Abstract
To identify novel fatty acid diol synthases, putative candidate sequences from Penicillium species were analyzed, and hydroxy fatty acid production by crude Penicillium enzyme extracts was assessed. Penicillium chrysogenum was found to produce an unknown dihydroxy fatty acid, a candidate gene implicated in this production was cloned and expressed, and the expressed enzyme was purified. The product obtained by the reaction of the purified enzyme with linoleic acid was identified as 8R,11S-dihydroxy-9,12(Z,Z)-octadecadienoic acid (8R,11S-DiHODE). The catalytic efficiency of this enzyme toward linoleic acid was the highest among the unsaturated fatty acids tested, indicating that this enzyme was a novel 8R,11S-linoleate diol synthase (8R,11S-LDS). A sexual stage in the life cycle of P. chrysogenum has recently been discovered, and 8R,11S-DiHODE produced by 8R,11S-LDS may constitute a precocious sexual inducer factor, responsible for regulating the sexual and asexual cycles of this fungus.
Keywords: 8R,11S-dihydroxy-9,12(Z,Z)-octadecadienoic acid; hydroxy fatty acid; precocious sexual inducer factor
Sexual and asexual life cycles are controlled by precocious sexual inducer (psi) factors (1), produced by psi-producing oxygenases (Ppo enzymes) in diverse fungi (2–6). Ppo enzymes include PpoA, PpoB, and PpoC. PpoA enzymes (5,8-diol synthases) from Aspergillus species, including Aspergillus clavatus (5), Aspergillus fumigatus (7), Aspergillus nidulans (2), Aspergillus niger (8), and Aspergillus terreus (9), have been shown to convert linoleic acid to 5S,8R-dihydroxy-9,12(Z,Z)-octadecadienoic acid (5S,8R-DiHODE). PpoB generates compounds with a hydroxyl group at the C8 position of an unsaturated fatty acid, for example, 8R-hydroxy-9,12(Z,Z)-octadecadienoic acid (8R-HODE), and PpoC gives rise to compounds with a hydroxyl group at the C10 position, for example, 10R-hydroxy-8,12(E,Z)-octadecadienoic acid (10R-HODE) (1–3, 6).
A. nidulans is a sexual fungus (2), whereas A. niger and A. clavatus are known as asexual fungi (8, 10). Although A. fumigatus was proposed as an asexual fungus, its sexual cycle was discovered (11). A. fumigatus, A. niger, and A. clavatus convert linoleic acid to not only 5S,8R-DiHODE but also the by-product 8R,11S-dihydroxy-9,12(Z,Z)-octadecadienoic acid (8R,11S-DiHODE) (5, 7, 8, 12). Agaricus bisporus converts linoleic acid to 8R,11S-DiHODE (13); however, involvement of an 8,11-diol synthase for 8R,11S-DiHODE production was speculated but not proved. Penicillium species were thought to reproduce exclusively asexually. However, sexual reproduction has recently been demonstrated in Penicillium roqueforti and Penicillium chrysogenum (14, 15). Exceptionally, A. bisporus was well known as a sexual fungus with sexual reproduction and multicellular fruiting bodies (13). 5S,8R-DiHODE has induced sexual production in fungi (16, 17), while 8R,11S-DiHODE, suggested as product of PpoB, has been mostly found in asexual fungi except for A. bisporus. Therefore, 8R,11S-DiHODE may be involved in the stimulation of asexual sporulation. However, the biological function of 8R,11S-DiHODE has not yet been clearly established. Moreover, researches on the psi factors present in Penicillium species have not been carried out, as they are yet to be identified in these organisms.
Dihydroxy fatty acids are converted from unsaturated fatty acids by fatty acid diol synthases, including Ppo enzymes. Fatty acid diol synthases are fusion proteins containing N-terminal fatty acid-heme peroxidase [dioxygenase (DOX)] and C-terminal cytochrome P450-heme thiolate (hydroperoxide isomerase) domains. N-terminal DOX domain converts unsaturated fatty acids to hydroperoxy fatty acids, including 8R-hydroperoxy-9,12(Z,Z)-octadecadienoic acid (8R-HPODE), which are subsequently isomerized to dihydroxy unsaturated fatty acids, such as 5S,8R-DiHODE, 7S,8S-dihydroxy-9,12(Z,Z)-octadecadienoic acid (7S,8S-DiHODE), and 8R,11S-DiHODE by the hydroperoxide isomerase activity of the C-terminal domain, which demonstrates different position specificities. The biochemical properties of 5S,8R- and 7S,8S-diol synthases have been characterized previously (18). However, 8R,11S-diol synthase, which converts linoleic acid to 8R,11S-DiHODE, has not yet been described.
In the present study, we isolated and characterized 8R,11S-linoleate diol synthase (8R,11S-LDS) from P. chrysogenum for the first time by assessing the hydroxy fatty acids produced by several Penicillium species, cloning the gene involved in the production of an unknown dihydroxy fatty acid according to sequence analysis of putative fatty acid diol synthase genes, identifying hydroxy fatty acids produced from unsaturated fatty acids by the expressed enzyme, and determining kinetic parameters for unsaturated fatty acids.
MATERIALS AND METHODS
Materials
Fatty acid standards, including palmitoleic acid, oleic acid, linoleic acid, conjugated linoleic acid, α-linolenic acid, γ-linolenic acid, eicosadienoic acid, dihomo-γ-linolenic acid, arachidonic acid, docosapentanoic acid, and docosahexaenoic acid, were purchased from Santa Cruz Biotechnology. Samples of 5S,8R-DiHODE, 8R-HODE, and 8R-HPODE were prepared from linoleic acid by the reactions of recombinant Escherichia coli cells expressing A. nidulans 5S,8R-diol synthase and its H1004A-C1006S variant (19). 7S,8S-DiHODE and 8R,11S-DiHODE were prepared from linoleic acid using recombinant E. coli cells expressing fatty acid diol synthase from Glomerella cingulate (unpublished observations) and crude enzyme extract from A. clavatus (5), respectively. The converted oxylipins were processed to >99% purity by a previously described method (20) and subsequently used as reference standards. Enantiomeric mixtures [8-2H]8-HODE and [8,11-2H2]8,11-DiHODE were prepared by oxidation with Dess-Martin periodinane and reduction with NaB2H4 as described previously (7).
Crude enzyme preparation
P. chrysogenum KACC 41892, Penicillium digitatum KCCM 60140, Penicillium oxalicum KCTC 6440, Penicillium marneffei KCCM 60287, and A. clavatus KCCM 60329 were used as the sources of crude enzymes to identify putative hydroxy linoleic acids produced from linoleic acid. Fungal spores were incubated on potato dextrose agar plates at 28°C for 5 days. Four agar pieces (10 × 10 mm) from the plate were used to inoculate a 100 ml baffled flask containing 25 ml of potato dextrose broth and 5 mM linoleic acid to induce protein expression, with shaking at 150 rpm for 5 days. Mycelia were harvested by filtration, washed three times with saline solution, and homogenized on ice for 10 min in 50 mM HEPES buffer (pH 7.5). Mycelial debris and unbroken mycelia were removed by centrifugation at 13,000 g for 30 min at 4°C, and the supernatant was filtered through a 0.45 μm filter. The filtrates were then used as crude Penicillium enzyme extracts.
Cloning and site-directed mutagenesis
P. chrysogenum and E. coli ER2566 were used as the sources of DNA template for fatty acid diol synthase gene and host cells, respectively. pET-21a(+) plasmid, which contained nucleotides encoding six histidine residues at C-terminal position, was used as an expression vector. Total RNA was isolated from the mycelia of P. chrysogenum using a Hybrid-R total RNA purification kit (GeneAll). Full-length cDNA was synthesized from total RNA by reverse transcription PCR using a reverse transcription kit (TaKaRa). The gene encoding fatty acid diol synthase and the full and partial genes of N-terminal domain or C-terminal domain were cloned using the Gibson assembly method (21). The sequences of the primers used for gene cloning were based on the DNA sequence of P. chrysogenum fatty acid diol synthase (GenBank accession number CAP97986). Primers were designed to amplify the expression vector and fatty acid diol synthase DNA fragments in supplementary Table 1. The amplified DNA fragments and linearized vector, which were obtained by PCR with Phusion High-Fidelity DNA Polymerase (New England Biolabs), were ligated using Gibson Assembly Master Mix (New England Biolabs). E. coli strain ER2566 was transformed with the ligation mixture and plated on Luria-Bertani (LB) agar containing 20 μg/ml ampicillin. An ampicillin-resistant colony was selected, and plasmid DNA from the transformant was isolated using a plasmid purification kit (Intron). Site-directed mutagenesis was performed using a QuikChange kit, according to the manufacturer’s protocol (Stratagene).
