Abstract
The covalent attachment of a 14-carbon aliphatic tail on a glycine residue of nascent translated peptide chains is catalyzed in human cells by two N-myristoyltransferase (NMT) enzymes using the rare myristoyl-CoA (C14-CoA) molecule as fatty acid donor. Although, NMT enzymes can only transfer a myristate group, they lack specificity for C14-CoA and can also bind the far more abundant palmitoyl-CoA (C16-CoA) molecule. We determined that the acyl-CoA binding protein, acyl-CoA binding domain (ACBD)6, stimulated the NMT reaction of NMT2. This stimulatory effect required interaction between ACBD6 and NMT2, and was enhanced by binding of ACBD6 to its ligand, C18:2-CoA. ACBD6 also interacted with the second human NMT enzyme, NMT1. The presence of ACBD6 prevented competition of the NMT reaction by C16-CoA. Mutants of ACBD6 that were either deficient in ligand binding to the N-terminal ACBD or unable to interact with NMT2 did not stimulate activity of NMT2, nor could they protect the enzyme from utilizing the competitor C16-CoA. These results indicate that ACBD6 can locally sequester C16-CoA and prevent its access to the enzyme binding site via interaction with NMT2. Thus, the ligand binding properties of the NMT/ACBD6 complex can explain how the NMT reaction can proceed in the presence of the very abundant competitive substrate, C16-CoA.
Keywords: protein acylation, membranes, phospholipids, binding protein, protein interaction, N-myristoyltransferase 2, coenzyme A
Members of the acyl-CoA binding domain (ACBD)-containing protein family are involved in the maintenance of diverse cellular functions and can interact with a multitude of proteins implicated in a variety of cellular functions such as neural stem cell self-renewal, neurodegeneration, stress resistance, lipid homeostasis, intracellular vesicle trafficking, organelle formation, viral replication, and apoptotic response. Several ACBD members have been reported to associate with proteins such as GABAA (1), Numb (2), DMT1 (3), TSPO, PPM1L (4), the mHtt/Rhes complex (5), giantin/golgin-160, HNF4α (6), SREBP1 (7), plant AtEBP (8), and several viral proteins (9–14). Plant ACBD members are also implicated in a multitude of functions such as embryogenesis and resistance to various stresses (15–17). However, the requirement and role of the acyl-CoA ligand bound to the conserved N-terminal ACBD for the interaction with other proteins is poorly understood. Human ACBD6 is a modular protein with the N-terminal ACBD linked to a nonconserved and nonessential carboxy-terminal domain carrying two ankyrin-repeat motifs (18). ACBD6 binds medium- to long-chain acyl-CoAs and several site-directed mutants have been obtained that affect this binding (19).
The role of ACBD6 in lipid metabolism of mammalian cells has not been established. Previous studies have shown that expression of the human ACBD6 protein was elevated in hematopoietic progenitor cells and erythrocyte precursors (18) and that ACBD6 mRNA was higher in hematopoietic progenitors, as compared with other lineage-committed cells (20, 21). In human cells infected with the bacterial pathogen Chlamydia trachomatis, ACBD6 protein was associated with cytosolic lipid droplets and was transferred to the parasitophorous vacuole resulting in its removal from the cytosol (22). Inside the vacuole, ACBD6 regulates the activity of a bacterial acyltransferase essential for lipid metabolism of the pathogen. A related fungal fatty acyl-CoA binding protein (ACBP) from the human pathogen Cryptosporidium parvum is required by the fungus for growth in human cells, as confirmed using chemical inhibition of acyl-CoA binding to this ACBD protein (23).
Co-translational N-myristoylation of the glycine +2 residue of nascent peptides, following removal of the initiator methionine by the human N-myristoyltransferase (NMT) enzymes (NMT1 and NMT2), can affect the association of the acyl-proteins with membranes (24–26). The addition of a 14-carbon long aliphatic tail can act as a lipid anchor and mediates oligomeric assembly and interaction with other proteins. Although amide linkage of the myristate to the glycine residue appears irreversible, a structural conformational property of some acyl-proteins, called the myristoyl-switch, can remove the aliphatic tail from the lipid bilayer and dissociate the proteins from their membrane-bound location (25, 27). The acyl-transferase activity of the NMT enzymes is specific toward myristoyl-CoA (C14-CoA) and is essential for the intracellular development of pathogens (28–36). Chemicals inhibiting myristoylation of proteins are potent drugs against parasitic protozoa and fungi. The catalytic reaction is a multistep process initiated by binding of C14-CoA to apo-NMT, which triggers a conformational change exposing the peptide binding site (26, 28). C14-CoA is then hydrolyzed and CoA is released. The aliphatic tail is transferred to the glycine +2 residue of the peptide, which is then released. NMT proteins can bind palmitoyl-CoA (C16-CoA), but a 16-carbon chain cannot be transferred to the glycine +2 residue by the myristoyl-transferase (26, 28, 29, 33). It is unclear how NMT enzymes are able to perform myristoylation in vivo using a very rare acyl-donor (C14-CoA) in the presence of a very abundant acyl-CoA competitor (C16-CoA). Thus, a process to prevent binding or trigger release of this competitor from the C14-CoA binding site of NMT seems essential to warrant its activity in vivo.
We determined that ACBD6 was associated with the two human NMT enzymes in human cells, with a stronger interaction with NMT2 than NMT1. The C-terminal domain of ACBD6 interacted with a C-terminal region of NMT2. Our results further show that ACBD6 protected the enzyme from competitive binding by C16-CoA and that lipid-bound ACBD6 stimulated NMT2 activity toward its substrate, C14-CoA. Together, these results establish a physiological role for the binding property of a member of the human family of acyl-CoA carriers, and determine how NMT enzymes can perform their function in vivo.
MATERIALS AND METHODS
Materials
The No-Weigh format of disuccinimidyl suberate (DSS) was purchased from Thermo Fisher Scientific. Ellman’s reagent [5,5′-dithio-bis-(2-nitrobenzoic acid)] (DTNB), acyl-CoAs, and fatty acids were from Sigma-Aldrich. All other compounds used were reagent grade. The peptide Hs pp60src#2-9 (GSNKSKPK, C-term amide; MW of 843.99) (37) was synthesized by YenZym Antibodies, LLC (South San Francisco, CA).
Cloning and site-directed mutagenesis
Cloning of human ACBD1 and ACBD6 was previously reported (18, 19). Human NMT2 isoform 1 (NP_004799) was cloned by PCR from NM_004808.2 into pBIND vector and human ACBD6 was cloned into the pACT vector (Check-Mate Mammalian two-hybrid system; Promega). As previously described (37, 38), the membrane targeting signal present in the first 97 residues of NMT2 was removed to produce an active cytosolic form of the protein in Escherichia coli (also known as ecNMT2, see below). Site-directed mutagenesis experiments were performed with the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. Primers were designed with the QuikChange® primer design program. The presence of the intended nucleotide change(s) and the absence of unwarranted mutations were verified by full-length sequencing of the constructs. N-terminally tagged green fluorescent protein (GFP) and hemagglutinin (HA) fusion of full-length ACBD6 and full-length NMT2 were obtained in the pAcGFP1-C1 and pIRESneo vector (Clontech Laboratories, Inc.), respectively.
Protein expression and purification
All proteins and mutant forms were produced as hexahistidine fusion forms. ACBD1 and ACBD6 were expressed in the E. coli host BL21(DE3) cells (Novagen) and ecNMT2 was produced in RosettaDE3 cells (Novagen). Proteins were purified by affinity metal chromatography with the addition of 0.2% Triton X-100 in all the buffers for the ecNMT2 protein. The purified proteins were stored at −80°C in Tris-HCl [50 mM (pH 8.0)], NaCl (0.1 M), EDTA (5 mM), and glycerol [10% (v/v), with 0.2% Triton X-100 for ecNMT2]. Prior to isothermal titration calorimetry (ITC) measurements, proteins were dialyzed in the ITC buffer (see below).
