum-D. villosum. Theor Appl Genet. 1987;73:337–342. doi: 10.1007/BF00262498. [DOI] [PubMed] [Google Scholar]
- 22.Linde Laursen IB, Frederiksen S. Comparison of the Giemsa C-banded karyotypes of Dasypyrum villosum (2×) and D. breviaristatum (4×) from Greece. Hereditas. 1991;114:237–244. doi: 10.1111/j.1601-5223.1991.tb00330.x. [DOI] [Google Scholar]
- 23.De Pace C, Delre V, Scarascia Mugnozza GT, Qualset CO, Cremonini R, Frediani M, et al. Molecular and chromosomal characterization of repeated and single copy DNA sequences in the genome of Dasypyrum villosum. Hereditas. 1992;116:55–65.
- 24.Li GR, Liu C, Wei P, Song XJ, Yang ZJ. Chromosomal distribution of a new centromeric Ty3-gypsy retrotransposon sequence in Dasypyrum and related Triticeae species. J Genet. 2012;91:343–348. doi: 10.1007/s12041-012-0181-3. [DOI] [PubMed] [Google Scholar]
- 25.Gustafson JP, Lukaszewski AJ, Bennett MD. Somatic deletion and redistribution of telomeric heterochromatin in the genus Secale and in Triticale. Chromosoma. 1983;88:293–298. doi: 10.1007/BF00292906. [DOI] [Google Scholar]
- 26.Dou QW, Tanaka H, Nakata N, Tsujimoto H. Molecular cytogenetic analyses of hexaploid lines spontaneously appearing in octoploid Triticale. Theor Appl Genet. 2006;114:41–47. doi: 10.1007/s00122-006-0408-x. [DOI] [PubMed] [Google Scholar]
- 27.Alkhimova AG, Heslop-Harrison JS, Shchapova AI, Vershinin AV. Rye chromosome variability in wheat-rye addition and substitution lines. Chromosome Res. 1999;7:205–212. doi: 10.1023/A:1009299300018. [DOI] [PubMed] [Google Scholar]
- 28.Bento M, Gustafson P, Viegas W, Silva M. Genome merger: from sequence rearrangements in triticale to their elimination in wheat-rye addition lines. Theor Appl Genet. 2010;121:489–497. doi: 10.1007/s00122-010-1325-6. [DOI] [PubMed] [Google Scholar]
- 29.Fu S, Yang M, Fei Y, Tan F, Ren Z, Yan B, et al. Alterations and abnormal mitosis of wheat chromosomes induced by wheat-rye monosomic addition lines. PLoS One. 2013;8:e70483. [DOI] [PMC free article] [PubMed]
- 30.Fu S, Chen L, Wang Y, Li M, Yang Z, Qiu L, et al. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep. 2015;5:10552. [DOI] [PMC free article] [PubMed]
- 31.Patokar C, Sepsi A, Schwarzacher T, Kishii M, Heslop-Harrison JS: Molecular cytogenetic characterization of novel wheat-Thinopyrum bessarabicum recombinant lines carrying intercalary translocations. Chromosoma 2015. DOI: 10.1007/s00412-015-0537-6. [DOI] [PubMed]
- 32.Dvořák J. Homology between Agropyron elongatum chromosomes and Triticum aestivum chromosomes. Can J Genet Cytol. 1980;22:237–259. doi: 10.1139/g80-029. [DOI] [Google Scholar]
- 33.Endo TR, Gill BS. The deletion stocks of common wheat. J Heredity. 1996;87:295–307. doi: 10.1093/oxfordjournals.jhered.a023003. [DOI] [Google Scholar]
- 34.Sears ER. Addition of the genome of H. villosa to T. aestivum. Am J Bot. 1953;40:168–174. doi: 10.2307/2438774. [DOI] [Google Scholar]
- 35.Lukaszewski AJ. A comparison of several approaches in the development of disomic alien addition lines of wheat. In: Miller TE, Koebner RMD, editors. Proceedings of the 7th international wheat genetics symposium. Cambridge, UK: Institute of Plant Sciences Research; 1988. pp. 363–367. [Google Scholar]
- 36.Chen PD, Liu DJ. Identification of H. villosa chromosomes in alien wheat addition of T. aestivum-H. villosa. In: Li ZS, editor. Proceedings of the 1st international symposium on chromosome engineering in plants. Xian, China: BookPublisher; 1986. pp. 31–33. [Google Scholar]
- 37.Niu Z, Klindworth DL, Friesen TL, Chao S, Jin Y, Cai X, et al. Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics. 2011;187:1011–21. [DOI] [PMC free article] [PubMed]
- 38.Guo J, He F, Cai JJ, Wang HW, Li AF, Wang HG, et al. Molecular and Cytological Comparisons of Chromosomes 7el1, 7el2, 7E(e), and 7E i Derived from Thinopyrum. Cytogenet Genome Res. 2015;145:68–74. [DOI] [PubMed]
- 39.Liu C, Qi L, Liu W, Zhao W, Wilson J, Friebe B, et al. Development of a set of compensating Triticum aestivum-Dasypyrum villosum Robertsonian translocation lines. Genome. 2011;54:836–44. [DOI] [PubMed]
- 40.Jiang HR, Dai DQ, Sun DF, Xiao SH. New artificial genetic resources of wheat: several polyploids of Triticum-Dasypyrum. Sci Agric Sin. 1992;25:89. [Google Scholar]
- 41.Han FP, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci U S A. 2006;103:3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang ZJ, Liu C, Feng J, Li GR, Deng KJ, Zhou JP, et al. Studies on genome relationship and species-specific PCR marker for Dasypyrum breviaristatum in Triticeae. Hereditas. 2006;143:47–54. [DOI] [PubMed]
- 43.Hu LJ, Li GR, Zeng ZX, Chang ZJ, Liu C, Zhou JP, et al. Molecular cytogenetic identification of a new wheat-Thinopyrum substitution line with stripe rust resistance. Euphytica. 2011;177:169–77.


