Abstract
Metabolic reprogramming has emerged as a hallmark of cancer. MicroRNAs are noncoding RNAs that posttranscriptionally repress the expression of target mRNAs implicated in multiple physiological processes, including apoptosis, differentiation, and cancer. MicroRNAs can affect entire biological pathways, making them good candidates for therapeutic intervention compared with classical single target approaches. Moreover, microRNAs may become more relevant in the fine-tuning adaptation to stress situations, such as oncogenic events, hypoxia, nutrient deprivation, and oxidative stress. Furthermore, artificial microRNAs can be designed to modulate the expression of multiple targets of a specific pathway. In this review, we describe the metabolic reprogramming associated to cancer, with a special interest in the altered lipid metabolism. Next, we describe specific features of microRNAs that make them relevant to target cancer cell metabolism. Finally, in an attempt to open new therapeutic windows, we emphasize two exciting scenarios for microRNA-mediated intervention that need to be further explored: 1) the cooperation between FA biosynthesis (lipogenesis) and FA oxidation as complementary partners for the survival of cancer cells; and 2) the regulation of the intracellular lipid content modulating both lipid storage into lipid droplets, and lipid mobilization through lipolysis and/or lipophagy.
Keywords: micro-ribonucleic acid, fatty acid oxidation, cancer therapy
A hallmark of cancer is the metabolic reprogramming to sustain cell proliferation and metastasis (1). Regardless of the oncogenic or tumor suppressor event implicated, cancer cells rely on a positive balance of energy and biosynthetic requirements. Moreover, cancer metabolism is the result of a combination of cell-autonomous genetic alterations and flexibility to adapt to the microenvironment (oxygen, pH, and nutrient availability) (2–4).
MicroRNAs are small single-stranded noncoding RNAs (19–25 nucleotides) that posttranscriptionally repress the expression of mRNAs implicated in nearly all cellular processes, such as cell cycle, apoptosis, autophagy, stemness, differentiation, angiogenesis, inflammation, drug resistance, stress response, transformation, and migration. MicroRNAs are frequently deregulated in cancer and so they are important biomarkers for diagnosis and prognosis of the cellular outcome (5, 6)
Importantly, individual microRNAs, combinations of microRNAs, or even artificial microRNAs can specifically be designed to affect entire biological pathways (7, 8), and this makes them good candidates for therapeutic intervention compared with classical single target approaches.
In this review, we briefly describe the altered cancer cell metabolism, and then we discuss the use of microRNAs as modulators of specific metabolic pathways in cancer (summarized in Fig. 1). We note the promising potentiallity of microRNAs for therapeutic intervention toward the altered lipid metabolism in cancer (summarized in Fig. 2). In an attempt to open new therapeutic windows, we emphasize two exciting scenarios for microRNA-mediated intervention that need to be further explored: 1) the cooperation between FA biosynthesis (lipogenesis) and FA oxidation (FAO) as a survival mechanism in cancer; and 2) the regulation of the intracellular lipid content in the cross-talk between lipid storage into lipid droplets (LDs) and lipid mobilization by lipolysis and/or lipophagy (Table 1).
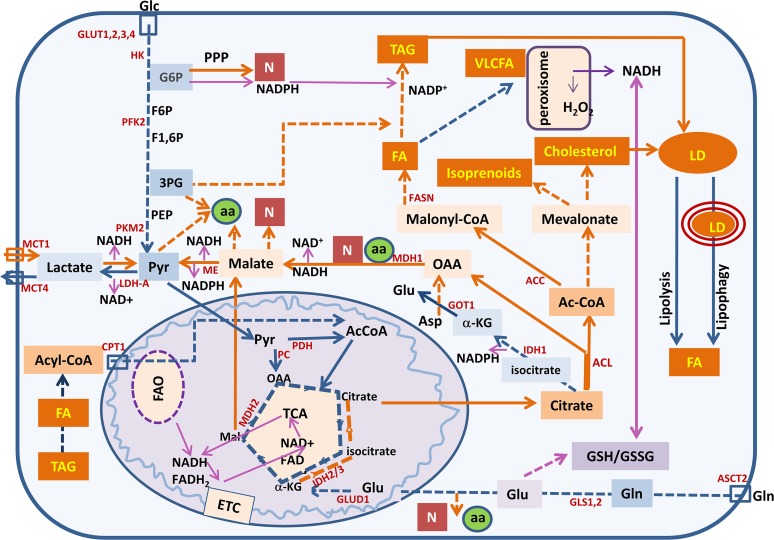
Fig. 1.
Summary of the metabolic pathways altered in cancer that are described in this review: enhanced aerobic glycolysis, increased flux to the PPP, increased glutaminolysis, and de novo lipogenesis and cholesterogenesis. These pathways support rapid proliferation and metastasis. In addition, cancer cells present an increased metabolic plasticity interconnecting pathways to adapt to the changing tumor microenvironment; lipolysis, lipophagy, and FAO contribute to provide energy and to face oxidative stress. GLUD1, glutamate dehydrogenase; GOT1, aspartate aminotransferase; MDH1, malate dehydrogenase 1; PDH, pyruvate dehydrogenase; α-KG, α-ketoglutarate; Asp, aspartate; Glc, glucose, Glu, glutamate; Gln, glutamine; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6P, fructose-1,6-bisphosphate; Mal, malate; 3PG, 3-phosphoglycerate; ETC, electron transporter chain; ASCT, neutral amino acid transporter; GLUT, glucose transporter; MCT, monocarboxylate transporter; aa, amino acids; N, nucleotides; VLCFA, very long chain FAs. (Blue lines, catabolic pathways; orange lines, anabolic pathways; purple lines, redox homeostasis).