Purified enzyme preparation
Recombinant E. coli cells were cultivated in a 2 liter flask containing 500 ml of LB medium and 20 μg/ml ampicillin at 37°C with shaking at 200 rpm. When the optical density at 600 nm of the bacterial culture reached 0.6, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.1 mM to induce enzyme expression. The culture was then incubated at 16°C with shaking at 150 rpm for 16 h to express the enzyme. Cells from the culture broth were harvested by centrifugation at 10,000 g for 30 min at 4°C and washed three times with 0.85% saline solution. The washed recombinant cells were subsequently resuspended in 50 mM phosphate buffer (pH 7.0) containing 300 mM NaCl and 1 mg/ml lysozyme. The resuspended cells were disrupted by sonication on ice for 10 min. The unbroken cells and cell debris were removed by centrifugation at 13,000 g for 20 min at 4°C, and the supernatant was passed through a 0.45 μm filter. The filtrate was applied to a HisTrap HP affinity chromatography column (GE Healthcare) equilibrated with 50 mM phosphate buffer (pH 7.0) on a fast-protein liquid chromatography (Bio-Rad) in a cold room at 4°C. The bound protein was eluted at 4°C using the same buffer containing 250 mM imidazole at a flow rate of 1 ml/min. The active fractions were collected and dialyzed against 50 mM 3-[4-(2-hydroxyethyl)-1-piperazinyl]-propane sulfonic acid (EPPS) buffer (pH 8.0) at 4°C for 16 h. After dialysis, the resulting solution was used as the purified enzyme.
Enzyme assays
Unless otherwise stated, the reactions were performed at 25°C for 5 min in 50 mM EPPS buffer (pH 8.0) containing 0.5 mM unsaturated fatty acid and 50 μg/ml purified enzyme or 5 mg/ml crude enzyme extract. The effect of pH and temperature on the activity of P. chrysogenum 8R,11S-LDS toward linoleic acid was investigated by varying the pH from 5.5 to 9.0, using 50 mM MES buffer (pH 5.5−6.0), 50 mM HEPES buffer (pH 6.0−8.0), 50 mM EPPS buffer (pH 8.0−8.5), and 50 mM 2-(cyclohexylamino)ethanesulfonic acid buffer (pH 8.6−9.0) at 25°C and by varying the temperature from 5°C to 65°C at pH 6.5. The effect of temperature on enzyme stability was investigated by varying the temperature from 15°C to 55°C in 50 mM HEPES buffer (pH 6.5). Enzyme activity was determined after incubating the solution at each temperature for 2 h. To determine the kinetic parameters of the DOX activity in 8R,11S-LDS, O2 uptake was measured by a Clark-type 5300A biological oxygen monitor (YSI) with 1 μg/ml purified enzyme and unsaturated fatty acids such as palmitoleic acid, oleic acid, linoleic acid, and α-linolenic acid (12.5 μM to 1 mM). The concentrations of linoleic acid and α-linolenic acid as substrates in the range of 50 to 500 μM were used to determine the kinetic parameters of whole enzyme using an HPLC system (Agilent 1100). The kinetic parameters, Km and kcat, were determined using a Hanes-Woolf plot based on the Michaelis-Menten equation.
Structural analysis
Homology modeling of C-terminal domains of 8R,11S-LDS from P. chrysogenum and 5S,8R-LDS from A. nidulans was performed using Build Homology Models module in the MODELER application of Discovery Studio (DS) 4.0 (Accerlys) based on the crystal structure of allene oxide synthase (AOS) from guayule [Protein Data Bank (PDB) entry 3DBM] as a template. Comparative modeling was used to generate the most probable structure of the query protein by aligning it with the template sequence, simultaneously considering spatial restraints, and local molecular geometry. The generated structure was improved by subsequent refinement of the loop conformations by assessing the compatibility of amino acid sequences with the known PDB structures using Protein Health module in DS 4.0. The geometry of the loop region was corrected using the Refine Loop/MODELER, and the best model was selected. The quality of the model was analyzed by PROCHECK (22). Hydrogen atoms were added to the model and minimized to have a stable energy conformation and to relax the conformation from close contacts. 8R-HPODE as a substrate was docked into the active-site pocket in the model of C-terminal domains of 8R,11S-LDS from P. chrysogenum and 5S,8R-LDS from A. nidulans using C-DOCKER module, and a sphere with a radius of 4.5 Å around the ligand-binding pocket of the enzyme was defined as the active site. Candidate poses were created using random rigid-body rotations, followed by simulated annealing. The structures of the protein, substrate, and their complexes were subjected to energy minimization using the CHARMM force field in DS 4.0 (23). Full-potential final minimization was used to refine the substrate poses. The energy-docked conformation of the substrate was retrieved for postdocking analysis using C-DOCKER module. The substrate orientation giving the lowest interaction energy was chosen for subsequent rounds of docking.
Analytical methods
The reaction products were extracted using an equal volume of ethyl acetate, the solvent was then removed with a rotary evaporator, and methanol was added to the dried extracts. Fatty acids and oxylipins were quantitatively analyzed using the HPLC system mentioned previously, with a UV detector at a detection wavelength of 202 nm and a reversed-phase Nucleosil C18 column (3.2 × 150 mm, 5 μm particle size; Phenomenex). The column was eluted at 35°C with a gradient of solvent A (acetonitrile-water-acetic acid, 50:50:0.1, v/v/v) and solvent B (acetonitrile-acetic acid, 100:0.1, v/v) as follows: 100% solvent A at a flow rate of 0.25 ml/min for 0–5 min; solvent A to solvent B for 5− 21 min at 0.25 ml/min; for 21–22 min at 0.4 ml/min; 100% solvent B at 0.4 ml/min for 22–27 min; solvent B to solvent A at 0.4 ml/min for 27–32 min; and 100% solvent A at 0.25 ml/min for 32–35 min. Chiral phase (CP)-HPLC and normal phase (NP)-HPLC were run using Chiralcel OD-H column (2.1 × 150 mm, 5 μm particle size; Daicel) and Zorbax RX SIL column (2.1 × 150 mm, 5 μm particle size; Agilent) with solvent systems of n-hexane/2-propanol/acetic acid (88:12:0.1, v/v/v) and n-hexane/diethyl ether/acetic acid (70:30:0.25), respectively.
LC/MS/MS analysis of oxylipins was performed using a Thermo-Finnigan LCQ Deca XP Plus ion trap mass spectrometer (Thermo Scientific). The instrument consisted of an LC pump, an autosampler, and a photodiode array detector. Ionization of the samples was carried out using ESI. The operation parameters were as follows: capillary temperature, 275°C; ion source voltage, 5 kV; nebulizer gas, 206.84 kPa; capillary voltage, 46 V in positive mode and 15 V in negative ionization mode; average scan time, 0.6 s; average time to change polarity, 1.2 s; and collision energy, ∼35% abundance of the precursor ion.