Affinity purification MS
Affinity purification followed by LC-MS/MS was used to identify candidate protein interactions of ACBD6 following identical methods to those previously published (11, 39). Briefly, ACBD6 was cloned into the pcDNA4TO expression vector encoding a C-terminal 2×-StrepTag and then transiently expressed in HEK293 cells following reported methods (39). The cells were harvested and lysed, and the Strep-tagged protein was captured on StrepTactin Sepharose under native conditions, and then eluted with 1× desthiobiotin (IBA Technology, Gottingen Germany) as reported (11). This protein sample was processed by in-solution digestion and analyzed by LC-MS/MS peptide sequencing as reported (11, 39).
MS data were searched using Protein Prospector software version 5.10.17 (40) against the Refseq NCBInr human database (downloaded July 29, 2015 from ftp://ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/mRNA_Prot/) containing 99,857 sequences, concatenated with 99,857 additional randomized decoy sequences for calculation of the false discovery rate (FDR) (41). The FDR was <1%, using a protein score of 22, a peptide score of 15, a protein expectation value of 0.01, and a peptide expectation value of 0.001. Modifications allowed were: fixed carbamidomethylation of Cys and the following variable modifications: oxidation of Met, start-Met cleavage, oxidation of the N terminus, acetylation of the N terminus, and pyroglutamate formation from Gln.
Protein identifications from ACBD6 affinity-purification experiments were performed over four biological replicate experiments. Proteins were reported as specific interacting proteins if observed in at least three of four replicate experiments with more than one unique peptide in at least one experiment. These proteins were also compared with a list of high-frequency background proteins from >2,000 similar affinity purification MS experiments with Strep-tagged proteins (11), and excluded if the background frequency was >0.10. MS data for proteins meeting these criteria are reported in supplementary Table 1.
Protein interaction assays
Mammalian two-hybrid interaction assays were performed in HeLa and HEK293 cells grown in 96-well plates with the CheckMate system (Promega). Hela cells were transfected with TurboFect reagent (Thermo Scientific) and HEK293 cells with TransIT-LT1 transfection reagent (Mirus Bio). Luminescence was quantified with the Dual-Luciferase reporter assay of Promega, according to the manufacturer’s instructions. The homobifunctional N-hydroxysuccinimide ester reagent, DSS, reacts with primary amine groups and was used to detect interaction of purified NMT2 and ACBD6 proteins. DSS was bought in individual sealed microtubes (Thermo Fisher Scientific) and stock solutions of 100 mM in DMSO (further diluted in DMSO as needed) were made fresh prior to each experiment. DSS was added in a 30-fold excess of the protein (mole/mole). Proteins were dialyzed in 10 mM potassium phosphate (pH 7.4) at 4°C. ACBD6 and NMT2 proteins were mixed at a 1:1 molar ratio (20 μM each) in 29 μl of 50 mM potassium phosphate (pH 7.4) at 37°C for 20 min. When acyl-CoAs were used, they were added at a final concentration of 50 μM. One microliter of DSS was then added to the mixture at a final concentration of 1 mM, and subsequently incubated for 30 min at room temperature. Untreated control reactions were performed with addition of 1 μl of DMSO and incubated as described for the DSS-treated samples. Reactions were stopped by quenching of DSS with 2 μl Tris-HCl (1 M, pH 7.4) for at least 15 min at room temperature. Mixtures were then boiled in SDS-PAGE loading buffer for 4 min and separated on denaturing SDS-polyacrylamide Tris-glycine gels. Proteins were detected by staining with GelCode Blue reagent (Thermo Fisher Scientific) or were transferred on nitrocellulose membrane and detected with a HRP-conjugated anti-histidine antibody (INDIA-HisProbe-HRP antibody; Thermo Fisher Scientific).
Co-immunoprecipitation experiments
Protein samples were made from HEK293 cells grown in 12-well plates. For each sample, three wells were cotransfected using the TurboFect reagent (Thermo Scientific) with the pair HA-ACBD6/GFP-NMT2 or HA-NMT2/GFP-ACBD6 for 48 h. Cells were lysed with lysis immunoprecipitation buffer (Thermo Scientific) and protease cocktail inhibitor (Thermo Scientific) in a final volume of 400 μl. Cleared lysates were stored at −80°C. For co-immunoprecipitation, 200 μg of each protein sample was precleared with 25 μl protein G-magnetic beads (NEBiolabs) for 1 h at 4°C. Following removal of the beads, 5 μg of mouse monoclonal anti-HA antibody (Sigma-Aldrich, H3663) was added and incubated for 1 hr at 4°C. Then, 25 μl of fresh protein G-magnetic beads was added and incubated overnight at 4°C. Beads were then washed three times with ice-cold PBS and with lysis IP buffer. Complexes were eluted by suspending the beads in 60 μl SDS-PAGE loading buffer and heating at 70°C for 10 min. After removal of the beads, 10 μl of each sample was analyzed by Western blot with rabbit polyclonal anti-GFP (Sigma-Aldrich, G1544) and with HRP-conjugated mouse anti-HA antibodies (Sigma-Aldrich, H6533)
ITC assays
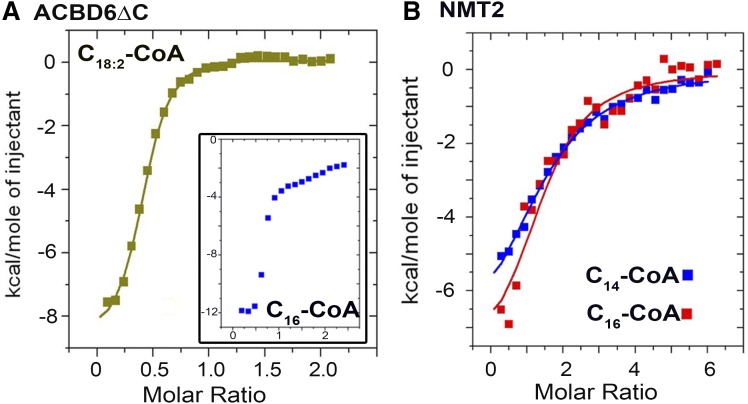
ITC measurements of the binding of acyl-CoAs and fatty acids were performed on a VP-ITC instrument (MicroCal, LLC) (18, 19). All experiments were performed in ammonium acetate (25 mM, pH 7.4), supplemented with 0.1% Triton X-100 to prevent precipitation of ecNMT2, at 30°C. The proteins were dialyzed in the same buffer and fresh 10 mM stocks of the ligands were prepared from powder with the dialyzing buffer. For NMT2 (Fig. 6B), measurements were performed with 28 injections of 10 μl of 165 μM ligand every 150 s. The chamber contained the protein at an initial concentration of 5.5 μM. For ACBD6ΔC measurements (Fig. 6A), protein was at a concentration of 30 μM and was injected with 600 μM of C16-CoA and 300 μM of C18:2-CoA. For data presented in Fig. 6C, proteins were at a concentration of 10 μM and C16-CoA at 100 μM. Control experiments were run by injecting buffer into the cell containing the protein and by injecting the ligand into the cell containing buffer. Heat generated from control runs was subtracted from the data of the experimental set performed under the same conditions
Fig. 6.
Acyl-CoA binding measurement assays. ITC measurements were performed in ammonium acetate (25 mM, pH 7.4), supplemented with 0.1% Triton X-100 when NMT2 was used, at 30°C. The ACBD6ΔC and ecNMT2 proteins were dialyzed in the same buffer and fresh 10 mM stocks of the acyl-CoA ligands were prepared from powder with the dialyzing buffer. Measurements were performed by 28 injections of ligand into the chamber containing the protein. In (A), ligand was at 100 μM and protein at 10 μM. Inset: Ligand was at 600 μM and protein at 30 μM. In (B), ligand was at 165 μM and protein at 5.5 μM.
