Abstract
Genetic studies of plasma TG levels have identified associations with multiple candidate loci on chromosome11q23.3, which harbors a number of genes, including BUD13, ZNF259, and APOA5-A4-C3-A1. This study aimed to examine whether these multiple candidate genes on the 11q23.3 regions exert independent effects on TG levels or whether their effects are confounded by linkage disequilibrium (LD). We performed a genome-wide association study and consequent fine-mapping analyses on TG levels in two Korean population-based cohorts: the Korea Association Resource study (n = 8,223) and the Healthy Twin study (n = 1,735). A total of 301 loci reached genome-wide significance level in pooled analysis, including 10 SNPs with weak LD (r2 < 0.06) clustered on 11q23.3: ApoA5 (rs651821, rs2075291); ZNF259 (rs964184, rs603446); BUD13 (rs11216126); Apoa4 (rs7396851); SIK3 (rs12292858); PCSK7 (rs199890178); PAFAH1B2 (rs12420127), and SIDT2 (rs2269399). When the inter-dependence between alleles was examined using conditional models, five loci on BUD13, ZNF259, and ApoA5 showed possible independent associations. A haplotype analysis using five SNPs revealed both hyper- and hypotriglyceridemic haplotypes, which are relatively common in Koreans (haplotype frequency 0.08–0.22). Our findings suggest the presence of multiple functional loci on 11q23.3, which might exert their effects on plasma TG level independently or through complex interactions between functional loci.
Keywords: genetic variant, genome-wide association study, polymorphisms, genetic epidemiology
Nonfasting TG levels show good correlation with remnant cholesterol levels, which is an emerging risk factor of ischemic vascular diseases (1). In addition, hypertriglyceridemia promotes insulin resistance (2). Plasma TG is a complex polygenic trait influenced by both genetic and environmental factors (3). Still, the genetic variation expected by the heritability estimates for plasma TG remains to be completely delineated (4, 5). Knowledge of the genetic variants regulating plasma TG has increased dramatically through genome-wide association studies (GWASs). GWASs have identified a number of “TG genes,” such as ApoA5 and LPL (6–8). As of November 2015, a total of 41 loci have been associated with the TG levels through GWASs in Europeans (8, 9). Still, much of the genetic variation expected by heritability of plasma TG remains unexplained, suggesting that more susceptibility loci may yet be identified (8–15).
GWAS findings indicated that the region of chromosome 11q23.3 that harbors BUD13-ZNF259-APOA5-A4-C3-A1 plays an important role in TG metabolism (8, 16–18). However, the reported candidate genetic variants in the 11q23.3 region vary by studies, and it is not clearly understood whether the discrepancy arises from differences in linkage disequilibrium (LD) structure tagging different causal alleles across populations or differences in study settings, such as genotype platforms. The purpose of this study is to further dissect this 11q23.3 region in Koreans, because previous studies have indicated multiple independent susceptibility loci of hypertriglyceridemia (13, 14), and to examine the possibility of the existence of more than one causal variant in this region.
MATERIALS AND METHODS
Subjects
Two population cohorts of Koreans with genome-wide genetic marker information were involved in this study; the Korea Association Resource study (KARE), consisting of 8,842 individuals, and the Healthy Twin study (HT) with 3,079 individuals (19, 20). The KARE served as discovery data for associated loci and validation was performed using the HT data.
The KARE is a prospective cohort study started in 2001 as a part of the Korean Genome Epidemiology Study in the rural Ansung and suburban Ansan areas, which recruited 10,038 healthy participants aged 40–69 years at baseline. Both base areas are located in Gyeonggi Province, close to Seoul, the capital of the Republic of Korea. Information on the health status and health-related behaviors of the participants was collected through a standardized questionnaire. Blood samples were drawn from an antecubital vein after more than 8 h of fasting. Serum TG level was measured using standard enzymatic method in a centralized laboratory. Details of the KARE are described in previous reports (13, 15). The genome-wide genetic marker information was available for a total of 8,842 individuals. After excluding individuals who had a diagnosis of diabetes or were using lipid-lowering agents (n = 619), 8,223 individuals (3,858 males and 4,365 females, with 82 related participants) were included for analysis.
The HT of Korea is also a component of the Korean Genome Epidemiology Study, a cohort of adult same-sex twin pairs older than age 30 and their first-degree family members, who have been recruited since 2005. Most protocols are shared between the KARE and the HT, and more detailed protocols are available in previous literature (20). A total of 3,079 study participants (n = 1,217 men, n = 1,862 women, from 17–81 years of age) were enrolled since 2005; 531 monozygotic twin pairs, 120 dizygotic twin pairs, and 1,777 non-twin family members in 661 families. The genotype data were available for a total of 1,857 individuals. Of these 1,857 individuals, those who reported using lipid-lowering drugs or had a diagnosis of diabetes (n = 122) were excluded, resulting in 1,735 individuals (672 males and 1,063 females) ranging in age from 17 to 81 years (mean = 44.1 years; SD = 13 years). Institutional Review Boards at Seoul National University, Seoul Samsung Hospital, and Busan Inje Baik Hospital approved the conduct of the cohort studies used in this study.
Genotyping, quality control, and imputation
Both studies performed genome-wide dense SNP marker analysis; the KARE using Affymetrix Genome-Wide Human SNP Array GeneChip version 5.0 and the HT using the Affymetrix Genome-Wide Human SNP Array GeneChip version 6.0. For the KARE, markers violating the Hardy-Weinberg equilibrium (P < 10E-06, 38,364 markers), genotype call rates below 95% (17,926 markers), and minor allele frequency (MAF) < 0.01 (92,050 markers) were deleted, leaving 352,228 markers for the subsequent analysis. For the HT, markers with Hardy-Weinberg equilibrium <0.001, MAF <0.01, and genotype missing rate >0.05 were excluded (n = 292,653); using family relationship, markers violating Mendelian consistency in more than three families (n = 11,456) or multimarker errors resembling close double-recombination were further deleted (n = 47,594); leaving 537,159 markers.