RESULTS
Assessment of the hydroxy fatty acids produced from linoleic acid by crude Penicillium enzyme extracts
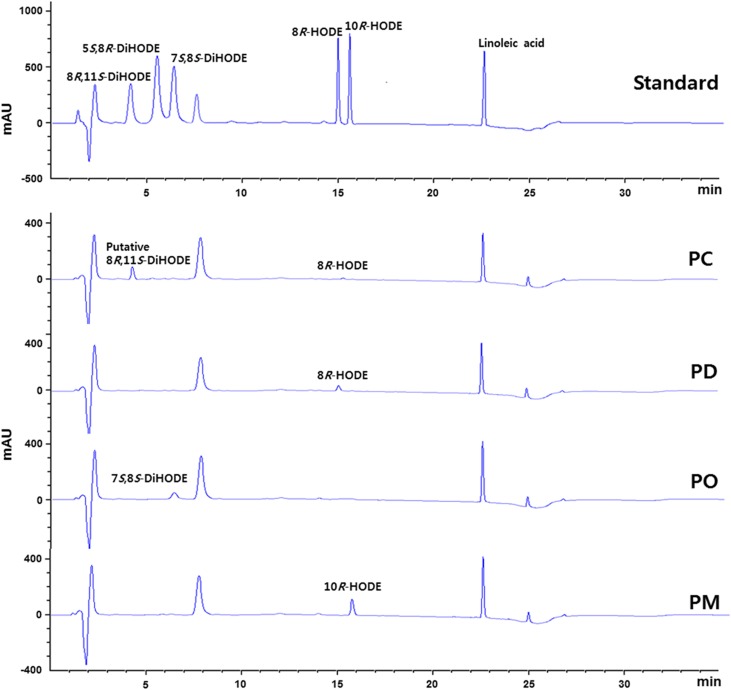
Crude enzyme extracts from P. chrysogenum, P. digitatum, P. marneffei, and P. oxalicum containing putative diol synthases were used for the conversion of linoleic acid to hydroxy fatty acids. The reaction products were analyzed by HPLC using a reversed phase Nucleosil C18 column and reference standards, including 5S,8R-DiHODE, 7S,8S-DiHODE, 8R,11S-DiHODE, 8R-HODE, and 10R-HODE. Crude enzyme extracts from P. digitatum, P. oxalicum, and P. marneffei converted linoleic acid to products, with the same retention times as those of 8R-HODE, 7S,8S-DiHODE, and 10R-HODE, respectively (Fig. 1). However, the retention time (4.3 min) of the compound produced by crude P. chrysogenum enzyme extract was the same as that of 8R,11S-DiHODE, suggesting that the compound was 8,11-DiHODE.
Fig. 1.
HPLC profiles for the conversions of linoleic acid to putative hydroxy fatty acids by crude enzyme extracts from P. chrysogenum (PC), P. digitatum (PD), P. oxalicum (PO), and P. marneffei (PM). Compounds used as standards, including 5S,8R-DiHODE, 7S,8S-DiHODE, 8R,11S-DiHODE, 8R-HODE, 10R-HODE, and linoleic acid, are shown. Crude P. chrysogenum (PC_1) enzyme extract produced a putative 8R,11S-DiHODE. The peak at about 7.7 min represents EPPS buffer.
Amino acid sequence analysis of putative diol synthases from Penicillium species
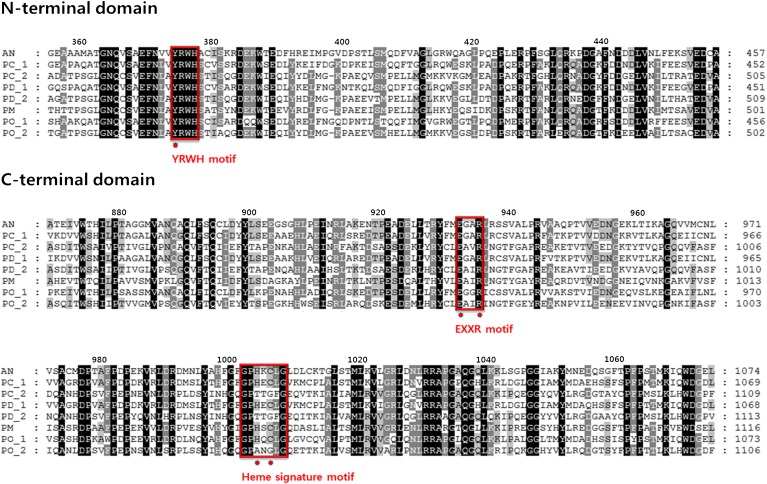
The production of monohydroxy and dihydroxy linoleic acids from linoleic acid by Penicillium species resulted from the reactions of the corresponding putative fatty acid diol synthases in these species. To analyze such enzymes, amino acid sequences of putative fatty acid diol synthases from Penicillium species, including PC_1 and PC_2 from P. chrysogenum, PD_1 and PD_2 from P. digitatum, PM from P. marneffei, and PO_1 and PO_2 from P. oxalicum, were aligned with that of PpoA from A. nidulans (Fig. 2). The YRWH motif of the N-terminal domain containing the catalytic residue Tyr374 of A. nidulans PpoA (2) and the Glu and Arg residues in EXXR motif of the C-terminal domain were found to be conserved across all of the Penicillium candidate proteins. However, the two crucial residues for the hydroperoxide isomerase activity in the C-terminal heme signature motif, His1004 and Cys1006 of A. nidulans PpoA, were conserved in P. chrysogenum PC_1, P. digitatum PD_1, P. marneffei PM, and P. oxalicum PO_1, but not in P. chrysogenum PC_2, P. digitatum PD_2, and P. oxalicum PO_2 (Fig. 2). These results suggest that PC_1, PD_1, PM, and PO_1 convert linoleic acid to dihydroxy linoleic acids, whereas PC_2, PD_2, and PO_2 convert linoleic acid to monohydroxy linoleic acids. As P. chrysogenum converted linoleic acid to putative 8,11-DiHODE, the gene encoding PC_1 was cloned, with the aim of identifying a novel 8,11-diol synthase.
Fig. 2.
Alignment of partial amino acid sequences of N- and C-terminal domains in putative diol synthase. Putative diol synthase sequences from P. chrysogenum (PC_1 and PC_2), P. digitatum (PD_1 and PD_2), P. marneffei (PM), and P. oxalicum (PO_1 and PO_2), and the sequence of PpoA from A. nidulans (AN) were aligned using ClustalW2. YRWH, EXXR, and heme signature motifs and crucial amino acids for the activities of N- and C-terminal domains in the motifs are represented by the red boxes and red dots, respectively. The GenBank accession numbers are as follows: AN, AAR88626; PC_1, CAP97986; PC_2, CAP94248; PD_1, EKV17116; PD_2, EKV06981; PM, EEA26582; PO_1, EPS31749; PO_2, EPS29986.
Gene cloning and purification of the putative fatty acid diol synthase from P. chrysogenum
A 3,225 bp gene encoding PC_1 from P. chrysogenum (1,074 amino acids), with the same sequence as that in GenBank (accession number CAP97986) (supplementary Fig. 1), and the full and partial genes encoding the N-terminal or C-terminal domain (supplementary Fig. 2) were cloned and expressed in E. coli, respectively. The N-terminal domain of the putative fatty acid diol synthase from P. chrysogenum (supplementary Fig. 3A) was identified as the amino acid residues of 1−669 by sequence alignment (supplementary Fig. 3B) with that of 5S,8R-LDS from A. fumigatus (4), which was the amino acid residues of 1−674 (supplementary Fig. 3C). The expressed protein of the residues of 1−669 was active, whereas the expressed protein of the residues of 93−642, which was predicted as the N-terminal peroxidase-like superfamily by NCBI, exhibited no activity for unsaturated fatty acids. Thus, the C-terminal domain was the amino acid residues of 670−1,074 as the rest of the enzyme except for the N-terminal domain; however, it exhibited no activity for 8R-HPODE.
The expressed protein exhibited the catalytic residue Tyr369 in its N-terminal domain and the crucial residues for hydroperoxide isomerase activity such as His999 and Cys1001 in its C-terminal domain. These residues were critical for the conversion of unsaturated fatty acid to dihydroxy fatty acid. The recombinant enzyme was purified as a soluble protein from crude E. coli extract by HisTrap affinity chromatography. The putative fatty acid diol synthase from P. chrysogenum was purified with a final purification of 6.4-fold, a yield of 21.2% compared with crude enzyme, and the specific activity of the purified enzyme for the conversion of linoleic acid to dihydroxy fatty acid was 1.43 μmol/min/mg.