NMT activity measurements
Real-time measurements of the released CoA from acyl-CoAs by purified NMT2 were performed at 30°C in a Cary Avian UV-Vis spectrophotometer (50 Bio; Varian) in the presence of DTNB. Measurements were made in dual-beam mode against blank buffer and normalized by subtracting values obtained in reactions performed in the absence of the target peptide. Reactions were performed in 500 μl of 20 mM sodium phosphate buffer at pH 8.0 with 1 mM EDTA and 100 μM Hs pp60src#2-9 peptide, 5–100 μM acyl-CoAs (C14-CoA, C16-CoA, C18:2-CoA), and 0.2 mM DTNB. Unless otherwise indicated, a 2:1 molar ratio of ACBD6/NMT2 was used. Titration of NMT2 was performed to determine the optimal concentration of the partially purified proteins to maintain linearity of the measurement over a period of 5 min (usually 2 μM ACBD6/1 μM NMT2). Kinetic calculations were performed with GraphPad Prism 6.
RESULTS
Interaction of NMT enzymes with ACBD6
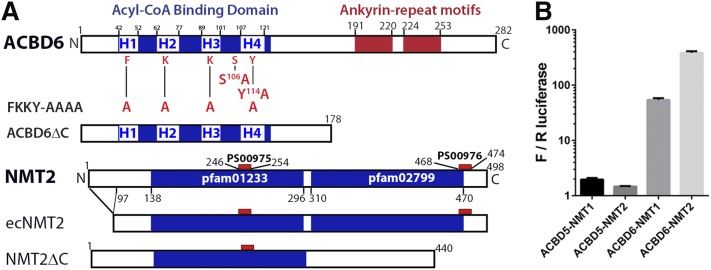
ACBD6 protein is a modular protein with an N-terminal conserved ACBD and two ankyrin-repeat motifs present in the nonconserved C-terminal domain (Fig. 1A). Affinity purification MS was performed using a C-terminally Strep-tagged ACBD6 construct, transiently expressed in HEK293 cells, in four biological replicates. From these experiments, the most specific interacting proteins were NMT1 and NMT2, identified with a total of 24 and 14 unique peptides, respectively (supplementary Table 1). NMT1 and NMT2 share high sequence identity (73%, ClustalW alignment), thus unique and shared peptide sequences were identified to each of these proteins in the MS data (supplementary Table 2). Sufficient unique peptide counts were obtained to both NMT1 and NMT2 in each experiment to confirm the presence of both proteins at comparable levels in this semi-quantitative analysis (supplementary Table 3). Two proteins, CAMK2D and UGGT1, were also identified at a lower specificity threshold, but NMT1 and NMT2 were the only proteins that reproducibly interacted with ACBD6 in all four experiments. The specificity of the association with ACBD6 was further confirmed with a mammalian two-hybrid interaction assay, which established a stronger binding of ACBD6 with NMT2 than with NMT1, as compared with the related ACBD5 protein (33% identical in the ACBD), which showed no interaction (Fig. 1B).
Fig. 1.
Acyl-CoA binding protein ACBD6 and NMT2. A: The predicted four helices (H1 to H4) of the ACBD and the two ankyrin-repeat motifs of ACBD6 are shown. Residues changed to alanine by site-directed mutagenesis in the mutated forms S106A, Y114A, and FKKY-AAAA (also known as FKKYmut) are shown. The form ACBD6ΔC lacked the ankyrin domains. Isoform 1 of NMT2 (NP_004799.1) is shown with the two C14-CoA:protein NMT conserved domains, pfam01233 and pfam02799, and the two signature motifs, PS00975 and PS00976. The recombinant form produced in E. coli (ecNMT2) lacking the first 97 residues and the truncated form (NMT2ΔC) lacking the last 58 residues are indicated. B: Luminescence interaction quantification assays were performed with the mammalian CheckMate system (Promega) in HEK293 cells. Compared with ACBD5, which did not interact with NMT proteins, association of ACBD6 with NMT1 and NMT2 was 25- and 250-fold stronger, respectively. Error bars represent the standard deviations of three measurements. We previously identified ZNF23 in a yeast two-hybrid screen using ACBD6 as bait (62), but that interaction could not be confirmed in mammalian cells (supplementary Fig. 1A).
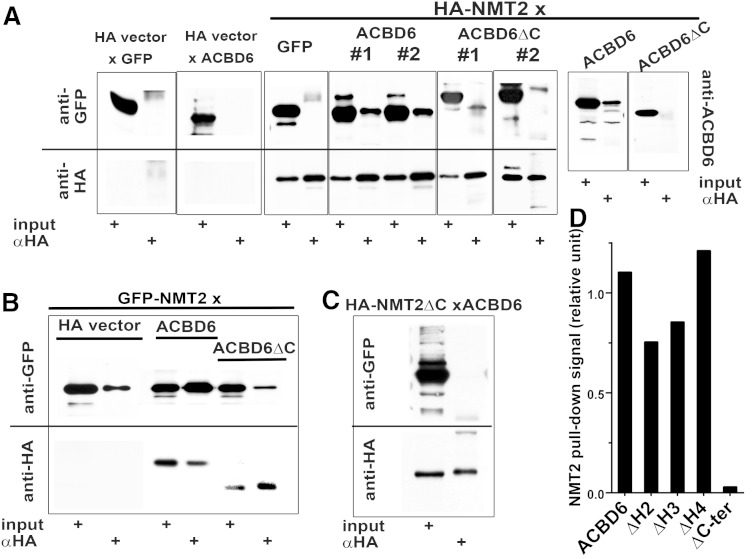
Association of NMT2 with ACBD6 was further investigated by co-immunoprecipitation of wild-type and mutant constructs of GFP-tagged ACBD6 by HA-NMT2, and of the inversely tagged GFP-NMT2 by HA-ACBD6 (Fig. 2A, B). A C-terminally truncated form of ACBD6 lacking the two ankyrin-repeat motifs (ACBD6ΔC, Fig. 1A) did not co-immunoprecipitate with NMT2 (Fig. 2A, B). The C terminus of NMT-2 includes residues that bind the target peptide substrate (Fig. 1A), and the C terminus itself may assist in catalysis by deprotonating the ammonium of the acceptor glycine residue (28). An NMT2 form lacking the last 58 residues, including the signature motif PS00976 (NMT2ΔC, Fig. 1A), did not interact with full-length ACBD6 either by co-immunoprecipitation or mammalian two-hybrid assay (Fig. 2C; supplementary Fig. 1A). Deletion mutants of conserved motifs of the ACBD of ACBD6 were also designed (Fig. 1, regions H2–4), and these did not prevent interaction with NMT2 (Fig. 2D). Thus, ACBD6 interacts via its C-terminal domain containing the ankyrin-repeat motifs with NMT2, and the NMT2 interaction with ACBD6 is dependent upon its C-terminal region.
Fig. 2.
ACBD6 and NMT2 interactions. Western blot analysis of co-immunoprecipitation samples obtained with a monoclonal anti-HA antibody from protein lysates of HEK293 cells cotransfected with GFP-tagged ACBD6 and HA-tagged NMT2 (A), GFP-tagged NMT2 and HA-tagged ACBD6 (B), and GFP-tagged ACBD6 and HA-tagged NMT2ΔC (C). GFP proteins were detected with a rabbit polyclonal anti-GFP antibody and HA proteins with an HRP-conjugated mouse monoclonal anti-HA antibody. As shown in (A), samples were also analyzed with a rabbit peptide-raised anti-ACBD6 antibody. Control cells were obtained by transfection of the two empty vectors (GFP and HA). For each sample, the signal obtained with 10% of the lysate before immunoprecipitation (input) is shown on the left of the signal obtained after pull-down (αHA). Note that the polyclonal anti-GFP antibody nonspecifically cross-reacted with a band that did not correspond to GFP-NMT2, GFP-ACBD6, or GFP-ACBD6ΔC. Duplicates of the co-immunoprecipitation of GFP-ACBD6 or GFP-ACBD6ΔC by HA-NMT2 are shown and indicated as samples #1 and #2 in (A). D: Densitometry quantification of the GFP signal obtained by Western blot analysis of co-immunoprecipitation samples obtained in cells cotransfected with GFP-NMT2 and the indicated truncated forms of ACBD6. Data were collected from a single representative blot.