We used the segmented haplotype estimation and imputation tool, SHAPEIT2, which scales linearly with the number of haplotypes to efficiently construct haplotypes (21) for determining phase, and then conducted imputation using IMPUTE2 (22). The 1000 Genomes Project’s haplotypes phase I in NCBI build 37 (hg19) of Asian ancestry were used as a reference panel, and the markers with imputation quality score greater than 0.9 were used for fine-mapping studies. The imputation process resulted in 4,166,520 and 4,174,873 markers in discovery (KARE) and replication (HT) samples.
Statistical analysis
The plasma TG level was adjusted for age, sex, and BMI using a linear regression model for association tests. For identical twins, we chose one twin randomly. For familial relationship, we corrected genetic correlation arising from polygenic background by decorrelating kinship coefficients calculated from the genetic markers. The genome-wide significant level for adjusting the type I error level was defined from original genotyped marker sets (352,228) to be P = 1.41E-07 after Bonferroni correction. All locations on a physical map are referred to build 37 of the human genome reference map ( hg19).
Dissecting the 11q23.3 region
Within the 11q23.3 region, we focused on chromosome 11:116,600,001-117,100,000 where previous GWAS findings were localized (16, 17, 23, 24). In order to identify the independent effects of candidate loci on TG, we used the following methods. First, we selected markers which showed genome-wide significance level in the discovery set and tested whether they were replicated in validation set. Second, for those selected markers, we examined LD (r2 < 0.06) and chose one top hit in each separate LD block (non-LD markers). Third, we performed a series of conditional analyses by adding one non-LD marker into the linear regression model as a covariate and tested all remaining non-LD SNPs for the association with TG level. If a locus showed persistent association with TG with adjustment of other top hit SNPs, we considered it as being a reasonable candidate for independent susceptibility loci regulating TG level. Finally, we conducted a haplotype analysis to further dissect the 11q23.3 region. For the haplotype analysis, we used HAPLOVIEW (version 4.0), which uses an accelerated expectation maximization algorithm to calculate haplotype frequencies (http://www.broad.mit.edu/mpg/haploview/). The effect of each haplotype on TG level was determined using the haplo.stats package (version 1.4.4) operated in the R language (version 2.14, available at http://www.r-project.org), which implements the expectation–maximization algorithm. In addition, the multiplicative interactions between two SNPs were tested by including both SNPs (assuming a general model) and an interaction term (product of two main effects) in a logistic regression model in PLINK (25).
RESULTS
The overall study design is provided in supplementary Fig. 1. The characteristics of the study participants are given in Table 1. The KARE participants were older and had higher BMI than those of the HT. The mean plasma TG level was higher in the KARE population (1.80 mmol/l) than in the HT participants (1.30 mmol/l).
TABLE 1.
Baseline characteristics of the discovery and replication studies
| Characteristics | Discovery (n = 8,223) | Replication (n = 1,735) |
| Age (years) | 51.86 ± 8.8 | 44.1 ± 13.0 |
| BMI (kg/m2) | 24.54 ± 3.12 | 23.68 ± 3.28 |
| TG (mmol/l) | 1.80 ± 1.15 | 1.30 ± 0.89 |
| HDL-C (mmol/l) | 0.50 ± 0.10 | 0.57 ± 0.14 |
| LDL-C (mmol/l) | 1.31 ± 0.37 | 1.25 ± 0.35 |
| TC (mmol/l) | 2.24 ± 0.41 | 2.14 ± 0.40 |
| Glucose (mmol/l) | 0.99 ± 0.25 | 1.05 ± 0.21 |
| Gender, Male, n (%) | 3,858 (46.9) | 672 (38.7) |
| Cigarette smoking | ||
| Never | 4,818 (59.4) | 1,219 (71.3) |
| Ex-smoker | 1,224 (15.1) | 180 (10.5) |
| Current smoker | 2,075 (25.6) | 310 (18.1) |
| Diabetes | ||
| Yes | 613 (6.93) | 87 (4.68) |
| No | 8,229 (93.1) | 1,770 (95.3) |
| Lipid-altering medication | ||
| Yes | 40 (0.45) | 40 (2.15) |
| No | 8,802 (99.5) | 1,817 (97.8) |
The lipid values were calculated after excluding individuals who had a diagnosis of diabetes or were using lipid-lowering agents. Values shown are numbers (frequencies) for categorical variables and mean ± SD for continuous variables. HDL-C, HDL cholesterol; LDL-C, LDL cholesterol; TC, total cholesterol.
The enrichment of markers for the 11q23.3 region (114,600,001–121,300,000 bp as GRCh38) by imputation increased the number of SNPs or single nucleotide variants from 1,103 to 10,690 (supplementary Table 1). When we performed fine-mapping analysis for this region using enriched markers, a total of 477 SNPs/single nucleotide variants reached genome-wide significance level (as P < 1.41 × 10−7) at discovery stage (supplementary Table 2), and among them, 301 markers were replicated (P < 0.05). When we conducted LD-clumping, 10 SNPs remained (r2 < 0.06) and were used for conditional analysis: rs11216126 (BUD13), rs964184 (ZNF259), rs603446 (ZNF259), rs2075291 (ApoA5), rs651821 (ApoA5), rs7396851 (Apoa4), rs12292858 (SIK3), rs12420127 (PAFAH1B2), rs2269399 (SIDT2), and rs199890178 (PCSK7) (supplementary Fig. 2).