Identification of the products obtained from the conversion of unsaturated fatty acids by the putative fatty acid diol synthase from P. chrysogenum
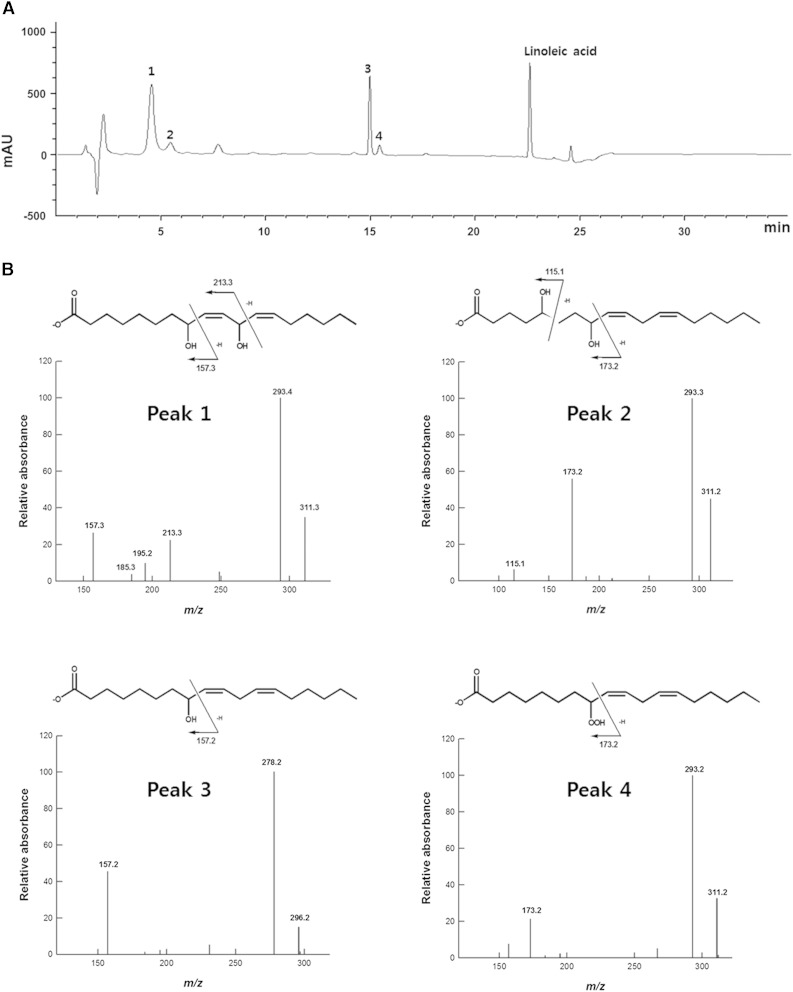
The products obtained from the conversion of linoleic acid by the putative fatty acid diol synthase from P. chrysogenum are represented in the HPLC profile given in Fig. 3A. Unknown peaks 1, 2, 3, and 4, which were assumed to be oxylipins, were analyzed by LC/MS/MS (Fig. 3B). The molecular anions of the products were represented by m/z 311 for peak 1 (4.3 min retention time in HPLC), m/z 311 for peak 2 (5.5 min), m/z 296 for peak 3 (14.9 min), and m/z 311 for peak 4 (15.3 min). Data regarding the fragments formed by cleavage of hydroxylated fatty acids have been reported previously (24). The m/z 157 and 213 for peak 1, m/z 115 and 173 for peak 2, m/z 157 for peak 3, and m/z 173 for peak 4 resulted from α-cleavage of the hydroxyl groups at the C8 and C11 positions, the C5 and C8 positions, and the C8 position, and the hydroperoxyl group at the C8 position, respectively. The m/z 293 for peak 1, m/z 293 for peak 2, m/z 278 for peak 3, and m/z 293 for peak 4 were derived from the loss of water from molecular anion of each product. Based on this analysis, the compounds represented by peaks 1, 2, 3, and 4 were identified as 8,11-DiHODE, 5,8-DiHODE, 8-HODE, and 8-HPODE, respectively. The LC/MS/MS spectrum of peak 1 consisted of the same pattern as that of the previously reported 8R,11S-DiHODE (7). 8,11-DiHODE in the HPLC profile was a major dihydroxy fatty acid product, whereas 5,8-DiHODE was a minor reaction product (Fig. 3A), suggesting that the enzyme was 8,11-diol synthase.
Fig. 3.
HPLC and LC/MS/MS analyses of the products obtained from the conversion of linoleic acid by the putative diol synthase from P. chrysogenum. A: HPLC profile of the products obtained from the conversion of linoleic acid. Unknown peaks 1, 2, 3, and 4 were assumed to be oxylipins. B: LC/MS/MS spectra and chemical structures of the products resulting from the conversion of linoleic acid. Peaks 1, 2, 3, and 4 were identified as 8,11-DiHODE, 5,8-DiHODE, 8-HODE, and 8-HPODE, respectively. The peak at ∼7.7 min represents EPPS buffer.
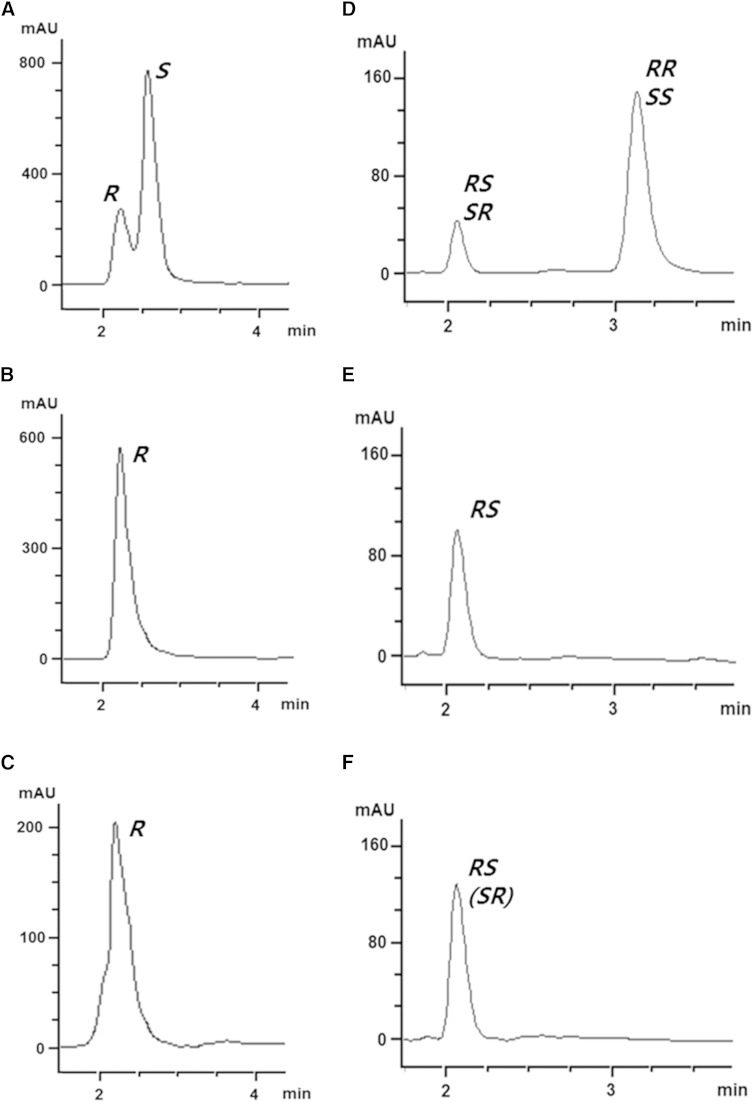
Enantiomeric mixtures [8-2H]8-HODE and [8,11-2H2]8,11-DiHODE were prepared to determine the stereoconfigurations. The chirality of 8-HODE produced by the putative fatty acid diol synthase from P. chrysogenum was confirmed by CP-HPLC with 8R-HODE produced by the H1004A-C1006S variant of 5S,8R-LDS from A. nidulans (19) and enantiomeric mixture [8-2H]8-HODE. The retention time of the earlier eluent of enantiomeric mixture [8-2H]8-HODE (Fig. 4A) was the same as those of 8R-HODE obtained from the H1004A-C1006S variant (Fig. 4B) and 8-HODE produced by the putative fatty acid diol synthase (Fig. 4C). Therefore, the later eluent of enantiomeric mixture [8-2H]8-HODE was 8S-HODE, and the product of the enzyme was 8R-HODE. The diastereoisomers of enantiomeric mixture [8,11-2H2]8,11-DiHODE were separated as two eluents by NP-HPLC. The retention time of the earlier eluent of enantiomeric mixture [8,11-2H2]8,11-DiHODE (Fig. 4D) was the same as those of 8R,11S-DiHODE (Fig. 4E), which was prepared by crude enzyme extract from A. clavatus, and 8,11-DiHODE, which was produced from linoleic acid by the putative fatty acid diol synthase from P. chrysogenum (Fig. 4F). The results indicated that the later eluent of enantiomeric mixture [8,11-2H2]8,11-DiHODE was 8R,11R- or 8S,11S-DiHODE and that 8,11-DiHODE was 8R,11S- or 8S,11R-DiHODE. Because 8R-HPODE produced by the putative fatty acid diol synthase from P. chrysogenum was a precursor of 8R,11S-DiHODE, the produced 8,11-DiHODE was 8R,11S-DiHODE.