ACBD6 and NMT2 form hetero-dimeric complexes
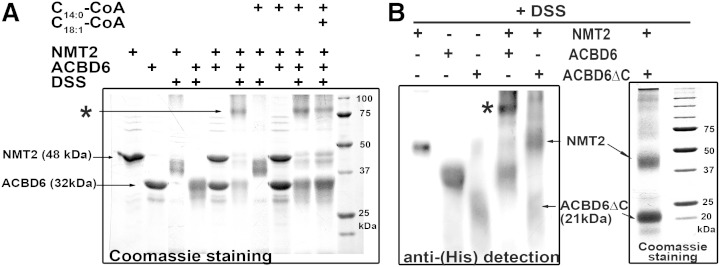
Association of the two purified proteins was investigated using covalent cross-linking with the reagent DSS, followed by analysis of the trapped complexes by SDS-PAGE. As previously reported (18, 19), ACBD6 (32 kDa) behaved as a monomeric protein, and no high molecular mass species were detected after DSS treatment (Fig. 3A). To produce and purify NMT2, a recombinant form was expressed in E. coli without the N-terminal membrane localization sequence (also known as ecNMT2 protein) (37, 38). Purified ecNMT2 (supplementary Fig. 1B) was primarily monomeric (48 kDa), and few bands of very high molecular mass were formed after DSS treatment (>100 kDa, not shown). When ACBD6 and ecNMT2 were mixed at a 1:1 molar ratio, most of the ecNMT2 monomeric form disappeared, and a new band was detected at approximately the calculated mass of a heterodimeric ACBD6/ecNMT2 complex (80 kDa). Addition of the NMT2 substrate, C14-CoA, and of the ACBD6 ligand, C18:1-CoA, did not prevent formation of the complex (Fig. 3A). In contrast, the truncated recombinant ACBD6ΔC mutant (21 kDa) did not form a complex with ecNMT2 (Fig. 3B).
Fig. 3.
DSS cross-linking of NMT2 and ACBD6. Purified hexahistidine-tagged NMT2 (also known as ecNMT2) and ACBD6 recombinant proteins (20 μM each) were incubated in 50 mM potassium phosphate (pH 7.4) at 37°C for 20 min. When indicated, C14-CoA and C18:1-CoA were added at a final concentration of 50 μM. DSS was added to a final concentration of 1 mM and incubated for 30 min at room temperature. Following quenching of DSS with 65 mM Tris-HCl (pH 7.4), proteins were loaded on denaturing SDS-PAGE gels. After electrophoresis, protein bands were detected with Gelcode Blue [(A) and right inset in (B)] or were transferred onto a PVDF membrane and detected with an anti-histidine antibody [left inset in (B)]. The position of the high molecular mass form observed with ACBD6/NMT2 is indicated with an asterisk in the panels. Note the absence of detection of an ACBD6ΔC/NMT2 complex in (B).
Ligand binding to ACBD6 stimulates the NMT reaction
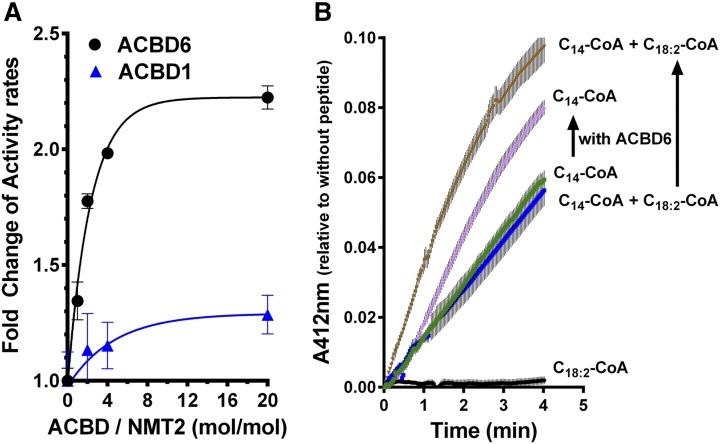
Activity of purified ecNMT2 enzyme was measured by the real-time detection of the release of CoA from C14-CoA in the presence of a synthetic peptide matching the amino terminal myristoylation sequence of c-Src protein (24) (supplementary Fig. 1C, D). Addition of increasing concentrations of ACBD6 protein resulted in the stimulation of activity of ecNMT2 (Fig. 4). No inhibitory effect was observed, even with a 20:1 ACBD6/ecNMT2 molar ratio. Under similar conditions, addition of purified ACBD1 had only a weak stimulatory effect. Compared with ACBD6, two ACBD6 mutant forms, Y114A and FKKY-AAAA (dubbed FKKYmut), with defects in acyl-CoA binding (19) failed to stimulate the reaction in the presence of C14-CoA, thus acyl-CoA binding of ACBD6 is required for the stimulatory effect on NMT2 activity (Fig. 5). The molecule C18:2-CoA, a ligand of ACBD6, was not an acyl donor for the NMT reaction (Figs. 4, 5). In the absence of ACBD6, addition of C18:2-CoA to C14-CoA had a weak inhibitory effect on the reaction (Fig. 5). However, in the presence of ACBD6, addition of C18:2-CoA resulted in increased stimulation of ecNMT2 activity compared with the activity obtained with only ACBD6. This stimulatory effect was not detected with the two mutant ACBD6 forms, Y114A and FKKYmut. Thus, compared with apo-ACBD6, acyl-CoA-bound ACBD6 appeared to be the form enhancing the processing of C14-CoA by NMT2, independent of its binding to C14-CoA.
Fig. 4.
ACBD6 stimulation of the NMT reaction. A: Real-time colorimetric measurements of the ecNMT2 activity in the presence of increasing concentrations of ACBD1 or ACBD6 were performed at 30°C. Rate values are presented relative to the rate obtained in their absence. B: Traces of absorbance at 412 nm of reactions are shown as a function of time in the presence of different acyl-CoA molecules. A molar ratio ACBD6 and ecNMT2 of 2:1 (2 and 1 μM) was used. The final concentration of C14-CoA was 15 μM and of C18:2-CoA was 50 μM. Reactions were performed in the absence or presence (upward arrow) of ACBD6. For each reaction, control measurements were performed in the absence of the target peptide and values were subtracted from the readings obtained in its presence (see supplementary Fig. 1C, D). Error bars represent the standard deviations of three measurements.
Fig. 5.