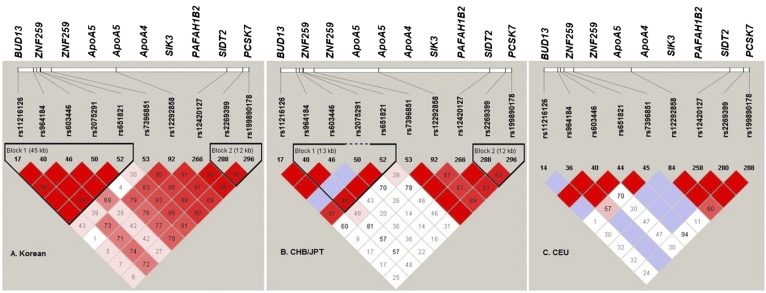
The LD block pattern between 301 markers on the 11q23.3 region showed six separate blocks in the Korean population (supplementary Fig. 3). The SNPs, rs11216126/BUD13, rs964184/ZNF259, rs603446/ZNF259, rs2075291/ApoA5, and rs651821/ApoA5, are located in block 1 (Fig. 1A). The LD profiles of the 11q23.3 region in Koreans were similar to those of other Asians (CHB/JPT), but Europeans (CEU) or Africans (YRI) showed much weaker and ill-defined LD blocks. Physical position and relation with LD structure of each SNP is shown in supplementary Fig. 3.
Fig. 1.
Haploview LD graph of the studied polymorphisms on 11q23.3 in Korean, 1000 Genome Pilot I CHB/JPT, and CEU populations. A standard color scheme is used to display LD with bright red color for high LD, white color for no LD, pink red, and blue for intermediate LD.
When we performed a two-way conditional analysis for 10 SNPs, markers on the proximal 11q23.3 region (BUD13, ZNF259, and ApoA5) did not materially affect or was affected their genome-wide significance levels by mutual adjustment; while the markers in the distal region (SIK3, PCSK7, PAFAH1B2, and SIDT2) showed a decrease in their significance levels (Table 2). The distal part loci genes showed a marked decrease in their signals below regional significance level (301 markers by Bonferroni correction; P = 0.00016) on mutual adjustment. Interestingly, the adjustment for the effect of rs2075291/ApoA5 resulted in stronger signals for rs964184/ZNF259 (from P = 1.3E-29 in its single marker models to P = 9.6E-40 in conditional analysis). Similarly, when adjusted for the effect of rs603446/ZNF259, the statistical significance of rs11216126/BUD13 became stronger (from 8.9E-15 to 4.6E-32).
TABLE 2.
Fixed effect models identified by step-wise linear regression analysis in the discovery and replication studies
| Additional Adjustment for SNP Markers (Two SNPs in the Model) | ||||||||||||||||||||||
| Single SNP | BUD13/rs11216126 | ZNF259/rs964184 | ZNF259/rs603446 | ApoA5/rs2075291 | ApoA5/rs651821 | Apoa4/rs7396851 | SIK3/rs12292858 | PAFAH1B2/rs12420127 | SIDT2/rs2269399 | PCSK7/rs199890178 | ||||||||||||
| Conditioned Markers | β | P | β | P | β | P | β | P | β | P | β | P | β | P | β | P | β | P | β | P | β | P |
| KARE (discovery) | ||||||||||||||||||||||
| BUD13/ rs11216126 | −0.11 | 8.9E-15 | — | NA | −0.11 | 7.4E-07 | −0.26 | 4.6E-32 | −0.13 | 1.4E-09 | −0.06 | 1.2E-05 | −0.17 | 2.1E-15 | −0.18 | 1.3E-17 | −0.18 | 4.5E-17 | −0.18 | 2.2E-17 | −0.16 | 5.4E-12 |
| ZNF259/ rs964184 | +0.16 | 1.3E-29 | +0.20 | 5.2E-21 | — | NA | +0.20 | 6.3E-20 | +0.28 | 9.6E-40 | −0.19 | 1.3E-07 | +0.21 | 7.7E-22 | +0.22 | 2.1E-24 | +0.22 | 1.9E-25 | +0.22 | 2.4E-25 | +0.22 | 2.9E-23 |
| ZNF259/ rs603446 | −0.12 | 8.2E-17 | −0.25 | 4.1E-32 | −0.16 | 1.4E-07 | — | NA | −0.13 | 3.0E-10 | −0.06 | 2.6E-05 | −0.15 | 7.7E-14 | −0.16 | 1.2E-13 | −0.16 | 3.5E-13 | −0.15 | 3.3E-13 | −0.15 | 2.4E-12 |
| ApoA5/ rs2075291 | +0.27 | 2.4E-32 | +0.37 | 7.7E-28 | +0.47 | 1.7E-44 | +0.37 | 3.3E-27 | — | NA | +0.19 | 1.1E-07 | +0.40 | 3.5E-33 | +0.38 | 1.0E-29 | +0.38 | 8.6E-31 | +0.38 | 9.2E-31 | +0.40 | 5.2E-31 |