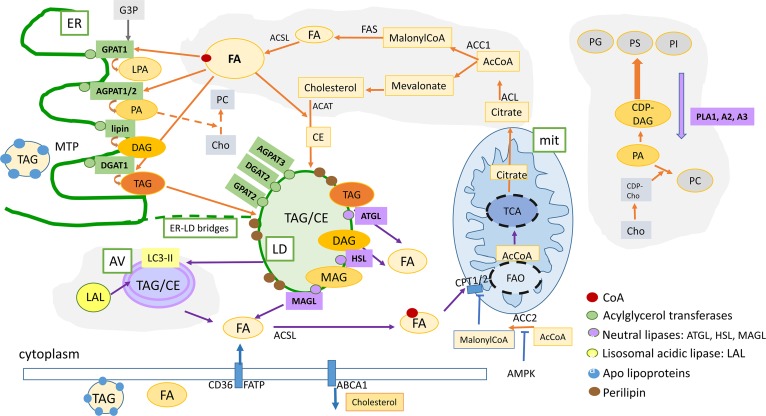
Fig. 2.
Summary of the lipid metabolism pathways that are described in this review: Lipogenesis and FAO can cooperate as partners to increase plasticity of cancer cells. De novo FA and cholesterol biosynthetic pathways and LD formation are indicated by the orange arrows. FAO contributes to increase survival under energy and oxidative stress (indicated by blue arrows). Intracellular lipid content is the result of a dynamic balance between neutral lipid storage in LDs (TAG and CE) and lipid mobilization by neutral lipases (lipolysis) or acidic lipases (lipophagy) that release FFA for FAO in mitochondria or substrates for lipid-related signaling molecules, as well as structural molecules for membrane biosynthesis. In addition, LDs contain an enriched proteome that regulates lipid mobilization and protects from toxic unfolded proteins, buffers excess of proteins, and regulates lipid mobilization when required. Lipophagy is an active process for lipid mobilization that contributes to increase the survival and fitness of cancer cells. Orange arrows, FA and lipid biosynthesis; blue arrows, lipid mobilization and FAO; green boxes, enzymes implicated in lipid biosynthesis; purple boxes, enzymes implicated in lipid mobilization [lipases (adipose TAG lipase, HSL, MAGL) and phospholipases (PLA1, PLA2, PLA3) are indicated in purple]. Lipophagy-related acidic lipases are indicated in yellow. mit, mitochondria; G3P, glycerol-3-phosphate; MAG, monoacylglycerol; CE, cholesterol esters.; PG, phosphatidylglycerol; PS, phosphatidylserine; PC, phosphatidylcholine, PE, phosphatidylethanolamine; PI, phosphatidylinositol; Cho, choline; CPT1A2, carnitine palmitoyltransferase 1A2; CD36, thrombospondin receptor; FATP, fatty acid transport protein.
TABLE 1.
Summary of the microRNAs cited and their targets
| MicroRNA | Target | Reference |
| MicroRNA-195-5p | GLUT3 | (87) |
| MicroRNA-223 | GLUT4 | (88) |
| MicroRNA-143 | HK2 | (89) |
| MicroRNA-15a/16-1 | Aldolase A | (90) |
| MicroRNA-122 | PK-M2 | (91) |
| MicroRNA-326 | PK-M2 | (93) |
| MicroRNA-124, -137, -140 | Warburg | (94) |
| MicroRNA-126 | PI3K pathways | (95) |
| MicroRNA-100, -193a-3p | mTOR pathway | (96, 97) |
| MicroRNA-21 | Phosphatase and tensin homolog | (98) |
| MicroRNA-451 | LKB1 | (99) |
| MicroRNA-143 | K-Ras | (101) |
| MicroRNA-429, -135a, -33b | cMyc | (104–106) |
| Let7-a | cMyc and K-Ras | (108) |
| MicroRNA-25, -30d, -504, -125b | p53 | (109, 110) |
| MicroRNA-34a | SIRT1 | (111) |
| MicroRNA-451 | LKB1/AMPK signaling | (99, 113) |
| MicroRNA-195 | CAB39 | (112) |
| MicroRNA-155 | VHL | (117) |
| MicroRNA-155 | mTOR pathway | (119) |
| MicroRNA-210 | ISCU1/2, COX10, SDHD | (120) |
| MicroRNAs-33a/b | CPT1a, HADHB, CROT, AMPKa1, SIRT6, IRS2 | (82, 131, 132) |
| MicroRNA-185, -342 | SREBP | (121) |
| MicroRNA-613 | LXRa | (122) |
| MicroRNA-122 | Upregulation of FAO | (123, 124) |
| MicroRNA-370 | CPT1a, AMACR | (125, 126) |
| MicroRNA-378/378* | Adipogenesis | (128) |
| MicroRNA-27 | PPARγ, C/EBPa | (129) |
| MicroRNA-122 | Upregulation of LDs and caveolae | (123) |
ISCU1/2, iron-sulfur cluster assembly homolog 1/2; COX10, cytochrome c oxidase assembly protein 10; SDHD, succinate dehydrogenase subunit D; LXRa, liver X receptor α HADHB, hydroxyl acyl-CoA dehydrogenase-3-ketoacyl-CoA thiolase; CROT, carnitine O-octanyltransferase; IRS2, insulin receptor substrate 2; VHL, Von Hippel-Lindau.