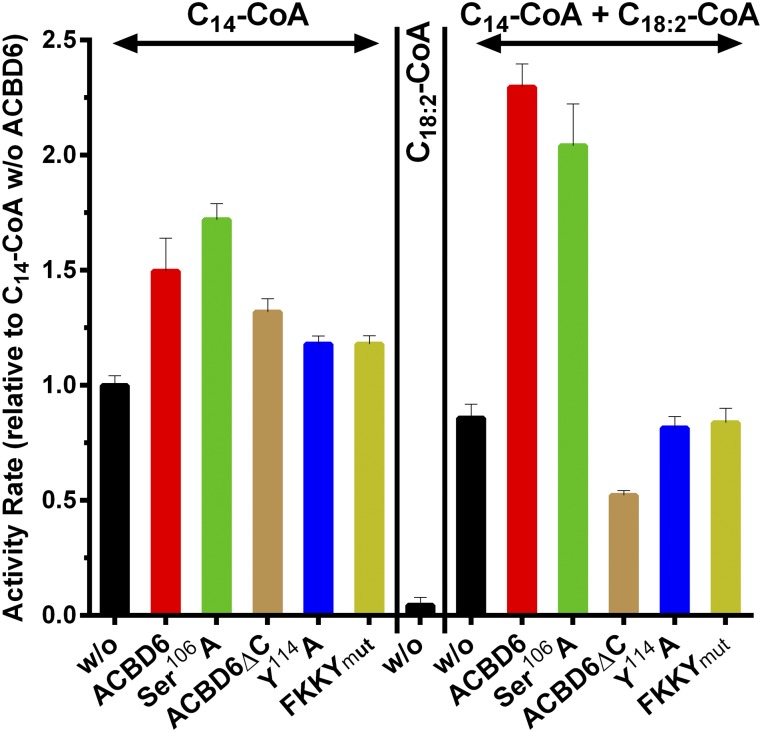
Difference in NMT2 activity in the presence of apoACBD6 and lipid-bound ACBD6. Measurements were performed as described in the legend of Fig. 4 with 15 μM C14-CoA, 50 μM C18:2-CoA, and 15 μM C14-CoA in the presence of 50 μM C18:2-CoA. The ACBD6 recombinant forms added to the reactions are shown in Fig. 1. Activity rate values are presented relative to the value obtained with 15 μM C14-CoA in the absence of ACBD6. Error bars represent the standard deviations of at least three measurements.
ACBD6 interaction is required for NMT2 activity stimulation
The requirement for protein interaction in the stimulatory effect of ACBD6 in the presence of its ligand on the NMT reaction was investigated. The carboxy-terminal domain of ACBD6 is dispensable for ligand binding (18) and we confirmed that the truncated ACBD6ΔC form binds C18:2-CoA (Fig. 6A). ACBD6ΔC did not interact with ecNMT2 (Fig. 2), suggesting that the ankyrin-repeat motifs were involved in the contact to the C-terminal motif of NMT2 (Fig. 2C; supplementary Fig. 1A). Compared with full-length ACBD6, ACBD6ΔC could not stimulate the NMT reaction and the ecNMT2 activity rate in the presence of the truncated form was similar to those obtained with the Y114A and FKKYmut mutants (Fig. 5). Thus, interaction of ABCD6 with NMT2 was required for the ligand-mediated increase in NMT2 activity.
N-terminal ACBD6 domain prevents binding of C16-CoA to NMT2
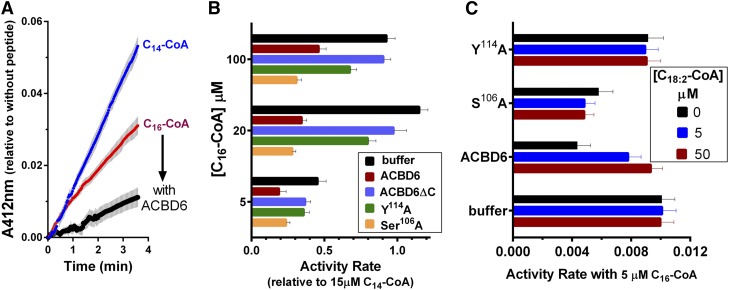
Purified ecNMT2 binds both C14-CoA and C16-CoA with similar affinity (Fig. 6B). Binding of C16-CoA to ecNMT2 resulted in cleavage of the ester bond and release of CoA (Fig. 7A). Confirmation of the strong binding affinity of NMT2 for this acyl-CoA competitor was obtained by the observation that 20 μM of C16-CoA produced a stronger signal than 15 μM of C14-CoA (Fig. 7B). Addition of ACBD6 resulted in the stimulation of the reaction with C14-CoA, but it decreased the processing of C16-CoA by ecNMT2 (Fig. 7A, B). The mutant S106A lacks one of the two phosphorylated serine residues of ACBD6 (42, 43), and binds to C16-CoA with stronger affinity than ACBD6. This mutant stimulated ecNMT2 activity with C14-CoA as ACBD6 (Fig. 5), but it could prevent the processing of C16-CoA at much higher concentration than ACBD6 (Fig. 7B). In contrast, the ACBD6 mutant, Y114A, which did not bind C16-CoA (19), could not prevent the processing of C16-CoA by NMT2, even at very low concentration (Fig. 7B).
Fig. 7.
Effect of ACBD6 on NMT2 activity in presence of C16-CoA. Measurements were performed as described in the legend of Fig. 4 with 1 μM ecNMT2 and 15 μM C14-CoA (blue). A: Competing ligand at 5 μM C16-CoA was added to the reaction alone (red) or in the presence of 2 μM ACBD6 (black). B: The indicated concentration of competing ligand C16-CoA was added in the presence of ACBD6 constructs at 2 μM. C: The first competing ligand, C16-CoA, was fixed at 5 μM, and increasing concentrations of C18:2-CoA were added as indicated in the presence of ACBD6 constructs at 2 μM. Activity rate values in (B) are presented relative to the value obtained with 15 μM C14-CoA in the absence of ACBD6. Error bars represent the standard deviations of at least three measurements.
NMT2 interaction to the C-terminal ACBD6 domain is required for competitor protection
As established above, binding of C16-CoA to ACBD6 prevented processing of the acyl-CoA competitor by ecNMT2. However, the effect of ACBD6 could not be accounted for by a model requiring exhaustion of free-competitor in the reaction mixture because as little as 1 μM of ACBD6 was enough to reduce the processing of 100 μM C16-CoA to half (Fig. 7B). As seen for the C18:2-CoA stimulation of C14-CoA processing by ecNMT2, ACBD6/NMT2 complex formation was also necessary to prevent usage of C16-CoA by ecNMT2. The truncated ACBD6ΔC form, which did not interact with ecNMT2 (Fig. 2A and 2B), but bound C16-CoA (Fig. 6A, inset), could not prevent processing of C16-CoA by ecNMT2. In fact, that form was even less efficient in protecting ecNMT2 than the mutant Y114A (Fig. 7B). These results suggested that in the hetero-dimeric complex, ACBD6-NMT2 formed through the interaction of their C-terminal domains, C16-CoA was prevented from accessing the NMT2 binding site through sequestration to the N-terminal ACBD of ACBD6.
Displacement of C16-CoA by linoleoyl-CoA
To confirm that protection of NMT2 from C16-CoA by ACBD6 was the result of C16-CoA binding to ACBD6, the ligand, C18:2-CoA, was added to the reaction. ACBD6 binds C18:2-CoA with greater affinity than C16-CoA (18, 19). At an equal concentration of C18:2-CoA and C16-CoA (5 μM), ACBD6 could no longer prevent processing of C16-CoA by ecNMT2 and activity rate doubled (Fig. 7C). The mutant Y114A, unable to bind either acyl-CoA species (19), had no detectable effect on ecNMT2 activity, irrespective of the C18:2-CoA concentrations. The mutant S106A, which binds C16-CoA with stronger affinity than ACBD6, limited use of C16-CoA by ecNMT2, even when a 10-fold excess of C18:2-CoA was added to the reaction. Thus, selectivity of binding of the ACB motif of the acyl-CoA carrier ACBD6 modulates the NMT reaction by controlling availability of acyl-CoA to NMT2 and stimulates its activity via protein contact with their respective C-terminal domains.