| ApoA5/ rs651821 | +0.22 | 1.5E-62 | +0.31 | 2.0E-50 | +0.47 | 1.8E-44 | +0.31 | 1.7E-49 | +0.27 | 6.7E-36 | — | NA | +0.32 | 7.2E-54 | +0.31 | 3.5E-54 | +0.32 | 1.2E-56 | +0.32 | 1.5E-56 | +0.32 | 2.5E-54 |
| Apoa4/ rs7396851 | +0.08 | 7.3E-11 | +0.10 | 1.2E-07 | +0.06 | 1.1E-03 | +0.09 | 7.3E-06 | +0.12 | 6.4E-11 | +0.03 | 3.4E-04 | — | NA | +0.09 | 2.4E-06 | +0.10 | 1.5E-07 | +0.10 | 1.7E-07 | +0.09 | 3.4E-06 |
| SIK3/ rs12292858 | −0.10 | 3.5E-11 | −0.15 | 4.4E-12 | −0.12 | 6.1E-07 | −0.12 | 3.1E-07 | −0.13 | 1.0E-07 | −0.09 | 1.3E-04 | −0.12 | 7.2E-08 | — | NA | −0.12 | 3.7E-04 | −0.12 | 6.6E-04 | −0.11 | 7.0E-03 |
| PAFAH1B2/ rs12420127 | −0.10 | 2.6E-09 | −0.16 | 1.5E-09 | −0.10 | 7.9E-05 | −0.10 | 5.6E-05 | −0.11 | 1.8E-05 | −0.07 | 2.4E-02 | −0.14 | 1.9E-07 | −0.06 | 0.141 | — | NA | −0.06 | 0.648 | −0.02 | 0.425 |
| SIDT2/ rs2269399 | −0.10 | 5.4E-09 | −0.15 | 3.2E-09 | −0.10 | 9.9E-05 | −0.10 | 8.2E-05 | −0.11 | 2.0E-05 | −0.05 | 2.9E-02 | −0.13 | 3.5E-07 | −0.06 | 0.134 | −0.11 | 0.415 | — | NA | −0.02 | 0.468 |
| PCSK7/ rs199890178 | −0.10 | 3.5E-10 | −0.13 | 1.6E-08 | −0.11 | 9.8E-06 | −0.11 | 1.3E-05 | −0.12 | 1.3E-06 | −0.07 | 1.1E-03 | −0.12 | 4.6E-06 | −0.06 | 4.1E-02 | −0.13 | 4.5E-03 | −0.13 | 4.9E-03 | — | NA |
| HT (validation) | ||||||||||||||||||||||
| BUD13/ rs11216126 | −0.16 | 5.0E-06 | — | NA | −0.08 | 2.0E-03 | −0.22 | 1.6E-10 | −0.08 | 1.2E-03 | −0.15 | 1.8E-05 | −0.17 | 1.0E-06 | −0.18 | 1.6E-07 | −0.18 | 2.7E-07 | −0.18 | 3.1E-07 | −0.08 | 3.5E-03 |
| ZNF259/ rs964184 | +0.06 | 4.1E-03 | +0.03 | 2.0E-02 | — | NA | +0.04 | 0.123 | +0.07 | 5.4E-03 | −0.03 | 2.1E-02 | +0.05 | 6.8E-03 | +0.04 | 7.0E-02 | +0.05 | 5.5E-02 | +0.04 | 4.1E-02 | +0.04 | 2.1E-02 |
| ZNF259/ rs603446 | −0.12 | 1.3E-03 | −0.18 | 3.6E-07 | −0.02 | 3.9E-03 | — | NA | −0.02 | 3.8E-03 | −0.07 | 2.9E-02 | −0.11 | 2.2E-03 | −0.11 | 2.7E-03 | −0.10 | 4.9E-03 | −0.12 | 1.8E-03 | −0.04 | 9.9E-02 |
| ApoA5/ rs2075291 | +0.38 | 8.5E-14 | +0.14 | 3.9E-04 | +0.11 | 2.1E-03 | +0.15 | 1.7E-04 | — | NA | +0.30 | 5.6E-11 | +0.13 | 7.2E-14 | +0.15 | 1.5E-04 | +0.15 | 1.0E-04 | +0.15 | 1.1E-04 | +0.13 | 1.1E-03 |
| ApoA5/ rs651821 | +0.19 | 4.7E-10 | +0.18 | 3.4E-10 | +0.27 | 2.8E-14 | +0.19 | 1.1E-10 | +0.18 | 2.4E-09 | — | NA | +0.18 | 4.8E-10 | +0.19 | 2.3E-11 | +0.20 | 1.1E-11 | +0.20 | 1.0E-11 | +0.20 | 6.4E-12 |
| Apoa4/ rs7396851 | +0.06 | 4.2E-02 | +0.06 | 3.4E-02 | +0.07 | 6.9E-02 | +0.07 | 2.6E-02 | +0.08 | 3.4E-02 | +0.04 | 1.2E-02 | — | NA | +0.07 | 1.4E-02 | +0.08 | 1.0E-02 | +0.08 | 1.0E-02 | +0.08 | 5.3E-02 |
| SIK3/ rs12292858 | −0.11 | 3.7E-03 | −0.10 | 6.2E-03 | −0.08 | 5.8E-03 | −0.08 | 4.6E-02 | −0.08 | 7.5E-03 | −0.04 | 2.4E-02 | −0.08 | 3.7E-02 | — | NA | −011 | 5.3E-02 | −0.11 | 5.5E-02 | −0.11 | 5.3E-02 |
| PAFAH1B2/ rs12420127 | −0.09 | 3.4E-02 | −0.07 | 0.102 | −0.06 | 4.7E-02 | −0.04 | 0.337 | −0.07 | 3.2E-02 | −0.05 | 0.261 | −0.07 | 0.139 | −0.01 | 0.331 | — | NA | −0.03 | 0.916 | −0.01 | 0.678 |
| SIDT2/ rs2269399 | −0.10 | 8.9E-02 | −0.07 | 0.116 | −0.05 | 3.4E-02 | −0.05 | 0.321 | −0.05 | 2.5E-02 | −0.05 | 0.216 | −0.06 | 0.138 | −0.01 | 0.984 | −0.07 | 0.803 | — | NA | −0.01 | 0.521 |
| PCSK7/ rs199890178 | −0.10 | 1.3E-02 | −0.05 | 6.7E-02 | −0.05 | 7.7E-02 | −0.05 | 9.7E-02 | −0.05 | 6.6E-02 | −0.04 | 3.6E-02 | −0.06 | 7.8E-02 | −0.05 | 0.264 | −0.07 | 0.191 | −0.06 | 0.253 | — | NA |