CANCER CELL METABOLIC REPROGRAMMING
Cancer cells alter their metabolism to provide the additional energy and anabolic demands of biosynthesis (nucleotides, lipids, proteins) that sustain cell proliferation and metastasis. Although proliferative metabolism is a hallmark of cancer, it is important to note that there is incredible biological diversity across cancer types and even heterogeneities within a single tumor. This affects not only the mechanism by which the metabolic reprogramming is achieved, but also the specific set of metabolic dependencies. For example, the exogenous glutamine dependency varies across different breast tumor subtypes (9).
In addition, cancer cells present increased metabolic plasticity that allows them to continuously adapt to changes in microenvironment. In this sense, some transformed cells depend on glutamine as an important carbon source for the replenishment of the tricarboxylic acid (TCA) cycle intermediates (anaplerosis) (10), but when glutamine metabolism is interrupted, they may rely on glucose-dependent anaplerosis of the TCA cycle (11).
This indicates that the altered tumor metabolism is the result of a combination of cell autonomous genetic alterations and flexibility to adapt to microenvironment changes (oxygenation, pH, and nutrient availability) (2–4).
Aerobic glycolysis in cancer is intimately coordinated with glutamine metabolism
The most prominent metabolic alteration in cancer proliferating cells is the increased glucose uptake and the use of aerobic glycolysis versus mitochondrial respiration regardless of the abundance of oxygen (Warburg effect) (1, 12). In spite of the lower energy generated by aerobic glycolysis, this metabolic preference sustains intermediates for biosynthesis of lipids, nucleotides, and amino acids, which are required to sustain cell proliferation. To compensate the low efficiency of ATP production compared with mitochondrial respiration, cancer cells upregulate glucose transporters (13) to increase the uptake of glucose. In addition, tumors upregulate the expression of glycolytic enzymes, such as hexokinase (HK)2, phosphofructokinase (PFK)1, and lactate dehydrogenase (LDH)A, among others (14).
A checkpoint in the glycolytic pathway is the pentose phosphate pathway (PPP). This pathway generates pentose sugars and NADPH (oxidative branch of the PPP). NADPH provides reducing equivalents that are needed for reductive biosynthesis (mainly de novo FAs and cholesterol biosynthesis) and for diminishing reactive oxygen species (ROS), which are produced in proliferating cells. A major mediator of glucose redirection to the PPP versus aerobic glycolysis is the transcription factor, transcriptional target TP53-induced glycolysis and apoptosis regulator (TIGAR), whose expression correlates with increased survival in various cancer types (15, 16). A second checkpoint in glycolysis is the differential expression of pyruvate kinase (PK) isoforms in cancer cells. PK catalyzes the rate-limiting ATP-generating step of glycolysis in which phosphoenolpyruvate (PEP) is converted to pyruvate (Pyr). The PK-M1 isoform is active forming tetramers that have high affinity for PEP, which is converted rapidly to Pyr (generating ATP as a result). By contrast, the PK-M2 isoform can switch from a tetrameric to a dimeric structure with decreased affinity for PEP. The PK-M2 isoform is normally expressed in embryonic tissues and, in adulthood, is substituted by the M1 isoform. Interestingly, cancer cells re-express the M2 isoform and, as a consequence, the glycolytic flux is slowed down and upstream glycolytic intermediates can be redirected to anabolic processes (17). In addition, PK-M2 is subjected to postranslational modifications, such as phosphorylation, glycosylation, ubiquitination, nitrosylation, methylation, acetylation, lipidation, and proteolysis, that influence its activity. Moreover, it has been described as a nuclear function of PK-M2 where it coactivates catenin to induce cMyc, Glut1, and LDHA, which contribute to the Warburg effect (18).
Aerobic glycolysis in cancer requires a coordinated adjustment of glutamine metabolism. Glutamine not only donates the nitrogen required for the synthesis of nucleotides, amino acids, and hexosamines, but it is also an important carbon source to fulfill TCA intermediates (19–23). Unlike normal cells, where the TCA cycle provides mainly ATP, in proliferating cells, the TCA cycle also provides intermediates for biosynthetic pathways. Glutamine is the main carbon source for the replenishment of TCA cycle intermediates. Glutamine-derived citrate can be converted to acetyl-CoA (AcCoA) and oxaloacetate (OAA) by ATP citrate lyase (ACL). AcCoA feeds de novo FA synthesis (20, 24, 25), and OAA can be metabolized to nucleotides and amino acid biogenesis.
Glutamine is also crucial for glutathione production and, in the reduced form (GSH), contributes to quench ROS produced in proliferating cells, and this has been reported to protect tumor cells from apoptosis (3, 26).
The TCA cycle produces reducing equivalents for the mitochondrial respiratory system and biosynthetic precursors, such as amino acids, citrate (for de novo FA synthesis), and OAA (implicated in gluconeogenesis). Thus, there is a constant efflux of intermediates from the TCA cycle that must be replenished to keep a functional cycle. The main anaplerotic enzymes are Pyr carboxylase (PC), that catalyzes the conversion of Pyr to OAA, and glutaminase (GLS), that catalyzes the deamination of glutamine to produce glutamate. In addition, there is increased evidence that cancer cells can use FAs as fuel in vivo. FA catabolism is carried out through the β-oxidation pathway in mitochondria, which is a process highly dependent on a functional oxidative phosphorylation at the TCA.