DISCUSSION
The acyl-transferase activity of the NMT proteins is highly specific toward C14-CoA. On the other hand, a variety of glycine-containing nascent peptide chains can bind to the C-terminal domain of NMT (33, 44–47), indicating that NMT is less selective toward its protein substrate. The catalytic reaction is a multistep process initiated by binding of C14-CoA to the N-terminal domain of apo-NMT, which triggers a conformational change exposing the peptide binding site (26, 28, 48). C14-CoA is then hydrolyzed and CoA is released. The aliphatic tail is transferred and covalently linked to the exposed glycine residue of the peptide substrate, releasing the acylpeptide. Whereas the transfer rates of acyl chains other than C14 are extremely slow compared with myristate (33, 44, 47), NMT proteins bind the very abundant C16-CoA molecule with the same affinity as C14-CoA, a minor acyl-CoA in cells (26, 28, 29, 33, 49). Thus, in the absence of a process to modulate binding or release of the acyl-CoA competitor from the C14-CoA binding site, compounds such as C16-CoA would inhibit the action of NMT enzymes in vivo. This is illustrated in Fig. 8, which provides a summary of data presented in this study, indicating an important role of acyl-CoA binding proteins in this process. Several other reports provide evidence that other cellular components modulate the NMT reaction to ensure that the correct acyl chain is linked to the protein substrate. Whereas NMT enzymes are highly specific in transferring a 14-carbon tail on nascent peptides, C14:1 and C14:2, rather than C14:0, are the fatty acids used to modify proteins such as the α-subunit of the G-protein photoreceptor in retinal photoreceptor cells (49–51). The acyl-CoA pools in the retina are not significantly different as compared with other tissues; hence a selective system needs to account for the unique acylation taking place in these cells (49). It can be hypothesized that acyl-CoA carriers with stronger affinity to unsaturated myristate control the acylation profile observed in this tissue.
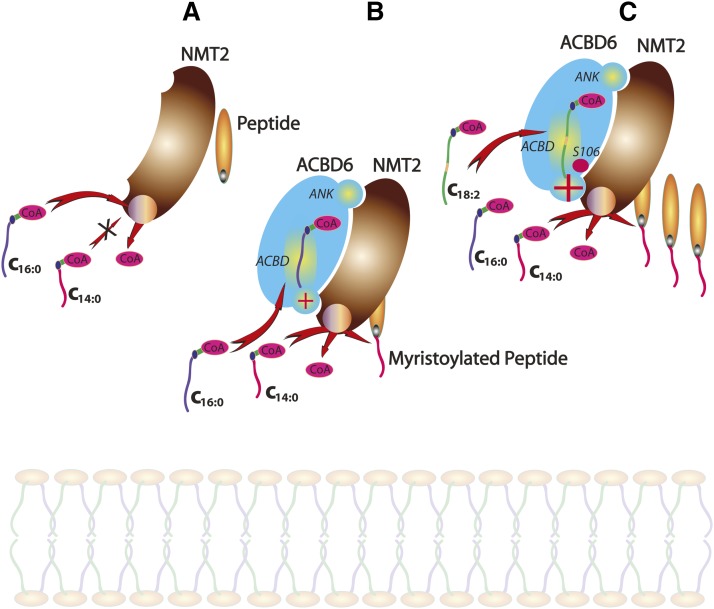
Fig. 8.
Proposed events leading to the protection of NMT2 activity by ACBD6. A: NMT2 will bind both C16-CoA (C16:0) and C14-CoA (C14:0), but can only use C14:0 to acylate a peptide. The high abundance of C16:0 in vivo effectively inhibits protein myristoylation. B: ACBD6 interacts with NMT2 through binding mediated by its ankyrin (ANK) motif to the C terminus of NMT2. The ACBD domain of ACBD6 sequesters C16:0-CoA. The complex formed by NMT2 and ACBD6 blocks access of C16:0-CoA to the acyltransferase site. ACBD6 prevents C16-CoA inhibition of the myristoylation reaction and stimulates the formation of myristoylated-peptide. A red plus sign in the speculated entry site of the ACBD6/NMT2 complex indicates stimulation. C: Protein myristoylation is further stimulated by the binding of linoleoyl-CoA (C18:2) to ACBD6. In vivo phosphorylation of the serine 106 residue (S106) in the ACBD of ACBD6 plays a role in this process. Phosphoserine106 is shown as a red filled circle next to the red plus sign indicating increased stimulation compared with the scheme depicted in (B).
In our study, the stimulation of the NMT activity of NMT2 in the presence of apo-ACBD6 and C14-CoA could be explained by the formation of the ACBD6/NMT2 complex (Fig. 8B). We speculate that in the ACBD6/NMT2 complex, the entry of the C16:0 acyl-chain to the binding site of the acyltransferase subunit is reduced in favor of the C14:0 acyl-chain. Mutant forms of ACBD6, which are unable to bind C16-CoA or are unable to interact with the enzyme, could not prevent NMT2 from using this acyl donor (Fig. 7). These findings suggested that protection of the NMT reaction was also the result of the local sequestration of C16-CoA by the acyl-CoA carrier subunit of the ACBD6/NMT2 complex. The highly improved preference for C14:0-CoA relative to C16:0-CoA in this complex is essential, given that these two compounds are at the low and high end of the concentration range, respectively (49, 52). Without this shift in specificity triggered by ACBD6, it is unlikely that the minor C14:0-CoA molecule could ever access the NMT2 binding site. Because ACBD6 can also bind C14-CoA, the stimulatory effect could also indicate the channeling of C14-CoA from the binding site of ACBD6 to NMT2 in the complex. In addition, the formation of the ACBD6/NMT2 complex could also alter the binding affinity of NMT2 toward C16-CoA and render its binding to NMT2 less favorable. A particular role should be considered for the serine 106 residue of ACBD6. This site is phosphorylated in vivo (42, 43) and, as shown in Fig. 7C, this residue is essential in controlling access of C16-CoA to NMT2. The binding of C16-CoA to the ACBD motif of phosphoACBD6 might trigger a substantial conformational change in the complex leading to blocking the entry or channeling of C16-CoA to the NMT2 binding site. In vivo, ACBD6 and NMT2 are exposed to a variety of acyl-CoAs at different (local) concentrations (49, 52). These acyl-CoAs that are bound to the ACBD6/NMT2 complex may further stimulate the NMT reaction toward C14:0-CoA (Fig. 8C). In support of this model, we established that activity of NMT2 toward C14:0-CoA was stimulated by binding of C18:2-CoA to ACBD6 (Fig. 5). The role of serine 106 is highlighted, as the ACBD6 serine mutant prevented C16-CoA access to NMT2 independently of C18:2-CoA (Fig. 7). Together, this suggests that even under condition of excess of C16-CoA, ACBD6 would protect the specificity of the NMT reaction as long as the formation of the complex with NMT2 or phosphorylation of ACBD6 was not prevented.
A variety of pathogens rely on protein acylation processes. Because binding of the peptides does not affect acyl-CoA binding to the N-terminal motif (33, 53, 54), compounds that can recognize the C-terminal binding domain of the NMT proteins of invading pathogens seem to be good candidates to specifically disrupt myristoylation without impacting acylation of the proteins of the host (28–32, 34–36). As examples, the development of Plasmodium falciparum in erythroid cells is blocked by several chemicals inhibiting the P. falciparum NMT enzyme (35–37, 55). The inhibition of myristoylation of the HIV Gag protein prevents its membrane association and blocks viral budding (56). Interestingly, both P. falciparum and Plasmodium vivax have an ACBD6 homolog which is expressed during development (57, 58). As defined in Chlamydia-infected human cells (22), the host ACBD6 and the parasite ACBD6 homolog could cross the vacuole membrane in association with lipid droplets. Disruption of such a process may offer another strategy to affect the activity of the parasite NMT and its association to ACBD6 (59–61).