| Meta-Analysis | ||||||||||||||||||||||
| BUD13/ rs11216126 | −0.12 | 3.50E-19 | — | N/A | −0.10 | 7.1E-09 | −0.25 | 7.9E-41 | −0.12 | 7.1E-12 | −0.08 | 8.0E-09 | −0.17 | 2.2E-20 | −0.18 | 2.5E-23 | −0.18 | 1.4E-22 | −0.18 | 7.2E-23 | −0.15 | 7.2E-14 |
| ZNF259/ rs964184* | 0.14 | 1.92E-30 | 0.17 | 1.8E-21 | — | N/A | 0.17 | 3.6E-19 | 0.24 | 1.7E-39 | −0.16 | 8.4E-09 | 0.18 | 6.4E-23 | 0.19 | 1.2E-23 | 0.19 | 9.2E-25 | 0.19 | 6.6E-25 | 0.19 | 1.6E-23 |
| ZNF259/ rs603446 | −0.12 | 5.08E-19 | −0.24 | 9.4E-38 | −0.14 | 2.1E-09 | — | N/A | −0.11 | 4.2E-12 | −0.06 | 2.2E-06 | −0.14 | 7.0E-16 | −0.15 | 1.3E-15 | −0.15 | 7.0E-15 | −0.14 | 2.4E-15 | −0.13 | 1.7E-12 |
| ApoA5/ rs2075291* | 0.29 | 9.00E-44 | 0.33 | 3.4E-30 | 0.41 | 1.6E-44 | 0.33 | 4.9E-30 | — | N/A | 0.21 | 4.0E-14 | 0.35 | 1.0E-44 | 0.34 | 1.6E-32 | 0.34 | 9.6E-34 | 0.34 | 1.2E-33 | 0.35 | 1.4E-32 |
| ApoA5rs651821* | 0.21 | 1.25E-70 | 0.29 | 5.8E-59 | 0.44 | 7.7E-57 | 0.29 | 1.5E-58 | 0.25 | 1.1E-43 | — | N/A | 0.30 | 3.5E-62 | 0.29 | 6.8E-64 | 0.30 | 1.2E-66 | 0.30 | 1.3E-66 | 0.30 | 1.3E-64 |
| Apoa4/ rs7396851 | 0.08 | 1.30E-11 | 0.09 | 1.2E-08 | 0.06 | 2.0E-04 | 0.09 | 5.6E-07 | 0.11 | 8.9E-12 | 0.03 | 1.7E-05 | — | N/A | 0.09 | 1.1E-07 | 0.10 | 5.0E-09 | 0.10 | 5.6E-09 | 0.09 | 4.9E-07 |
| SIK3/ rs12292858 | −0.10 | 4.80E-13 | −0.14 | 1.1E-13 | −0.11 | 1.3E-08 | −0.11 | 4.2E-08 | −0.12 | 2.6E-09 | −0.08 | 9.9E-06 | −0.11 | 8.2E-09 | — | N/A | −2.02 | 5.3E-05 | −0.12 | 9.8E-05 | −0.11 | 1.1E-03 |
| PAFAH1B2/rs12420127 | −0.10 | 3.05E-10 | −0.14 | 6.6E-10 | −0.09 | 1.0E-05 | −0.09 | 4.9E-05 | −0.10 | 1.7E-06 | −0.07 | 1.2E-02 | −0.13 | 8.7E-08 | −0.05 | 8.1E-02 | — | N/A | −0.05 | 6.5E-01 | −0.02 | 3.7E-01 |
| SIDT2/ rs2269399 | −0.10 | 1.84E-09 | −0.14 | 1.6E-09 | −0.09 | 9.8E-06 | −0.09 | 6.5E-05 | −0.10 | 1.5E-06 | −0.05 | 1.2E-02 | −0.12 | 1.5E-07 | −0.05 | 1.7E-01 | −0.10 | 4.0E-01 | — | N/A | −0.02 | 3.5E-01 |
| PCSK7/ rs199890178* | −0.10 | 1.60E-11 | −0.12 | 3.7E-09 | −0.10 | 2.0E-06 | −0.10 | 3.2E-06 | −0.11 | 2.4E-07 | −0.06 | 1.2E-04 | −0.11 | 9.6E-07 | −0.06 | 2.0E-02 | −0.12 | 1.8E-03 | −0.12 | 2.4E-03 | — | N/A |
P (significance) and β (regression coefficient) for linear regression. Single SNP models are adjusted for age, sex, and BMI; in other models, one additional SNP on each gene was considered simultaneously. Information for the SNP ID (rs) and chromosomal position is based on NCBI genome build 37 (hg19).
*Imputed SNP. P-values in bold face achieve genome-wide significance.
We targeted the proximal part of the 11q23.3 for haplotype analysis, which showed possible independent associations and paradoxical behaviors on conditional analysis. Five SNPs (rs11216126/BUD13, rs964184/ZNF259, rs603446/ZNF259, rs651821/ApoA5, and rs2075291/ApoA5) were chosen. As shown in Table 3, five haplotypes were constructed: the ACCCT haplotype as reference; two haplotypes, ACCAC (β = 0.17, P = 1.1E-05) and ACGCC (β = 0.10, P = 1.4E-05), showed significantly raised TG levels, while the other two haplotypes, CCCCT (β = −0.05, P = 1.5E-05) and ATCCT (β = −0.07, P = 1.0E-05), showed reduced TG concentrations in this population.