Lipogenesis and cholesterogenesis
Cancer cells show specific alterations in lipid metabolism (23) that may affect numerous cellular processes, including cell growth, proliferation, differentiation, malignancy, and motility (27, 28). The majority of adult mammalian tissues satisfy their lipid requirements through the uptake of FFAs and lipoproteins. FA and cholesterol biosynthesis are mainly restricted to liver and adipose tissues. However, numerous studies have confirmed that neoplastic tissues show aberrant activation of de novo lipogenesis (29). In fact, overexpression of lipogenic enzymes is reported as a common characteristic of many cancers (27, 30, 31), and inhibition of different enzymes within the FA biosynthetic pathway can block cancer cell growth (28, 32, 33). FAs can be converted to triacylglycerols (TAGs) for storage and to phospholipids for membrane biogenesis. In addition, they can produce second messengers and hormones implicated in signaling pathways or they can be completely oxidized to CO2 for energy production.
Several studies have shown that key enzymes involved in lipid metabolism are differentially expressed in normal versus tumor tissues, and some of them have been proposed as prognosis biomarkers in different types of cancer (30).
De novo lipid biosynthesis is directly supported by the generation of AcCoA and NADPH from glucose and glutamine metabolism. Large amounts of NADPH are consumed during de novo lipid synthesis and several pathways are required for the replenishment of NADPH in proliferative cells. Glucose-6-phosphate dehydrogenase that redirects glucose-6-phosphate to the oxidative branch of the PPP to produce NADPH is frequently upregulated in malignant cancer cells (34, 35). In addition, NADP-dependent isocitrate dehydrogenase (IDH)1 and malic enzyme (ME), that generate NADPH, are found to be highly expressed in tumor cells (36, 37).
Reductive carboxylation of glutamate to isocitrate can be further used to sustain FA synthesis during hypoxia (38).
Synthesis of FAs starts with the utilization of citrate to produce AcCoA and OAA by ACL. Then, AcCoA carboxylase (ACC) converts AcCoA to malonyl-CoA. FASN condenses AcCoA molecules sequentially to produce saturated FAs, which are further desaturated by stearoyl-CoA desaturase. FASN is frequently overexpressed in breast, prostate, and other types of cancer (29, 39–41). ACL has been reported to be required for tumor formation and transformation (40, 42), and inhibition of ACC induces apoptosis of prostate cancer (PCa) cells (43).
The mevalonate pathway for cholesterol synthesis has been reported to be upregulated in PCa (44), and inhibition of HMG-CoA reductase, the rate limiting enzyme of the mevalonate pathway, induces cell cycle arrest in breast cancer (45). High expression levels of genes of the mevalonate pathway correlate with poor prognosis in breast cancer due to the accumulation of cholesterol that alters membrane fluidity properties contributing to the promotion of transformation (41).
Sterol regulatory element-binding proteins (SREBPs) are the main transcription factors for the expression of the majority of enzymes involved in FA (SREBP1c) and cholesterol synthesis (SREBP2). SREBPs are controlled by oncogenes and tumor suppressors, and they are frequently overexpressed in cancer (29). Oncogenic activation of the PI3K/Akt pathway activates SREBP to promote glucose uptake and its use in lipid synthesis. Loss of retinoblastoma increases the expression of SREBPs by E2F transcription factors and, on the contrary, AMPK activation stimulates β-oxidation through direct phosphorylation and inhibition of ACC and SREBP functions (46).
FAO
Regardless of the relevance of de novo lipogenesis and cholesterol synthesis in cancer, the mobilization of intracellular FAs to be oxidized in mitochondria (FAO) is starting to get relevance for cancer cell metabolism. PPARs are ligand-activated nuclear receptors that influence transcriptional activity-regulating lipid and glucose metabolism, inflammatory responses, regenerative mechanisms, cell differentiation, and cell cycle (47–49). Three major isoforms of PPARs (PPARα, PPARβ/δ, and PPARγ) have been characterized, and they are expressed in highly metabolically active tissues, such as skeletal muscles, heart, adipose tissue, and liver. Differential activation of PPARs reflects the specific biological activity of the three PPAR subtypes. PPARα is mainly expressed in the liver, where it activates FA catabolism. PPARβ/δ participates in FAO, mostly in skeletal and cardiac muscles, and it also regulates blood glucose and cholesterol levels and keratinocyte differentiation. PPARγ is mostly involved in the regulation of adipogenesis, energy balance, and lipid homeostasis. PPARα and PPARβ/δ play a critical role in the sensing of FAs and the activation of the FAO program (50).
While most tumors exhibit high rates of glucose uptake, others exhibit increased dependency on FAO as the main energy source. FAO is the dominant bioenergetic pathway in PCa (51, 52) and enhanced mitochondrial β-oxidation of FAs has been linked to tumor promotion in pancreatic cancer (53). Pharmacological inhibition of α-methylacyl-CoA racemase (AMACR), implicated in the oxidation of branched chain FAs, induces cell growth arrest and apoptosis in PCa (54, 55) and androgen upregulation of carnitine palmitoyltransferase (CPT)1 in PCa leads to increased mitochondrial FAO and ROS production that it has been associated with cell proliferation (56). Highly metastatic breast cancer cells rely partially on FAO induced by AMPK activation (54, 57). Although FAO-associated ROS production seems to be beneficial for cancer cells because it promotes proliferation and/or protects from apoptosis as indicated, sometimes it is not. In this regard, there are situations in which oxidative stress may be detrimental for cancer cell progression, and survival depends on an increased capacity to diminish ROS.