Fig. 4.
Steric analysis of 8-HODE and 8,11-DiHODE. CP-HPLC and NP-HPLC were run using Chiralcel OD-H column (2.1 × 150 mm, 5 μm particle size; Daicel) with n-hexane/2-propanol/acetic acid (88:12:0.1, v/v/v) and Zorbax RX SIL column (2.1 × 150 mm, 5 μm particle size; Agilent) with n-hexane/diethyl ether/acetic acid (70:30:0.25), respectively. A: CP-HPLC profile of enantiomeric mixture [8-2H]8-HODE. B: CP-HPLC analysis of 8R-HODE, which was prepared by H1004A-C1006S variant 5,8-LDS from A. nidulans. C: CP-HPLC profile of 8-HODE, which was converted by H999A-C1001S variant 8,11-LDS from P. chrysogenum. D: NP-HPLC profile of diastereoisomers of enantiomeric mixture [8,11-2H2]8,11-DiHODE. E: NP-HPLC profile of 8R,11S-DiHODE, which was prepared by crude enzyme extract from A. clavatus. F: NP-HPLC analysis of 8,11-DiHODE, which was produced by the putative diol synthase from P. chrysogenum.
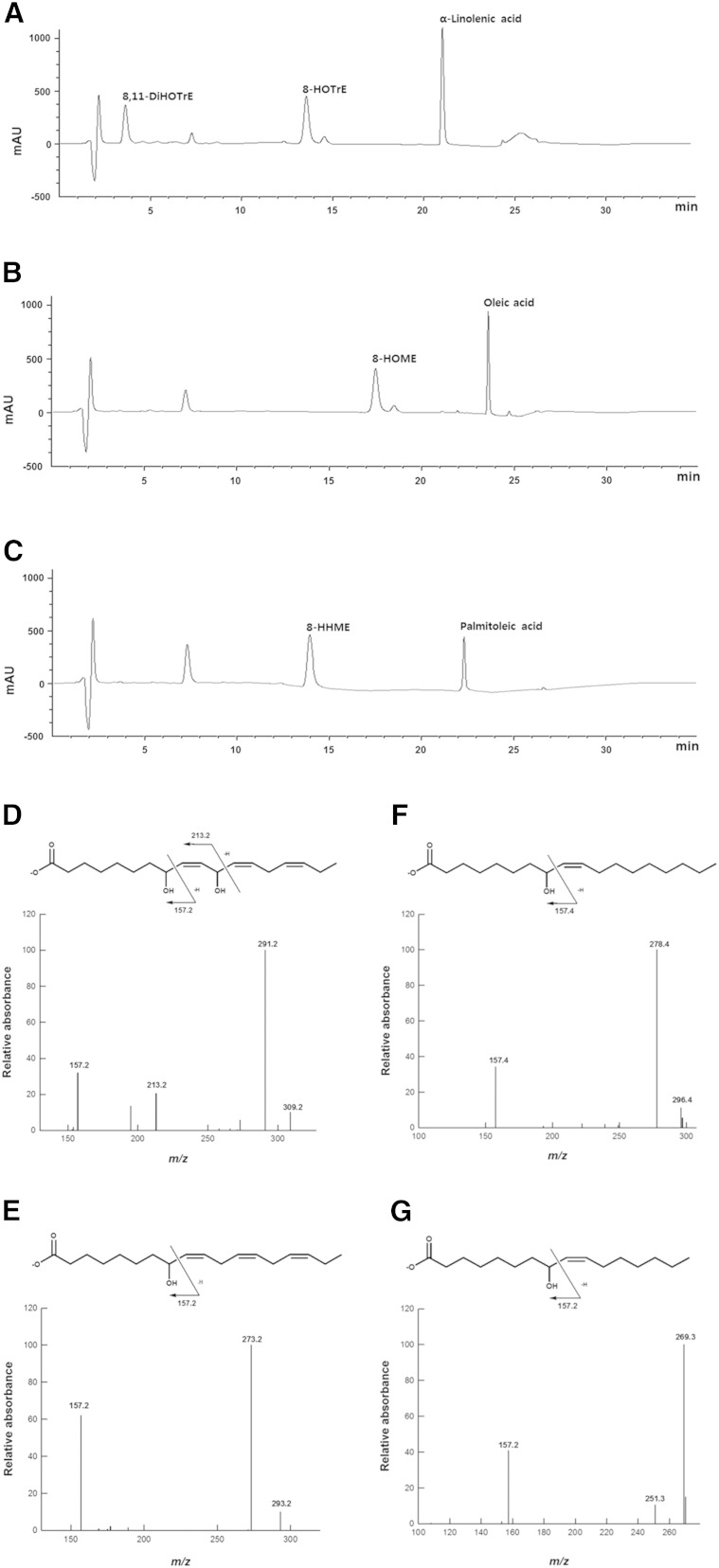
The products obtained from the conversion of α-linolenic acid, oleic acid, and palmitoleic acid by P. chrysogenum 8R,11S-diol synthase are shown in the HPLC profiles (Fig. 5A–C, respectively). The molecular anion of the compound produced from α-linolenic acid by the enzyme was represented by a peak at m/z 309, while peaks at m/z 157 and 213 resulted from cleavage of the hydroxyl groups at the C8 and C11 positions, respectively (Fig. 5D). These peaks represent the compound as 8,11-dihydroxy-9,12,15(Z,Z,Z)-octadecatrienoic acid (8,11-DiHOTrE). The molecular anion of the products obtained from the conversion of α-linolenic acid, oleic acid, and palmitoleic acid were represented by peaks at m/z 293 (Fig. 5E), 296 (Fig. 5F), and 269 (Fig. 5G), respectively. A peak at m/z 157 in each profile resulted from cleavage of the hydroxyl group at the C8 position for each product. Thus, these compounds were identified as 8-hydroxy-9,12,15(Z,Z,Z)-octadecatrienoic acid (8-HOTrE), 8-hydroxy-9(Z)-octadecenoic acid (8-HOME), and 8-hydroxy-9(Z)-hexadecenoic acid (8-HHME), respectively.
Fig. 5.
HPLC and LC/MS/MS analyses of the products obtained from the conversions of α-linolenic acid, oleic acid, and palmitoleic acid by the putative diol synthase from P. chrysogenum. HPLC profiles of the products obtained from the conversion of α-linolenic acid (A), oleic acid (B), and palmitoleic acid (C). LC/MS/MS spectra and chemical structures of 8,11-DiHOTrE (D), 8-HOTrE (E), 8-HOME (F), and 8-HHME (G) obtained from the conversion of α-linolenic acid, oleic acid, and palmitoleic acid. The peak at ∼7.7 min represents EPPS buffer.
Biochemical properties of 8R,11S-diol synthase from P. chrysogenum
The expressed protein was visualized by SDS-PAGE as a single band of ∼120 kDa (supplementary Fig. 1), which was consistent with the calculated value of 120,998 Da based on the 1,074 amino acid residues plus 6 histidine residues. Truncated N-terminal and C-terminal domains were expressed in cell debris as the bands of ∼74 and 45 kDa, respectively, in SDS-PAGE. However, the C-terminal domain protein was not observed as soluble enzyme in crude extract, and thus it was not purified by HisTrap affinity chromatography (supplementary Fig. 2). The activities of truncated N-terminal domain for linoleic acid and truncated C-terminal domain for 8R-HPODE were evaluated by whole cells containing each domain protein or crude enzyme extracts. Truncated N-terminal domain exhibited activity, whereas truncated C-terminal domain showed no activity. However, the activity of truncated N-terminal domain reduced to 60% of the DOX activity of whole enzyme. Therefore, the DOX activity of whole enzyme was measured to determine the activity of N-terminal domain.
Heme occupancy of the wild-type and H999A, C1001S, and H999A-C1001S variant enzymes of 8R,11S-diol synthase from P. chrysogenum was determined by measuring the ratio of heme absorption to protein absorption (A410/A280) with a UV-visible spectroscopy (supplementary Fig. 4). The H999A, C1001S, and H999A-C1001S variant enzymes showed 0.8-, 0.6-, 0.4-fold lower heme occupancy than the wild-type enzyme, respectively. The hydroperoxide isomerase activity of the double-site H999A-C1001S variant was completely abolished, whereas those of the H999A and C1001S variants remained. The ratios of heme absorption to protein absorption of the wild-type and variant enzymes were closely related with the hydroperoxide isomerase activities.