TABLE 3.
Haplotype association of five significant SNPs from BUD13-ZNF259-A5 in proximity to the 11q23.3 region with plasma TG level
| Haplotype | Haplotype frequency | β (95% CI) | P |
| ACCAC | 0.079 | 0.17 (0.14 to 0.20) | 1.1E-10−5 |
| ACGCC | 0.218 | 0.10 (0.08 to 0.12) | 1.4E-10−5 |
| ACCCT | 0.276 | Reference | Reference |
| CCCCT | 0.202 | −0.05 (−0.08 to 0.03) | 1.5E-10−5 |
| ATCCT | 0.224 | −0.05 (−0.07 to 0.03) | 1.0E-10−5 |
The “haplo.glm” function implemented in the “haplo.stats” R package was used to calculate the coefficient β and P values for each haplotype compared with the reference haplotype, which was set as the ACCCT haplotype. The same covariates used for genotype analysis were applied in haplotype analysis. Haplotype analysis was performed in 8,223 unrelated individuals in the KARE. Alleles in haplotypes were presented in order of polymorphisms rs11216126/BUD13, rs964184/ZNF259, rs603446/ZNF259, rs2075291/ApoA5, and rs651821/ApoA5. CI, confidence interval. Changes of nucleotides are underlined.
Next, we investigated whether SNP-SNP interactions existed between the five SNPs in ApoA5, ZNF259, and BUD13 on TG level. The strongest interactions were found between rs651821/ApoA5 and rs2075291/ApoA5 (β = 34.46, P = 1.0E-08) in a synergistic manner. Additionally, there was suggestive evidence of positive interactions between rs2075291/ApoA5 and rs964184/ZNF259 (β = 20.59, P = 4.0E-03), and rs11216126/BUD13 and rs603446/ZNF259 (β = 12.04, P = 7.6E-03); rs11216126/BUD13 and rs651821/ApoA5 (β = −1.02, P = 7.7E-03) showed negative interaction for TG level (supplementary Table 3).
DISCUSSION
Our findings from conditional analysis suggest that multiple variants in the 11q23.3 genomic region may be independently associated with TG, and one more locus in the distal region might be another candidate. When we fixed each top hit marker in the series conditional models, most loci showed a decrease in strength of association, but some loci showed even stronger association, particularly in the models between loci in the proximal 11q23.3 region. This paradoxical increase in the significance level from subsequent haplotype analysis corresponds with the negative interaction effects in gene-gene interaction analysis. The haplotype analysis in the proximal region shows two haplotypes that increase TG level. These two hypertriglyceridemic haplotypes, ACCAC and ACGCC , are characterized by the presence of allele C of rs651821/ApoA5, but have different changes in ZNF259 (rs964184 C>G) or ApoA5 (rs2075291 C>A). Conventional genetic analysis based on single SNPs might underestimate or even mislead an association when multiple functional variants are present on a haplotype including the locus, particularly when the directions of the functional loci conflict (27). The substitutions of allele C>G at the rs603446/ZNF259 and A>C at the rs11216126/BUD13 are expected to lower TG based on our findings. Despite weaker LD between the SNPs (r2 < 0.06), alleles with different directions cosegregate in the haplotype which might dilute the effect of each SNP; hypotriglyceridemic allele G at rs603446/ZNF259 cosegregates with hypertriglyceridemic allele A of rs11216126/BUD13; likewise, the TG lowering allele C at rs1121626/BUD13 coexists with the C allele of rs603446/ZNF259 which increases TG. Thus, previous reports on these loci might represent diluted effects. We believe that the functional variants captured by the haplotype might have different mutational origin, if we consider the most common ACCCT haplotype as a wild-type. Additionally, two haplotypes were found to be protective of hypertriglyceridemia (CCCCT and ATCCT), and these protective haplotypes might have resulted from two diverging changes from the wild-type haplotypes. In a similar vein, this finding also suggests that the variants captured by these protective haplotypes might be independent. The presence of both harmful and protective haplotypes in the 11q23.3 region is supported by the results of interaction analysis. Findings from interaction analyses suggest that two variants, both tagged by ApoA5 (rs651821 and rs2075291) gene, have strong synergistic effects to increase TG level. Additionally, interaction analysis indicates that the joint effect between ApoA5 (rs2075291) and ZNF259 (rs964184), albeit less significant, plays a role in increasing the TG level. A negative interaction was also found between ApoA5 and BUD13, which is consistent with the results from conditional models (paradoxical increase in significance levels) or haplotype analysis (both protective and harmful haplotypes). In summary, we believe that the findings from different analyses converge on the presence of multiple functional loci on the 11q23.3 region. Our findings also suggest the proximal part might have a more complex network, with both agonistic and antagonistic regulations, than the distal part.
We did not include SNPs in the distal region of 11q23.3 in haplotype or interaction analysis, but our findings of conditional analysis suggest that another variant in the distal region (SIK3, PCSK7, PAFAH1B2, and SIDT2) affects plasma TG as well. Thus, all four loci in the distal 11q23.3 region show very similar patterns of changes in the conditional analysis; associations with TG are not substantially affected, or even become stronger, by adjusting proximal region variants, while the associations decrease dramatically upon considering another locus within the distal part of 11q23.3. Thus, we consider that the possibility of another candidate variant on the distal region (SIK3-PCSK7-PAFAH1B2-SIDT2 gene complex) is not sufficient, at least by this study alone.