Importantly, FAO provides an alternative pathway to support cancer cell survival when glucose becomes limiting and/or when there is an increased necessity to face oxidative stress. FAO produces AcCoA, NADH, and FADH2 in each cycle. On one hand, AcCoA can enter TCA to produce more NADH and FADH2 to fuel the electron transport chain for ATP production. On the other hand, AcCoA condenses with OAA to produce citrate that can be further redirected to NADPH, generating reactions in cytoplasm via IDH1 and ME activities (58). The transcription factor, NF-E2 p45-related factor 2 (Nrf2), a master regulator of the response against oxidative stress, induces FAO to regulate redox homeostasis, allowing adaptation and survival (52).
There are specific situations in which cancer cells require extra ATP and/or NADPH, and then FAO is crucial for cancer cell survival. For example, increased FAO is essential for the survival of cells from solid tumors that are undergoing loss of attachment (LOA). During LOA, the loss of cells to extracellular matrix interactions induces an apoptotic process known as anoikis. LOA inhibits the uptake of glucose and glycolysis, as well as the glycolytic flux to the PPP, which results in diminished levels of ATP and NADPH; the resulting metabolic stress triggers cell death or anoikis. In this specific scenario, treatment with antioxidants inhibits anoikis due to the reactivation of FAO for ATP production (59).
FAO counteracts oxidative stress in leukemia and glioma cells by increasing the pool of NADPH in the cells, and inhibition of β-oxidation induces apoptosis (60, 61).
In addition, FAO has been described to preserve the hematopoietic stem cell pool and this is dependent on the specific role of promyelocytic leukemia protein in the activation of PPARδ, which increases asymmetric divisions in hematopoietic stem cells by a mechanism that is not well-known (62). Similarly, it has been proposed that this mechanism may also be relevant for the maintenance of the pool of leukemia-initiating cells. A role for promyelocytic leukemia protein promoting the deacetylation and activation of PPARγ coactivator 1A has been associated with aggressive breast cancer (63).
In Fig. 1, we summarize the main metabolic pathways altered in cancer: enhanced aerobic glycolysis, increased flux to the PPP, increased glutaminolysis, and de novo lipogenesis and cholesterogenesis. These pathways support rapid proliferation and metastasis.
Regulation of FA storage versus FA mobilization (lipolysis and lipophagy)
The increased lipogenesis in cancer cells requires mechanisms to accommodate their lipid content intracellularly and, importantly, there must exist an equilibrium between lipid storage and lipid mobilization (64). Indeed, alterations on this equilibrium are responsible for several human diseases, such as lipodystrophies (inability to store lipids), cancer cachexia (due to the increased lipolysis), and excessive accumulation of FAs in LDs related to obesity.
FAs and their derivatives are thought to be rapidly incorporated into neutral lipids and phospholipids. LDs are complex and dynamic organelles that store and supply lipids for energy metabolism, membrane synthesis, and the production of essential lipid-derived molecules. Parton and collaborators have written an excellent review that helps in understanding the biogenesis and lipid-related pathways that control LD biology (65). Briefly, LD biogenesis initiates at the cytoplasmic face of the endoplasmic reticulum (ER) with the esterification of acyl-CoA with glycerol-3-phosphate to generate lysophosphatidic acid (LPA) by the activity of glycerol-3-phosphate acyltransferase. LPA is then converted into phosphatidic acid (PA) by the incorporation of an acyl-CoA molecule by 1-acylglycerol-3-phosphate-O-acyltransferase. Next, lipin dephosphorylates PA to give rise to diacylglycerol (DAG). Finally, DAG acyltransferase adds the last acyl-CoA to generate TAG. Importantly, intermediates of the TAG biosynthetic pathway also participate in the synthesis of membrane phospholipids and lipid signals that promote cell growth, proliferation, and differentiation. LDs may have a regulatory role, by interaction with intracellular compartments, to provide specific lipids and/or sequester misfolded and functional proteins. Moreover, membrane bridges between the ER and LDs have been observed, and this can contribute to the relocalization of TAG synthesis enzymes (66).
LDs consist of a neutral lipid core surrounded by a phospholipid monolayer that is mainly composed of phosphatidylcholine. There is a complex proteome associated with LDs that includes structural molecules, enzymes, and signaling molecules. Among the LD-associated proteins are members of the cell death-inducing DFF45-like effector family and several components of sterol biosynthesis, acyl-CoA metabolism (ACSL3), and TAG biosynthesis. Structural proteins, such as perilipins or caveolins, are critical for the integrity of the LDs to avoid coalescence and to protect them from lipolysis.
It is clear that the increased of de novo lipogenesis observed in cancer cells requires complementary lipolytic mechanisms to remodel the lipid content and to make the stored lipids available for the synthesis of complex lipids and phospholipids (67), and for ATP and NADPH production when required. This cooperation is reflected by the fact that several enzymes of lipolysis, such as adipose TAG lipase or monoacylglycerol lipase (MAGL), have been related to tumorigenesis (68).