The NMT process in vivo is more complex than explored in this study. For example, the generation of the acyl-donor and acyl-acceptor of the reaction are localized processes that require movement of the fatty acyl molecules through membranes and binding to carriers such as FABP, their activation to acyl-CoAs by acyl-CoA synthetases, and their transport by ACBD proteins to the NMT enzyme. ACBD proteins are involved in a variety of cellular mechanisms through association with a multiplicity of proteins. However, to our knowledge, the role of the binding of the acyl-CoA ligand to the ACBD function has not been established in these processes. The modulation of the NMT reaction by ligand binding to ACBD6 in the enzymatic complex formed with NMT2 provides one of the first examples of a physiological role for the acyl-CoA binding property of a member of the human ACBD family.
Supplementary Material
Footnotes
Abbreviations:
- ACBD
- acyl-CoA binding domain
- C14-CoA
- myristoyl-CoA
- C16-CoA
- palmitoyl-CoA
- DSS
- disuccinimidyl suberate
- DTNB
- 5,5′-dithio-bis-(2-nitrobenzoic acid) or Ellman’s reagent
- FDR
- false discovery rate
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- ITC
- isothermal titration calorimetry
- NMT
- N-myristoyltransferase
MS for this work was provided through the University of California at San Francisco Mass Spectrometry Facility (A.L. Burlingame, director), supported by National Institutes of Health Grant 8P41GM103481. J.L.D. is supported by the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Knudsen J., Mandrup S., Rasmussen J. T., Andreasen P. H., Poulsen F., and Kristiansen K.. 1993. The function of acyl-CoA-binding protein (ACBP)/diazepam binding inhibitor (DBI). Mol. Cell. Biochem. 123: 129–138. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y., Atkins J. B., Rompani S. B., Bancescu D. L., Petersen P. H., Tang H., Zou K., Stewart S. B., and Zhong W.. 2007. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 129: 163–178. [DOI] [PubMed] [Google Scholar]
- 3.Okazaki Y., Ma Y., Yeh M., Yin H., Li Z., Yeh K. Y., and Glass J.. 2012. DMT1 (IRE) expression in intestinal and erythroid cells is regulated by peripheral benzodiazepine receptor-associated protein 7. Am. J. Physiol. Gastrointest. Liver Physiol. 302: G1180–G1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinoda Y., Fujita K., Saito S., Matsui H., Kanto Y., Nagaura Y., Fukunaga K., Tamura S., and Kobayashi T.. 2012. Acyl-CoA binding domain containing 3 (ACBD3) recruits the protein phosphatase PPM1L to ER-Golgi membrane contact sites. FEBS Lett. 586: 3024–3029. [DOI] [PubMed] [Google Scholar]
- 5.Sbodio J. I., Paul B. D., Machamer C. E., and Snyder S. H.. 2013. Golgi protein ACBD3 mediates neurotoxicity associated with Huntington’s disease. Cell Reports. 4: 890–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrescu A. D., Payne H. R., Boedecker A., Chao H., Hertz R., Bar-Tana J., Schroeder F., and Kier A. B.. 2003. Physical and functional interaction of acyl-CoA-binding protein with hepatocyte nuclear factor-4 alpha. J. Biol. Chem. 278: 51813–51824. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y., Patel V., Bang S., Cohen N., Millar J., and Kim S. F.. 2012. Maturation and activity of sterol regulatory element binding protein 1 is inhibited by acyl-CoA binding domain containing 3. PLoS One. 7: e49906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H. Y., and Chye M. L.. 2004. Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol. Biol. 54: 233–243. [DOI] [PubMed] [Google Scholar]
- 9.Dorobantu C. M., Ford-Siltz L. A., Sittig S. P., Lanke K. H., Belov G. A., van Kuppeveld F. J., and van der Schaar H. M.. 2015. GBF1- and ACBD3-independent recruitment of PI4KIIIbeta to replication sites by rhinovirus 3A proteins. J. Virol. 89: 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan J., Liu J., Culty M., and Papadopoulos V.. 2010. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): an emerging signaling molecule. Prog. Lipid Res. 49: 218–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greninger A. L., Knudsen G. M., Betegon M., Burlingame A. L., and DeRisi J. L.. 2013. ACBD3 interaction with TBC1 domain 22 protein is differentially affected by enteroviral and kobuviral 3A protein binding. MBio. 4: e00098–e00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong Z., Yang X., Yang G., and Zhang L.. 2014. Hepatitis C virus NS5A competes with PI4KB for binding to ACBD3 in a genotype-dependent manner. Antiviral Res. 107: 50–55. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa-Sasaki K., Sasaki J., and Taniguchi K.. 2014. A complex comprising phosphatidylinositol 4-kinase IIIbeta, ACBD3, and Aichi virus proteins enhances phosphatidylinositol 4-phosphate synthesis and is critical for formation of the viral replication complex. J. Virol. 88: 6586–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki J., Ishikawa K., Arita M., and Taniguchi K.. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 31: 754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao H., Zhou M., McIntosh A., Schroeder F., and Kier A. B.. 2003. ACBP and cholesterol differentially alter fatty acyl CoA utilization by microsomal ACAT. J. Lipid Res. 44: 72–83. [DOI] [PubMed] [Google Scholar]
- 16.Xiao S., and Chye M. L.. 2009. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol. Biochem. 47: 479–484. [DOI] [PubMed] [Google Scholar]
- 17.Xiao S., and Chye M. L.. 2011. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog. Lipid Res. 50: 141–151. [DOI] [PubMed] [Google Scholar]
- 18.Soupene E., Serikov V., and Kuypers F. A.. 2008. Characterization of an acyl-coenzyme A binding protein predominantly expressed in human primitive progenitor cells. J. Lipid Res. 49: 1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soupene E., and Kuypers F. A.. 2015. Ligand binding to the ACBD6 protein regulates the acyl-CoA transferase reactions in membranes. J. Lipid Res. 56: 1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y. C., Clark T. A., Gee S. L., Kang J. A., Schweitzer A. C., Wickrema A., and Conboy J. G.. 2009. The transcriptome of human CD34+ hematopoietic stem-progenitor cells. Proc. Natl. Acad. Sci. USA. 106: 8278–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto M. L., Clark T. A., Gee S. L., Kang J. A., Schweitzer A. C., Wickrema A., and Conboy J. G.. 2009. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood. 113: 3363–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soupene E., Wang D., and Kuypers F. A.. Remodeling of host phosphatidylcholine by Chlamydia acyltransferase is regulated by acyl-CoA binding protein ACBD6 associated with lipid droplets. MicrobiologyOpen.Epub ahead of print. January 21, 2015; doi:10.1002/mbo3.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritzler J. M., and Zhu G.. 2012. Novel anti-Cryptosporidium activity of known drugs identified by high-throughput screening against parasite fatty acyl-CoA binding protein (ACBP). J. Antimicrob. Chemother. 67: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patwardhan P., and Resh M. D.. 2010. Myristoylation and membrane binding regulate c-Src stability and kinase activity. Mol. Cell. Biol. 30: 4094–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Resh M. D. 2013. Covalent lipid modifications of proteins. Curr. Biol. 23: R431–R435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright M. H., Heal W. P., Mann D. J., and Tate E. W.. 2010. Protein myristoylation in health and disease. J. Chem. Biol. 3: 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resh M. D. 2006. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2: 584–590. [DOI] [PubMed] [Google Scholar]
- 28.Bhatnagar R. S., Futterer K., Waksman G., and Gordon J. I.. 1999. The structure of myristoyl-CoA:protein N-myristoyltransferase. Biochim. Biophys. Acta. 1441: 162–172. [DOI] [PubMed] [Google Scholar]