The effect of pH on the enzymatic production of 8R-HODE or 8R,11S-DiHODE from linoleic acid was investigated at 25°C. The maximum rates of 8R-HODE production by DOX activity and 8R,11S-DiHODE production by whole enzyme were observed at pH 8.0 and 6.5, respectively (supplementary Fig. 5). The effect of temperature on enzyme activity was assessed at pH 6.5, and the maximum activities of both DOX and whole enzyme were observed at 25°C (supplementary Fig. 6). The thermal stability of the enzyme was examined by measuring activity after incubation for 2 h at temperatures ranging from 15°C to 55°C. As temperature increased, 8R,11S-DiHODE production by whole enzyme decreased and stopped at 55°C (supplementary Fig. 7), indicating that the activity of C-terminal domain was completely abolished. However, 8 R-HODE production by DOX activity was exhibited below 65°C. Thus, the N-terminal domain demonstrated greater thermal stability than the C-terminal domain.
The production of monohydroxy fatty acids from substrates by the enzyme followed the order α-linolenic acid > linoleic acid > palmitoleic acid > oleic acid (Table 1). However, no activity was observed for conjugated linoleic acid, γ-linolenic acid, eicosadienoic acid, dihomo-γ-linolenic acid, arachidonic acid, docosapentaenoic acid, and docosahexaenoic acid. Dihydroxy fatty acids were only produced by P. chrysogenum 8R,11S-diol synthase when α-linolenic acid and linoleic acid were used as substrates, and the specific production using linoleic acid was higher than that using α-linolenic acid. The Michaelis-Menten constants (Km), turnover numbers (kcat), and catalytic efficiencies (kcat/Km) of P. chrysogenum 8R,11S-diol synthase for unsaturated fatty acids are presented in Table 2. The kcat order of DOX activity for unsaturated fatty acids (α-linolenic acid > linoleic acid > palmitoleic acid > oleic acid) was the same as that observed for its specific activity for the production of monohydroxy fatty acids. However, kcat/Km values of DOX activity were in the order linoleic acid > palmitoleic acid > oleic acid > α-linolenic acid, while the order of Km of DOX activity was exactly the reverse order of kcat/Km. The catalytic efficiency of whole enzyme for linoleic acid was higher than that for α-linolenic acid.
TABLE 1.
Specific activity of P. chrysogenum 8R,11S-diol synthase toward unsaturated fatty acids
| Specific Production (μmol/min/mg) | |||
| Substrate | Product | Monohydroxy Fatty Acid | Dihydroxy Fatty Acid |
| Palmitoleic acid (16:1Δ9Z) | 8-HHME | 9.6 ± 0.2 | NA |
| Oleic acid (18:1Δ9Z) | 8-HOME | 9.0 ± 0.1 | NA |
| Linoleic acid (18:2Δ9Z,12Z) | 8-HODE, 8,11-DiHODE | 10.8 ± 0.1 | 1.4 ± 0.0 |
| Conjugated linoleic acid (18:2Δ9E,12E) | NA | NA | NA |
| α-Linolenic acid (18:3Δ9Z,12Z,15Z) | 8-HOTrE, 8,11-DiHOTrE | 14.9 ± 0.0 | 1.1 ± 0.0 |
| γ-Linolenic acid (18:3Δ6Z,9Z,12Z) | NA | NA | NA |
| Eicosadienoic acid (20:2Δ11Z,14Z) | NA | NA | NA |
| Dihomo-γ-linolenic acid (20:3Δ8Z,11Z,14Z) | NA | NA | NA |
| Arachidonic acid (20:4Δ5Z,8Z,11Z,14Z) | NA | NA | NA |
| Docosapentaenoic acid (22:5Δ7Z,10Z,13Z,16Z,119Z) | NA | NA | NA |
| Docosahexaenoic acid (22:6Δ4Z,7Z,10Z,13Z,16Z,19Z) | NA | NA | NA |
NA, no activity.
TABLE 2.
Kinetic parameters of P. chrysogenum 8R,11S-diol synthase toward unsaturated fatty acids
| DOX Activity | Whole Enzyme Activitya | |||||
| Substrate | Km (μM) | kcat (min−1) | kcat/Km (μM−1 min−1) | Km (μM) | kcat (min−1) | kcat/Km (μM−1 min−1) |
| Palmitoleic acid | 20.0 ± 0.5 | 4,910 ± 226 | 259 | NA | NA | NA |
| Oleic acid | 34.3 ± 1.2 | 4,528 ± 56 | 134 | NA | NA | NA |
| Linoleic acid | 18.9 ± 0.6 | 5,392 ± 151 | 285 | 86.1 ± 1.8 | 622 ± 7.2 | 7.2 |
| α-Linolenic acid | 69.9 ± 0.3 | 7,907 ± 113 | 113 | 138 ± 3.2 | 829 ± 16.8 | 6.0 |
NA, no activity.
The activity for the conversion of fatty acid to dihydroxy fatty acid by the sequential reactions of N-terminal DOX domain and C-terminal hydroperoxide isomerase domain.
DISCUSSION
The C-terminal crucial residues for the hydroperoxide isomerase activity (His1004 and Cys1006) of A. nidulans 5,8-LDS (PpoA) (2) were conserved across P. chrysogenum PC_1, P. digitatum PD_1, P. marneffei PM, and P. oxalicum PO_1 but differed from those of P. chrysogenum PC_2, P. digitatum PD_2, and P. oxalicum PO_2 (Fig. 2). These results suggest that PC_1, PD_1, PM, and PO_1 can convert linoleic acid to dihydroxy linoleic acids, while PC_2, PD_2, and PO_2 are only able to convert it to monohydroxy linoleic acids. The crude enzyme extracts from P. digitatum and P. oxalicum converted linoleic acid to a monohydroxy linoleic acid (8R-HODE) and a dihydroxy linoleic acid (7S,8S-DiHODE), respectively. Thus, P. digitatum PD_2 and P. oxalicum PO_1 may be involved in this activity. Although P. marneffei PM containing the crucial residues His and Cys for hydroperoxide isomerase activity was expected to produce a dihydroxy linoleic acid, the enzyme produced monohydroxy linoleic acid (10R-HODE), indicating that the His and Cys residues in the C-terminal domain may not be critical for 10R-DOX activity. We proceeded to clone the gene P. chrysogenum PC_1 containing one catalytic residue for DOX activity (Tyr369) and two crucial residues for hydroperoxide isomerase activity (His999 and Cys1001) because PC_1 might be responsible for the production of the putative 8,11-DiHODE, which was produced without 10R-HODE accumulation by crude P. chrysogenum enzyme extract (Fig. 1).
The products obtained from the conversion of linoleic acid by the putative fatty acid diol synthase from P. chrysogenum were identified by LC/MS/MS as 8,11-DiHODE, 5,8-DiHODE, 8-HODE, and 8-HPODE (Fig. 3), and the 8,11-DiHODE and 8-HODE products were identified as 8R,11S-DiHODE and 8R-HODE, respectively, by steric analysis. When a high concentration of the putative fatty acid diol synthase (5 mg/ml) was used to completely convert linoleic acid to dihydroxy linoleic acids for the exact evaluation of dihydroxy fatty acids, the enzyme produced 8R,11S-DiHODE (97% of DiHODE) as a main product with the by-product 5,8-DiHODE (supplementary Fig. 8), indicating that the enzyme was 8R,11S-diol synthase. We replaced the crucial residues for the hydroperoxide isomerase activity in the C-terminal domain of this enzyme, His999 and Cys1001, with alanine and serine, respectively. As a result, decrease in heme occupancies of the variant enzymes led to a decrease of their activities, and a double-site variant (H999A-C1001S) completely abolished hydroperoxide isomerase activity (supplementary Fig. 4). The double-site variant of P. chrysogenum 8R,11S-diol synthase produced only a monohydroperoxy fatty acid, 8R-HPODE, without producing dihydroxy fatty acids, in the same manner as that of the H1004A-C1006S variant of A. nidulans 5S,8R-diol synthase (19). The 8R-HPODE produced was converted to 8R-HODE by the addition of cysteine as a reducing agent (supplementary Fig. 9). These results demonstrated that 8R,11S-diol synthase from P. chrysogenum converted linoleic acid to 8R,11S-DiHODE via 8R-HPODE and shared a common mechanism with other fatty acid diol synthases (5). Among the substrates tested, both DOX activity and whole enzyme activity exhibited the highest catalytic efficiency for linoleic acid (Table 2). Therefore, this enzyme is characterized as an 8R,11S-LDS.