Recently, a specific function of autophagy in the lipid mobilization has been described that may have important consequences in the regulation of the cellular lipid content (69). Indeed, in cells lacking an essential autophagy gene, LDs accumulate in the cytoplasm due to defective catabolism, suggesting that it takes a functional autophagic pathway to break them down. Lipophagy is achieved by components of the autophagy machinery, such as microtubule-associated protein light chain 3 that has been found to be associated to LDs (70). Lysosomes contain a single lysosomal acid lipase (LAL) whose deficiency has been associated with Wolman disease (71), which is characterized by the storage of lipids in various organs. In response to nutrient starvation, it has been shown that the upregulation of lipophagy in hepatocytes contributes to the catabolism of LDs (69).
Similar to lipolysis, lipophagy releases FAs to sustain FAO and to supply lipids for membrane biogenesis or cell signaling. Moreover, lipophagy has been reported to support resistance to cell death by metabolizing toxic FFAs (72).
In mammals, transcription factor EB (TFEB) exerts the most global control over autophagy (66), as it regulates autophagosome biogenesis and substrate targeting for lysosome degradation. LAL is one of the TFEB targets, suggesting that TFEB activation may promote lysosomal degradation of lipids (73). Furthermore, TFEB regulates the expression of genes involved in the uptake of FAs across the plasma membrane [by CD36 and FA binding proteins (FABPs)], and master regulators of lipid catabolism, such as as PGC1α and PPARα genes (74). Importantly, mammalian target of rapamycin (mTOR)C1 blocks autophagy and FAO by the inhibition of TFEB and PPARα, respectively. Recently, it has been proposed that signals that trigger lipophagy may start from within the lysosome, as mTORC1 and members of the ERK pathway have been found associated to the lysosomal membrane and autophagosomes, respectively (75).
Moreover, lipidomic studies in aged and obese mice have shown alterations in the lysosomal membrane composition and accumulation of lipids in the lysosomal lumen that may impair mTORC1 function and may affect the lysosomal degradative capacity (76).
Consequently, regulation of the overall lipid content by the interplay between lipid storage and lipid mobilization (by lipolysis and, more recently described, lipophagy) may impact cancer cell metabolism.
Figure 2 summarizes lipid metabolism interconnected pathways that are altered in cancer, such as lipogenesis and FAO, as well as processes regulating the overall intracellular lipid content based on LD formation and lipid mobilization by neutral (lipolysis) and/or acidic hydrolases (LAL).
Tumor microenvironment
Most of the work done in relation to target cancer metabolism has been focused on the identification of oncogenes and tumor suppressors that mediate metabolic reprogramming. However, cancer cell metabolic reprogramming must be accompanied by flexibility and the capacity to adapt to substrate and oxygen availability. Although aerobic glycolysis is a hallmark of cancer, a wide variety of tumors rely on a functional mitochondrial metabolism (77), and they trigger adaptative mechanisms (e.g., mitochondrial biogenesis and the formation of an extensive mitochondrial network) to optimize their oxidative phosphorylation according to the substrate supply and their energy demand. A nice example of the incredible plasticity of cancer cells to support proliferation and survival has recently been described. Cancer cells with mitochondrial defects rely on glutamine reductive carboxylation to generate isocitrate that requires, simultaneously, oxidation of α-ketoglutarate provided by the carboxylation of Pyr from glycolysis (78).
Finally, it has been shown that stromal adjacent glycolytic cells support lactate production to be oxidized by the tumor cells (reverse Warburg effect) in lung and colorectal adenocarcinoma (79). A similar mechanism occurs in ovarian cancer, where adipocytes provide lipids to malignant cells to be oxidized in mitochondria (80).
MicroRNAs IN CANCER
Introduction
MicroRNAs are small noncoding RNAs (19–25 nucleotides) that posttranscriptionally repress the expression of target mRNAs. They are located in intergenic regions or within introns or exons of genes, and they are transcribed as independent units or as part of the gene (81). The canonical pathway of microRNA biogenesis is an evolutionarily conserved process. Briefly, it starts by the transcription by RNA polymerase II of a primary-microRNA (pri-miR), which contains a hairpin stem of 33 base pairs, a terminal loop, and two single stranded flanking regions upstream and downstream of the hairpin. This pri-miR is processed in the nucleus by the nuclear microprocessor complex formed by the RNase III enzyme, Drosha, and the DiGeorge critical region 8 (DGCR8) protein that release a precursor-microRNA of around 60 to 100 nucleotides. Exportin 5 Ran-GTPase exports the precursor-microRNA to the cytoplasm where it is further processed by Dicer, releasing a mature microRNA of around 19–25 nucleotides with a two nucleotide tail at each 3′ end (17).
Finally, the functional strand of the mature microRNA is loaded together with Argonaute (Ago2) proteins into the RNA-induced silencing complex to silence target mRNAs through mRNA cleveage, translational repression, or deadenylation, whereas the called passenger strand is degraded. The last two to eight nucleotides at the 5′ end of the microRNA (seed sequence) are key for the identification of the target mRNA (7).
Due to their mechanism of action, microRNAs may affect cell metabolism by modulating the expression of specific enzymes, transporters, oncogenes, and tumor suppressors, as well as key transcription factors (82).
MicroRNAs are frequently found to be deregulated in cancer and, therefore, they are important biomarkers for diagnosis and prognosis of the cellular outcome (5, 6, 83). Moreover, as microRNAs can be either down- or upregulated in cancer cells, the therapeutic strategies should be directed either to overexpress or to knock down the corresponding microRNA.