- 29.Bhatnagar R. S., Schall O. F., Jackson-Machelski E., Sikorski J. A., Devadas B., Gokel G. W., and Gordon J. I.. 1997. Titration calorimetric analysis of AcylCoA recognition by myristoylCoA:protein N-myristoyltransferase. Biochemistry. 36: 6700–6708. [DOI] [PubMed] [Google Scholar]
- 30.Brannigan J. A., Smith B. A., Yu Z., Brzozowski A. M., Hodgkinson M. R., Maroof A., Price H. P., Meier F., Leatherbarrow R. J., Tate E. W., et al. 2010. N-myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J. Mol. Biol. 396: 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desmeules P., Penney S. E., Desbat B., and Salesse C.. 2007. Determination of the contribution of the myristoyl group and hydrophobic amino acids of recoverin on its dynamics of binding to lipid monolayers. Biophys. J. 93: 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goncalves V., Brannigan J. A., Whalley D., Ansell K. H., Saxty B., Holder A. A., Wilkinson A. J., Tate E. W., and Leatherbarrow R. J.. 2012. Discovery of Plasmodium vivax N-myristoyltransferase inhibitors: screening, synthesis, and structural characterization of their binding mode. J. Med. Chem. 55: 3578–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuckeroth R. O., Glaser L., and Gordon J. I.. 1988. Heteroatom-substituted fatty acid analogs as substrates for N-myristoyltransferase: an approach for studying both the enzymology and function of protein acylation. Proc. Natl. Acad. Sci. USA. 85: 8795–8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards E. W., Hamm M. W., and Otto D. A.. 1991. The effect of palmitoyl-CoA binding to albumin on the apparent kinetic behavior of carnitine palmitoyltransferase I. Biochim. Biophys. Acta. 1076: 23–28. [DOI] [PubMed] [Google Scholar]
- 35.Tate E. W., Bell A. S., Rackham M. D., and Wright M. H.. 2014. N-myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology. 141: 37–49. [DOI] [PubMed] [Google Scholar]
- 36.Wright M. H., Clough B., Rackham M. D., Rangachari K., Brannigan J. A., Grainger M., Moss D. K., Bottrill A. R., Heal W. P., Broncel M., et al. 2014. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat. Chem. 6: 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goncalves V., Brannigan J. A., Thinon E., Olaleye T. O., Serwa R., Lanzarone S., Wilkinson A. J., Tate E. W., and Leatherbarrow R. J.. 2012. A fluorescence-based assay for N-myristoyltransferase activity. Anal. Biochem. 421: 342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S., and Sharma R. K.. 2014. An improved method and cost effective strategy for soluble expression and purification of human N-myristoyltransferase 1 in E. coli. Mol. Cell. Biochem. 392: 175–186. [DOI] [PubMed] [Google Scholar]
- 39.Greninger A. L., Knudsen G. M., Betegon M., Burlingame A. L., and Derisi J. L.. 2012. The 3A protein from multiple picornaviruses utilizes the Golgi adaptor protein ACBD3 to recruit PI4KIIIbeta. J. Virol. 86: 3605–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., and Burlingame A. L.. 2008. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics. 7: 2386–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elias J. E., and Gygi S. P.. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 4: 207–214. [DOI] [PubMed] [Google Scholar]
- 42.Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P.. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 105: 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., and Nigg E. A., et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3: ra3. [DOI] [PubMed] [Google Scholar]
- 44.Rudnick D. A., Lu T., Jackson-Machelski E., Hernandez J. C., Li Q., Gokel G. W., and Gordon J. I.. 1992. Analogs of palmitoyl-CoA that are substrates for myristoyl-CoA:protein N-myristoyltransferase. Proc. Natl. Acad. Sci. USA. 89: 10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., and Gordon J. I.. 1987. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc. Natl. Acad. Sci. USA. 84: 2708–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., and Gordon J. I.. 1988. Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263: 1784–1790. [PubMed] [Google Scholar]
- 47.Towler D. A., Eubanks S. R., Towery D. S., Adams S. P., and Glaser L.. 1987. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J. Biol. Chem. 262: 1030–1036. [PubMed] [Google Scholar]
- 48.Rudnick D. A., McWherter C. A., Rocque W. J., Lennon P. J., Getman D. P., and Gordon J. I.. 1991. Kinetic and structural evidence for a sequential ordered Bi Bi mechanism of catalysis by Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J. Biol. Chem. 266: 9732–9739. [PubMed] [Google Scholar]
- 49.DeMar J. C. Jr., and Anderson R. E.. 1997. Identification and quantitation of the fatty acids composing the CoA ester pool of bovine retina, heart, and liver. J. Biol. Chem. 272: 31362–31368. [DOI] [PubMed] [Google Scholar]
- 50.Dizhoor A. M., Ericsson L. H., Johnson R. S., Kumar S., Olshevskaya E., Zozulya S., Neubert T. A., Stryer L., Hurley J. B., and Walsh K. A.. 1992. The NH2 terminus of retinal recoverin is acylated by a small family of fatty acids. J. Biol. Chem. 267: 16033–16036. [PubMed] [Google Scholar]
- 51.Kokame K., Fukada Y., Yoshizawa T., Takao T., and Shimonishi Y.. 1992. Lipid modification at the N terminus of photoreceptor G-protein alpha-subunit. Nature. 359: 749–752. [DOI] [PubMed] [Google Scholar]
- 52.Knudsen J., Neergaard T. B., Gaigg B., Jensen M. V., and Hansen J. K.. 2000. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. J. Nutr. 130(2S Suppl): 294S–298S. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S., and Sharma R. K.. 2015. N-terminal region of the catalytic domain of human N-myristoyltransferase 1 acts as an inhibitory module. PLoS One. 10: e0127661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J., Tao Y., Zhang M., Howard M. H., Gutteridge S., and Ding J.. 2007. Crystal structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoyl-CoA and inhibitors reveal the functional roles of the N-terminal region. J. Biol. Chem. 282: 22185–22194. [DOI] [PubMed] [Google Scholar]
- 55.Rackham M. D., Brannigan J. A., Moss D. K., Yu Z., Wilkinson A. J., Holder A. A., Tate E. W., and Leatherbarrow R. J.. 2013. Discovery of novel and ligand-efficient inhibitors of Plasmodium falciparum and Plasmodium vivax N-myristoyltransferase. J. Med. Chem. 56: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindwasser O. W., and Resh M. D.. 2002. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl. Acad. Sci. USA. 99: 13037–13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bozdech Z., Mok S., Hu G., Imwong M., Jaidee A., Russell B., Ginsburg H., Nosten F., Day N. P., White N. J., et al. 2008. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc. Natl. Acad. Sci. USA. 105: 16290–16295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roobsoong W., Roytrakul S., Sattabongkot J., Li J., Udomsangpetch R., and Cui L.. 2011. Determination of the Plasmodium vivax schizont stage proteome. J. Proteomics. 74: 1701–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackson K. E., Klonis N., Ferguson D. J., Adisa A., Dogovski C., and Tilley L.. 2004. Food vacuole-associated lipid bodies and heterogeneous lipid environments in the malaria parasite, Plasmodium falciparum. Mol. Microbiol. 54: 109–122. [DOI] [PubMed] [Google Scholar]
- 60.Palacpac N. M., Hiramine Y., Mi-ichi F., Torii M., Kita K., Hiramatsu R., Horii T., and Mitamura T.. 2004. Developmental-stage-specific triacylglycerol biosynthesis, degradation and trafficking as lipid bodies in Plasmodium falciparum-infected erythrocytes. J. Cell Sci. 117: 1469–1480. [DOI] [PubMed] [Google Scholar]
- 61.Vielemeyer O., McIntosh M. T., Joiner K. A., and Coppens I.. 2004. Neutral lipid synthesis and storage in the intraerythrocytic stages of Plasmodium falciparum. Mol. Biochem. Parasitol. 135: 197–209. [DOI] [PubMed] [Google Scholar]
- 62.Soupene E., Rothschild J., Kuypers F. A., and Dean D.. 2012. Eukaryotic protein recruitment into the Chlamydia inclusion: implications for survival and growth. PLoS One. 7: e36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.