It is beyond the scope of our study to pinpoint the functional variants or to figure out exactly how many genes are functioning in TG regulation. It would be, however, a logical inference from our findings that at least one harmful and one protective variant in the proximal region are participating in TG regulation. ApoA5 and ZNF259 are the most probable genes hosting these variants. Studies involving people of different ancestries indicate that variants on ApoA5 and ZNF259 (rs651821, rs2075291, and rs964184) are associated with plasma TG levels (6, 16, 24, 28). Particularly, rs2075291 (ApoA5) is a nonsynonymous mutation in the coding region (Gly185Cys) mainly found in Asian populations. This “Asian-specific” phenomenon is in part caused by the difference in MAF across populations (only 1% MAF in Caucasians and 7% in Koreans) (supplementary Table 4). Individuals homozygous for rs2075291 (reported as TT genotype, in fact AA as forward strand) in China were reported to develop severe hypertriglyceridemia with mean TG levels of 25.89 ± 5.05 mmol/l (29). The rare allele at SNP site rs964184 (ZNF259) has been reported to be associated with TG in both European and Asian populations (8, 11, 16). The MAF of rs964184 in the Korean population is 0.218, while it is less than 1% in Caucasians (supplementary Table 4).
Many of the top-hit SNPs in our study have been reported in previous studies on TG genetics. Two GWASs conducted in Korea, the KARE project and the Health Examinee (HEXA), both showed that the rs603446/ZNF259 is strongly associated with TG (13). SNP rs7396851 (Apoa4) was also associated with plasma TG in an isolated Pacific Islander Kosrae population (30).
LD structure comparisons across populations show that LD blocks in the 11q23.3 region in Koreans and Asians are stronger and larger than other populations (supplementary Fig. 3). The strong LD structure of Koreans confers advantage in reliably constructing haplotypes which tag functional loci. In Europeans, who have weaker LD across the 11q23.3 region, multiple independent associations with TG level are not shown. If rare variants influence the TG level near the ApoA5 gene region, poor LD structure in the European ancestry population may not allow SNP markers to tag the association (supplementary Fig. 3). Here, we show that haplotypes “ACCAC” and “ACGCC” were associated with increased TG in our Korean study samples (Table 3). However, these haplotypes are not reconstructed in the 1000 Genomes Pilot I CEU population because the genotyping on rs2075291 was not available.
We reviewed public databases to see whether the markers on the 11q23.3 region have been reported to modify the gene expression level, throughout literature and gene-expression quantitative loci (eQTL) databases. The recent report of Yao et al. (31) suggested cis-regulation between rs964184 (ZNF259) and PCSK7, SIDT2, TAGLN, and BUD13, and trans-regulation of ZNF259 (rs964184) with TMEM165, YPEL5, PPM1B, and OBFC2A, suggesting that the effect of rs964184 on TG is mediated through PCSK7. Further, this is in line with an analysis of our previous eQTL study in 10 metabolic traits conducted in Korean populations; rs651821 was defined as the eQTL-SNP hot spot of TAGLN with significant eQTL excess (32). It was also reported that rs603446 (ZNF259) has cis-regulatory effects over the expression of adiposity-associated genes in NCBI’s eQTL database (26).
Considering the results of conditional analysis, LD structure, haplotype analysis, gene-gene interaction analysis, and previous evidence from functional or genetic association studies, it would be a reasonable inference to conclude that the 11q23.3 region, including the APOA5-A4-C3-A1 gene cluster, harbors multiple functional variants that influence the TG levels.
Supplementary Material
Footnotes
Abbreviations:
- CEU
- Central European-like Utah
- CHB
- Han Chinese in Beijing
- QTL
- expression quantitative loci
- GWAS
- genome-wide association study
- HT
- Healthy Twin study
- JPT
- Japanese in Tokyo
- KARE
- Korea Association Resource study
- LD
- linkage disequilibrium
- MAF
- minor allele frequency
- YRI
- Yoruba in Ibadan
This study was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI #HI14C0064) and Post-Genome Technology Development Program (10050164, Developing Korean Reference Genome) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Freiberg J. J., Tybjaerg-Hansen A., Jensen J. S., and Nordestgaard B. G.. 2008. Nonfasting triglycerides and risk of ischemic stroke in the general population. JAMA. 300: 2142–2152. [DOI] [PubMed] [Google Scholar]
- 2.Reaven G. M. 1988. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 37: 1595–1607. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S., Liu X., Yu Y., Hong X., Christoffel K. K., Wang B., Tsai H. J., Li Z., Tang G., Xing H., et al. 2009. Genetic and environmental contributions to phenotypic components of metabolic syndrome: a population-based twin study. Obesity (Silver Spring). 17: 1581–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snieder H., van Doornen L. J., and Boomsma D. I.. 1999. Dissecting the genetic architecture of lipids, lipoproteins, and apolipoproteins: lessons from twin studies. Arterioscler. Thromb. Vasc. Biol. 19: 2826–2834. [DOI] [PubMed] [Google Scholar]
- 5.Isaacs A., Sayed-Tabatabaei F. A., Aulchenko Y. S., Zillikens M. C., Sijbrands E. J., Schut A. F., Rutten W. P., Pols H. A., Witteman J. C., Oostra B. A., et al. 2007. Heritabilities, apolipoprotein E, and effects of inbreeding on plasma lipids in a genetically isolated population: the Erasmus Rucphen Family Study. Eur. J. Epidemiol. 22: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J., Ban M. R., Kennedy B. A., Anand S., Yusuf S., Huff M. W., Pollex R. L., and Hegele R. A.. 2008. APOA5 genetic variants are markers for classic hyperlipoproteinemia phenotypes and hypertriglyceridemia. Nat. Clin. Pract. Cardiovasc. Med. 5: 730–737. [DOI] [PubMed] [Google Scholar]