By assessing the correlation between the expression of individual microRNAs and their targets, it appears that deregulation of “key” microRNAs may result in the global alterations of entire pathways (84), and this makes microRNAs important players that affect cell homeostasis in both normal and cancer cells.
MicroRNAs are good candidates to face cancer cell metabolism for several reasons. First, due to the diversity of mechanisms that trigger the metabolic reprogramming in cancer, a combination of microRNAs and/or artificial microRNAs can be designed to regulate a specific pathway. Second, microRNAs can be thought of as modulators rather than absolute effectors, and so they may affect altered pathways in cancer without strong toxic effects in normal cells. Third, cancer metabolism is the result of a combination of cell autonomous genetic alterations and microenvironment adaptation. It is under stress conditions (oncogenic events, hypoxia, oxidative stress) where microRNAs may become more relevant in fine-tuning the adaptation to modifying situations, which is crucial for cancer cell survival. Fourth, glucose- and glutamine-reprogrammed metabolism in cancer ends up with an increase of de novo FA biosynthesis and microRNA-mediated targeting of de novo lipogenesis, as well as restrained tumor growth by the anaplerotic pathways of the TCA cycle. Fifth, cancer cells are more efficient in FAO activation in response to nutrient and oxidative stress compared with normal cells, and so it should be expected that cancer cells are more sensitive to FAO inhibition.
In an attempt to open new therapeutic windows based on microRNAs, we propose microRNA-mediated targeting not only of de novo lipogenesis as the ending point of the metabolic reprogramming, including fueling sources of the TCA, but also FAO as a complementary process. In addition, microRNAs may contribute to alter the overall lipid content by disrupting the cross-talk between LD formation and lipid mobilization by lipolysis and/or lipophagy.
MicroRNAs in the net of oncogenic and tumor suppressor pathways that regulate aerobic glycolysis and glutaminolysis in cancer
MicroRNAs contribute to the fine-tuning regulation of cancer-associated glycolytic pathways by targeting specific metabolic enzymes, oncogenes, and/or tumor suppressors, as well as transcription factors and master regulators of cell metabolism (22, 85, 86).
Several microRNAs have been reported to affect the glycolytic flux by regulating the expression of glucose transporters or specific enzymes of glycolysis. MicroRNA-195-5p, that has been found downregulated in bladder cancer, targets GLUT3 (87) and microRNA-223 targets the glucose transporter, GLUT4 (88). HK2 glycolytic enzyme is upregulated in tumors, and microRNA-143 that targets HK2 has been found to be downregulated in cancer cells (89). The microRNA-15a/16-1 cluster downregulates aldolase A glycolytic enzyme (90). Recently, it has been reported that microRNA-122 targets PK-M2, a key regulator of cancer metabolism (91) and overexpression of this microRNA resensitizes 5-fluorouracil-resistant colon cancer cells to 5-fluorouracil through the inhibition of PK-M2 in vitro and in vivo (92). MicroRNA-326 has been reported to be downregulated in glioblastoma cells and this correlates with the upregulation of PK-M2 (93). MicroRNA-124, microRNA-137, and microRNA-340 regulate colorectal cancer growth via inhibition of the Warburg effect (94).
In addition, microRNAs regulate the expression of master regulators of cell metabolism. Growth factors and PI3K/Akt signaling activate mTOR1, which upregulates the expression of glycolytic enzymes (GLUT1, LDHB, and PK-M2) and the hypoxia induction factor (HIF)-1. MicroRNA-126 inhibits PI3K activity, which indirectly represses mTOR and tumor growth (95). MicroRNA-100 actively inhibits mTOR in clear cell ovarian cancer (96) and microRNA-193a-3p exerts a similar effect in hepatocellular carcinoma (HCC) (97). MicroRNA-21 inhibits phosphatase and tensin homolog tumor suppressor, which indirectly activates mTOR in HCC (98), and microRNA-451 has a similar effect on glioma cells by directly targeting the LKB1 activator of AMPK (99).
Oncogenic K-Ras activates the Raf/MEK/ERK and PI3K/Akt pathways promoting tumor growth and survival. In addition, K-Ras enhances cancer metabolism through HIF-1 activation (100). MicroRNA-143 inhibits colorectal cancer growth by a direct effect on K-Ras expression (101). Conversely, microRNA-18* and microRNA-217 promote anchorage-independent growth by repressing K-Ras expression (102, 103).
Oncogenic cMyc increases both glucose and glutamine uptake to support cancer cell proliferation. MicroRNAs targeting cMyc have been found to be downregulated in cancer [microRNA-429 (104), -135a (105), and -33b (106)]. Transformed B cells overexpressing cMyc exhibit increased glutaminolysis under hypoxia to support oxidative TCA metabolism and NADPH generation via ME (3). This effect is partially mediated via cMyc-dependent suppression of microRNA-23-a/b, which inhibits mitochondrial GLS expression (107). On the contrary, overexpression of let7-a inhibits growth of lung cancer in nude mice by suppression of cMyc and K-Ras oncogenes (108).
P53 tumor suppressor inhibits glycolysis and enhances mitochondrial respiration. MicroRNA-25, microRNA-30d, microRNA-504, and microRNA-125b directly target p53 (109, 110) and microRNA-34a indirectly activates p53 by downregulating sirtuin (SIRT)1 that negatively regulates p53 through deacetylation (111).