P. chrysogenum 8R,11S-LDS converted α-linolenic acid, linoleic acid, palmitoleic acid, and oleic acid to the monohydroxy fatty acids 8-HOTrE, 8-HODE, 8-HHME, and 8-HOME, respectively. Moreover, it produced the dihydroxy fatty acids 8R,11S-DiHODE and 8,11-DiHOTrE from linoleic acid and α-linolenic acid, respectively (Table 1). Based on this activity, we observed that unsaturated fatty acids in which the first cis double bond was located at the C9 and C10 positions were converted to hydroxy fatty acids. Unsaturated fatty acids containing one double bond and two or more double bonds were converted to monohydroxy fatty acids and dihydroxy fatty acids via monohydroxy fatty acids, respectively. Whether monohydroxy or dihydroxy fatty acids are formed by P. chrysogenum 8R,11S-LDS may therefore depend on the position and number of carbon-carbon double bonds in the unsaturated fatty acid substrate.
Some fungi such as A. fumigatus, A. niger, and A. clavatus convert linoleic acid to the by-product 8R,11S-DiHODE as well as 5S,8R-DiHODE (5, 8, 12); however, the enzymes involved in 8R,11S-DiHODE production have not been identified. It was previously assumed that PpoB was responsible for 8R,11S-diol synthase activity in A. fumigatus. However, the biosynthesis of 8R,11S-DiHODE by A. fumigatus has only been observed in cytosolic fractions of A. fumigatus, and the enzyme involved in 8R,11S-DiHODE biosynthesis (PpoB) is not found (12). Thus, the 8,11-LDS isolated from P. chrysogenum in the present study constitutes a novel fatty acid diol synthase.
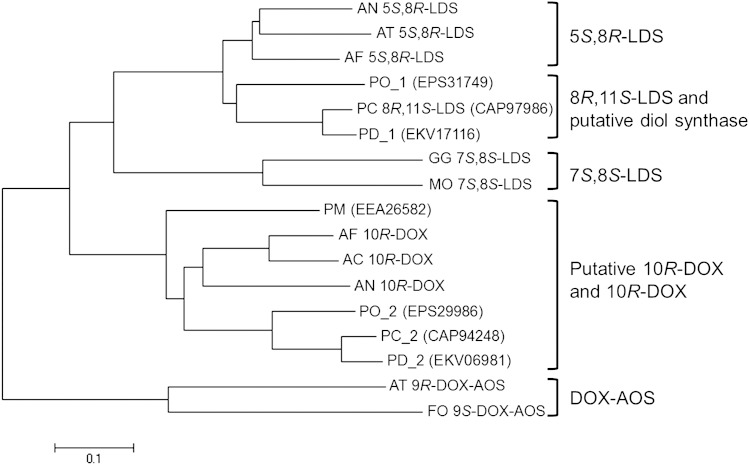
Amino acid sequences of fatty acid diol synthase-related fungal proteins, including that of 8R,11S-LDS from P. chrysogenum, were used to construct a phylogenetic tree (Fig. 6). Five groups were recovered, with each protein being categorized as a 5S,8R-LDS (2, 4, 9), 7S,8S-LDS (25, 26), 10R-DOX (3–5), DOX-AOS (9, 27), or 8R,11S-LDS and putative fatty acid diol synthase. P. marneffei PM, P. oxalicum PO_2, P. chrysogenum PC_2, and P. digitatum PD_2 clustered together in the putative 10R-DOX and 10R-DOX group. The putative fatty acid diol synthase group containing P. chrysogenum 8R,11S-LDS was more closely related to the LDS groups than those containing the 10R-DOX and DOX-AOS enzymes, with the 5S,8R-LDS group being its nearest phylogenetic neighbor. The amino acid sequence of 8R,11S-LDS from P. chrysogenum exhibited 74, 70, and 69% identity with 5S,8R-LDSs from A. fumigatus, A. nidulans, and A. terreus, respectively; 45% and 44% identity with 7S,8S-LDSs from Magnaporthe oryzae and Gaeumannomyces graminis, respectively; 44, 44, and 43% identity with 5S,8R-LDSs from A. fumigatus, A. clavatus, and A. nidulans, respectively; and 35% and 33% identity with DOX-AOSs from A. terreus and Fusarium oxysporum, respectively. Sequence analysis indicated that 8R,11S-LDS from P. chrysogenum is closest to 5S,8R-LDS from A. fumigatus.
Fig. 6.
Phylogenetic tree based on amino acid sequences of diol synthase-related fungal proteins. The tree was constructed using MEGA6, based on the full amino acid sequences of selected proteins from five groups. The bar below the tree represents an estimation of the replacement rate per residue. The putative diol synthase group may constitute an 8R,11S-LDS group. Characterized proteins are indicated by the following enzyme names: 5S,8R-LDS, 8R,11S-LDS, 7S,8S-LDS, 10R-DOX, 9R-DOX-AOS, and 9S-DOX-AOS. GenBank accession numbers are as follows: AN 5S,8R-LDS, EAA65132; AT 5S,8R-LDS, AGA95448; AF 5S,8R-LDS, EDP50447; PC 8R,11S-LDS, CAP97986; GG 7S,8S-LDS, AAD49559; MO 7S,8S-LDS, EHA52010; AF 10R-DOX, ABV21633; AC 10R-DOX, EAW09782; AN 10R-DOX, AAT36614; AT 9R-DOX-AOS, AGH14485; FO 9S-DOX-AOS, EGU88194. AN, Aspergillus nidulans; AT, Aspergillus terreus; AF, Aspergillus fumigatus; PO, Penicillium oxalicum; PC, Penicillium chrysogenum; PD, Penicillium digitatum; GG, Gaeumannomyces graminis; MO, Magnaporthe oryzae; PM, Penicillium marneffei; AC, Aspergillus clavatus; FO, Fusarium oxysporum.
The homology models for the active-site in the C-terminal domains of P. chrysogenum 8R,11S-LDS and A. nidulans 5S,8R-LDS were generated based on the known structure of AOS from guayule (PDB entry 3DBM) as a template. A ligand docking study with 8R-HPODE was conducted using these homology models to identify the specific residues involved in substrate binding. The proposed active-site residues within a sphere of 4.5 Å radius near to the docked 8R-HPODE with the heme of the C-terminal domain of P. chrysogenum 8R,11S-LDS were identified as Lys729, Val738, Ala739, Val744, Arg839, Glu905, Arg908, and Phe976, and those of A. nidulans 5S,8R-LDS were Lys734, Lys738, Glu910, and Phe981 (supplementary Fig. 10). Among the proposed active-site residues, the different residue between the two enzymes was Val738 in P. chrysogenum 8R,11S-LDS and Lys738 in A. nidulans 5S,8R-LDS. These residues may be potential amino acids to be determinant of substrate specificity. However, determination of the protein structure with the substrate bound as well as site-directed mutagenesis experiments involving the active-site residues predicted to be involved in substrate binding are necessary to provide further evidence of this conclusion because of low amino acid identity with the template structure.
CONCLUSION
A putative fatty acid diol synthase gene from P. chrysogenum was cloned and expressed, and the expressed protein was characterized. The enzyme was found to be a novel 8R,11S-LDS by identifying its products and determining its substrate specificity. Discovery of this novel fatty acid diol synthase may provide additional information on the difference with other-type fatty acid diol synthases and reproduction of Penicillium species. Discovery of this novel fatty acid diol synthase may assist in the characterization of other enzymes and cast light on reproduction in Penicillium species.