- 7.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kettunen J., Tukiainen T., Sarin A. P., Ortega-Alonso A., Tikkanen E., Lyytikainen L. P., Kangas A. J., Soininen P., Wurtz P., Silander K., et al. 2012. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 44: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansen C. T., Wang J., Lanktree M. B., Cao H., McIntyre A. D., Ban M. R., Martins R. A., Kennedy B. A., Hassell R. G., Visser M. E., et al. 2010. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 42: 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y. J., Go M. J., Hu C., Hong C. B., Kim Y. K., Lee J. Y., Hwang J. Y., Oh J. H., Kim D. J., Kim N. H., et al. 2011. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 43: 990–995. [DOI] [PubMed] [Google Scholar]
- 14.Park M. H., Kim N., Lee J. Y., and Park H. Y.. 2011. Genetic loci associated with lipid concentrations and cardiovascular risk factors in the Korean population. J. Med. Genet. 48: 10–15. [DOI] [PubMed] [Google Scholar]
- 15.Cho Y. S., Go M. J., Kim Y. J., Heo J. Y., Oh J. H., Ban H. J., Yoon D., Lee M. H., Kim D. J., Park M., et al. 2009. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 41: 527–534. [DOI] [PubMed] [Google Scholar]
- 16.Braun T. R., Been L. F., Singhal A., Worsham J., Ralhan S., Wander G. S., Chambers J. C., Kooner J. S., Aston C. E., and Sanghera D. K.. 2012. A replication study of GWAS-derived lipid genes in Asian Indians: the chromosomal region 11q23.3 harbors loci contributing to triglycerides. PLoS One. 7: e37056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko A., Cantor R. M., Weissglas-Volkov D., Nikkola E., Reddy P. M., Sinsheimer J. S., Pasaniuc B., Brown R., Alvarez M., Rodriguez A., et al. 2014. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat. Commun. 5: 3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui G., Li Z., Li R., Huang J., Wang H., Zhang L., Ding H., and Wang D. W.. 2014. A functional variant in APOA5/A4/C3/A1 gene cluster contributes to elevated triglycerides and severity of CAD by interfering with microRNA 3201 binding efficiency. J. Am. Coll. Cardiol. 64: 267–277. [DOI] [PubMed] [Google Scholar]
- 19.Sung J., Cho S. I., Lee K., Ha M., Choi E. Y., Choi J. S., Kim H., Kim J., Hong K. S., Kim Y., et al. 2006. Healthy Twin: a twin-family study of Korea–protocols and current status. Twin Res. Hum. Genet. 9: 844–848. [DOI] [PubMed] [Google Scholar]
- 20.Gombojav B., Song Y. M., Lee K., Yang S., Kho M., Hwang Y. C., Ko G., and Sung J.. 2013. The Healthy Twin Study, Korea updates: resources for omics and genome epidemiology studies. Twin Res. Hum. Genet. 16: 241–245. [DOI] [PubMed] [Google Scholar]
- 21.Delaneau O., Zagury J. F., and Marchini J.. 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 10: 5–6. [DOI] [PubMed] [Google Scholar]
- 22.Howie B. N., Donnelly P., and Marchini J.. 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aung L. H., Yin R. X., Wu D. F., Wang W., Liu C. W., and Pan S. L.. 2014. Association of the variants in the BUD13–ZNF259 genes and the risk of hyperlipidaemia. J. Cell. Mol. Med. 18: 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Hu B., Wang Y., Wu D., Jin L., and Wang X.. 2014. Influences of APOA5 variants on plasma triglyceride levels in Uyghur population. PLoS One. 9: e110258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schüpbach T., Xenarios I., Bergmann S., and Kapur K.. 2010. FastEpistasis: a high performance computing solution for quantitative trait epistasis. Bioinformatics. 26: 1468–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tryka K. A., Hao L., Sturcke A., Jin Y., Wang Z. Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M., et al. 2014. NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 42: D975–D979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan A., Sim X., Ng H. J., Manning A., Rivas M. A., Highland H. M., Locke A. E., Grarup N., Im H. K., Cingolani P., et al. 2015. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 11: e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guardiola M., Cofan M., de Castro-Oros I., Cenarro A., Plana N., Talmud P. J., Masana L., Ros E., Civeira F., and Ribalta J.. 2015. APOA5 variants predispose hyperlipidemic patients to atherogenic dyslipidemia and subclinical atherosclerosis. Atherosclerosis. 240: 98–104. [DOI] [PubMed] [Google Scholar]
- 29.Pullinger C. R., Aouizerat B. E., Movsesyan I., Durlach V., Sijbrands E. J., Nakajima K., Poon A., Dallinga-Thie G. M., Hattori H., Green L. L., et al. 2008. An apolipoprotein A-V gene SNP is associated with marked hypertriglyceridemia among Asian-American patients. J. Lipid Res. 49: 1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe J. K., Maller J. B., Pe’er I., Neale B. M., Salit J., Kenny E. E., Shea J. L., Burkhardt R., Smith J. G., Ji W., et al. 2009. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 5: e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao C., Chen B. H., Joehanes R., Otlu B., Zhang X., Liu C., Huan T., Tastan O., Cupples L. A., Meigs J. B., et al. 2015. Integromic analysis of genetic variation and gene expression identifies networks for cardiovascular disease phenotypes. Circulation. 131: 536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong K. W., Jeong S. W., Chung M., and Cho S. B.. 2014. Association between expression quantitative trait loci and metabolic traits in two Korean populations. PLoS One. 9: e114128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.