Supplementary Material
Footnotes
Abbreviations:
- AOS
- allene oxide synthase
- CP
- chiral phase
- DiHODE
- dihydroxy-9,12(Z,Z)-octadecadienoic acid
- DiHOTrE
- dihydroxy-9,12,15(Z,Z,Z)-octadecatrienoic acid
- DOX
- dioxygenase
- EPPS
- 3-[4-(2-hydroxyethyl)-1-piperazinyl]-propane sulfonic acid
- HHME
- hydroxy-9(Z)-hexadecenoic acid
- 8R-HODE
- 8R-hydroxy-9,12(Z,Z)-octadecadienoic acid
- 10R-HODE
- 10R-hydroxy-8,12(E,Z)-octadecadienoic acid
- HOME
- hydroxy-9(Z)-octadecenoic acid
- HOTrE
- hydroxy-9,12,15(Z,Z,Z)-octadecatrienoic acid
- 8R-HPODE
- 8R-hydroperoxy-9,12(Z,Z)-octadecadienoic acid
- LDS
- linoleate diol synthase
- NP
- normal phase
- PDB
- Protein Data Bank
- Ppo
- psi-producing oxygenase
- psi
- precocious sexual inducer
This work was supported by grants from the Bio-industry Technology Development Program, the Korea Healthcare Technology R&D Project, the Ministry for Health & Welfare, Republic of Korea (No. 2012-009).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Mazur P., Nakanishi K., Atef Ebrahim El-Zayat A., and Champe S.. 1991. Structure and synthesis of sporogenic psi factors from Aspergillus nidulans. J. Chem. Soc. Chem. Commun. 1991: 1486–1487. [Google Scholar]
- 2.Brodhun F., Gobel C., Hornung E., and Feussner I.. 2009. Identification of PpoA from Aspergillus nidulans as a fusion protein of a fatty acid heme dioxygenase/peroxidase and a cytochrome P450. J. Biol. Chem. 284: 11792–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodhun F., Schneider S., Gobel C., Hornung E., and Feussner I.. 2010. PpoC from Aspergillus nidulans is a fusion protein with only one active haem. Biochem. J. 425: 553–565. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann I., Jerneren F., Garscha U., and Oliw E. H.. 2011. Expression of 5,8-LDS of Aspergillus fumigatus and its dioxygenase domain. A comparison with 7,8-LDS, 10-dioxygenase, and cyclooxygenase. Arch. Biochem. Biophys. 506: 216–222. [DOI] [PubMed] [Google Scholar]
- 5.Jernerén F., Garscha U., Hoffmann I., Hamberg M., and Oliw E. H.. 2010. Reaction mechanism of 5,8-linoleate diol synthase, 10R-dioxygenase, and 8,11-hydroperoxide isomerase of Aspergillus clavatus. Biochim. Biophys. Acta. 1801: 503–507. [DOI] [PubMed] [Google Scholar]
- 6.Mazur P., Meyers H. V., Nakanishi K., Atef Ebrahim El-Zayat A., and Champe S. P.. 1990. Structural elucidation of sporogenic fatty acid metabolites from Aspergillus nidulans. Tetrahedron Lett. 31: 3837–3840. [Google Scholar]
- 7.Garscha U., and Oliw E. H.. 2007. Steric analysis of 8-hydroxy- and 10-hydroxyoctadecadienoic acids and dihydroxyoctadecadienoic acids formed from 8R-hydroperoxyoctadecadienoic acid by hydroperoxide isomerases. Anal. Biochem. 367: 238–246. [DOI] [PubMed] [Google Scholar]
- 8.Wadman M. W., de Vries R. P., Kalkhove S. I., Veldink G. A., and Vliegenthart J. F.. 2009. Characterization of oxylipins and dioxygenase genes in the asexual fungus Aspergillus niger. BMC Microbiol. 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann I., Jerneren F., and Oliw E. H.. 2013. Expression of fusion proteins of Aspergillus terreus reveals a novel allene oxide synthase. J. Biol. Chem. 288: 11459–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Huang Y., Fang M., Zhang Y., Zheng Z., Zhao Y., and Su W.. 2002. Brefeldin A, a cytotoxin produced by Paecilomyces sp. and Aspergillus clavatus isolated from Taxus mairei and Torreya grandis. FEMS Immunol. Med. Microbiol. 34: 51–57. [DOI] [PubMed] [Google Scholar]
- 11.O’Gorman C. M., Fuller H., and Dyer P. S.. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 457: 471–474. [DOI] [PubMed] [Google Scholar]
- 12.Jernerén F., and Oliw E. H.. 2012. The fatty acid 8,11-diol synthase of Aspergillus fumigatus is inhibited by imidazole derivatives and unrelated to PpoB. Lipids. 47: 707–717. [DOI] [PubMed] [Google Scholar]
- 13.Wadman M. W., van Zadelhoff G., Hamberg M., Visser T., Veldink G. A., and Vliegenthart J. F.. 2005. Conversion of linoleic acid into novel oxylipins by the mushroom Agaricus bisporus. Lipids. 40: 1163–1170. [DOI] [PubMed] [Google Scholar]
- 14.Böhm J., Hoff B., O’Gorman C. M., Wolfers S., Klix V., Binger D., Zadra I., Kürnsteiner H., Pöggeler S., Dyer P. S., et al. . 2013. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc. Natl. Acad. Sci. USA. 110: 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ropars J., Dupont J., Fontanillas E., Rodriguez de la Vega R. C., Malagnac F., Coton M., Giraud T., and Lopez-Villavicencio M.. 2012. Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti. PLoS One. 7: e49665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calvo A. M., Gardner H. W., and Keller N. P.. 2001. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 276: 25766–25774. [DOI] [PubMed] [Google Scholar]
- 17.Dagenais T. R., Chung D., Giles S. S., Hull C. M., Andes D., and Keller N. P.. 2008. Defects in conidiophore development and conidium-macrophage interactions in a dioxygenase mutant of Aspergillus fumigatus. Infect. Immun. 76: 3214–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann I., and Oliw E. H.. 2013. 7,8- and 5,8-Linoleate diol synthases support the heterolytic scission of oxygen-oxygen bonds by different amide residues. Arch. Biochem. Biophys. 539: 87–91. [DOI] [PubMed] [Google Scholar]
- 19.Jeong Y-J., Seo M-J., Shin K-C., and Oh D-K.. 2015. Production of 8-hydroxy-9,12(Z,Z)-octadecadienoic acid from linoleic acid by recombinant cells expressing H1004A-C1006S variant of Aspergillus nidulans diol synthase. J. Mol. Catal. B Enzym. 115: 35–42. [Google Scholar]
- 20.Seo M. J., Shin K. C., and Oh D. K.. 2014. Production of 5,8-dihydroxy-9,12(Z,Z)-octadecadienoic acid from linoleic acid by whole recombinant Escherichia coli cells expressing diol synthase from Aspergillus nidulans. Appl. Microbiol. Biotechnol. 98: 7447–7456. [DOI] [PubMed] [Google Scholar]
- 21.Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. III, and Smith H. O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6: 343–345. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R. A., Moss D. S., and Thornton J. M.. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231: 1049–1067. [DOI] [PubMed] [Google Scholar]
- 23.Al-Balas Q., Hassan M., Al-Oudat B., Alzoubi H., Mhaidat N., and Almaaytah A.. 2012. Generation of the first structure-based pharmacophore model containing a selective “zinc binding group” feature to identify potential glyoxalase-1 inhibitors. Molecules. 17: 13740–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliw E. H., Su C., Skogstrom T., and Benthin G.. 1998. Analysis of novel hydroperoxides and other metabolites of oleic, linoleic, and linolenic acids by liquid chromatography-mass spectrometry with ion trap MSn. Lipids. 33: 843–852. [DOI] [PubMed] [Google Scholar]
- 25.Brodowsky I. D., Hamberg M., and Oliw E. H.. 1992. A linoleic acid (8R)-dioxygenase and hydroperoxide isomerase of the fungus Gaeumannomyces graminis. Biosynthesis of (8R)-hydroxylinoleic acid and (7S,8S)-dihydroxylinoleic acid from (8R)-hydroperoxylinoleic acid. J. Biol. Chem. 267: 14738–14745. [PubMed] [Google Scholar]
- 26.Jernerén F., Sesma A., Franceschetti M., Hamberg M., and Oliw E. H.. 2010. Gene deletion of 7,8-linoleate diol synthase of the rice blast fungus: studies on pathogenicity, stereochemistry, and oxygenation mechanisms. J. Biol. Chem. 285: 5308–5316. [Erratum. 2010. J. Biol. Chem. 285: 20422.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann I., and Oliw E. H.. 2013. Discovery of a linoleate 9 S-dioxygenase and an allene oxide synthase in a fusion protein of Fusarium oxysporum. J. Lipid Res. 54: 3471–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.