AMPK is a highly conserved master regulator of numerous cellular processes, including energy homeostasis, cytoskeletal dynamics, modulation of cell growth rates, and cell death pathways. AMPK deficiencies have been shown to enhance cell growth and proliferation, and this correlates with enhancement of tumorigenesis. Conversely, activation of AMPK is associated with tumor growth suppression via inhibition of the mTOR signal pathway. MicroRNA-195 has been reported to target LKB-1-associated protein, CAB39 (112), leading to upregulation of the mTOR pathway, and upregulation of microRNA-451 has been shown to target LKB1/AMPK signaling in glioma cells (99, 113). In response to energy stress, the AMPK pathway is activated and it induces mitochondrial respiration and inhibits cell proliferation. Many cancer cells present decreased AMPK signaling, which promotes enhanced aerobic glycolysis (Warburg effect).
In addition, AMPK inhibits SREBP activation and suppresses ACC (114), stimulating FAO.
MicroRNAs in the adaptation to microenvironment in cancer
In the in vivo scenario, tumor microenvironments are highly heterogeneous due to glucose and oxygen gradients, and metabolic transitions can be crucial for tumor physiology. Hypoxia-inducible transcription factors (HIFs) are stable and active during hypoxia, allowing the cells to switch to an anaerobic metabolism. However, in several tumors, these transcription factors are active even in normoxia (115). HIF-1 plays a key role in the reprogramming of cancer metabolism by activating the transcription of glucose transporters and glycolytic enzymes, transcription of Pyr dehydrogenase kinase 1, which drives Pyr away from the mitochondria (116), and BNIP3, which triggers selective mitochondrial autophagy. Recently, it has been reported that microRNA-155 targets Von Hippel-Lindau, which results in increased HIF activity (117). MicroRNA-155 presents a hypoxia-responsive element, which contributes to the resistance of tumors to radiation therapies (118). Moreover, hypoxia induces microRNA-155 that has been reported to inhibit multiple targets of the mTOR pathway, increasing autophagic activity in human nasopharyngeal cancer and cervical cancer cells (119).
MicroRNA-210 has been shown to inhibit mitochondrial respiration in breast and colon cancer by targeting the iron-sulfur cluster assembly homolog 1/2, cytochrome c oxidase assembly protein 10, and succinate dehydrogenase subunit D (120).
In addition, cancer cells must adapt their metabolism to a more effective utilization of the available substrates. In the absence of glutamine, alternative anaplerotic substrates are needed to maintain a functional TCA cycle. Hepatoma cells can increase flux through PC to proliferate and maintain oxidative metabolic activity (11) when glutamine is depleted.
Targeting TCA anaplerosis by microRNAs constitutes an attractive therapeutic opportunity to limit the efflux of anabolic precursors in cancer. In this sense, PC, aspartate aminotransferase, propionyl-CoA carboxylase, GLS, glutamate dehydrogenase, and alanine aminotransferase are promising targets for microRNA-mediated intervention in cancer.
MicroRNAs DISRUPTING LIPID HOMEOSTASIS IN CANCER
MicroRNAs in the cross-talk between lipogenesis and FA catabolism
Glucose and glutamine reprogrammed metabolism in cancer ends up with an increase of de novo FA biosynthesis independently of the availability of the extracellular lipids. This contributes not only to increase cancer cell proliferation and survival under oxidative and energy stress, but also to stimulate signaling pathways that can promote invasion and metastasis (28). Moreover, the hostile microenvironment of solid tumors has been shown to activate several signaling pathways that end up promoting FASN expression. Consequently, microRNA-mediated targeting of de novo lipogenesis, including the anaplerotic pathways of the TCA, will restrain tumor growth. MicroRNA-185 and microRNA-342 have been described as regulators of lipogenesis and cholesterogenesis by targeting SREBP transcription factor in prostate cancer (PCa) cells (121). MicroRNA-613 suppresses lipogenesis by directly targeting liver X receptor α in HCC HepG2 cells, suggesting that it may be a novel target for regulating lipid homeostasis (122).
Regarding de novo lipogenesis, there are specific situations in which cancer cells require an extra FAO activation, as indicated previously, and then cancer cells should be expected to be more sensitive to FAO inhibition. In this sense, downregulation of microRNA-122 correlates with an increase of FAO observed in HCC (123, 124). MicroRNA-370 targets carnitine pamitoyl transferase (CPT1a) protein, which mediates the transport of long FAs across the mitochondrial membrane and, consequently, affects the rate of β-oxidation at mitochondria (125). Importantly, inhibition of CPT1a has been reported to sensitize human leukemia cells to chemotherapy (60). Recently, it has been reported that overexpression of microRNA-26a reduces the expression levels of alpha-methylacyl-CoA racemase (AMACR), which drives the isomeric conversion of FAs entering the β-oxidation pathway AMACR in PCa (126). MicroRNAs-33a/b are coexpressed within their host genes, the SREBF genes, and they contribute to diminish the expression of acyl-CoA transferases involved in FAO, such as CPT1a, hydroxyl acyl-CoA dehydrogenase-3-ketoacyl-CoA thiolase, and carnitine O-octanyltransferase.
Finally, although lipogenesis and FAO activities seem to oppose one another, these two processes can be thought of as partners to increase the plasticity of cancer. Thus, there are situations in which both processes may occur simultaneously to support each other. Indeed FASN, a lipogenic enzyme, which is upregulated in colorectal cancer, is crucial to maintain energy homeostasis via increasing the mitochondrial β-oxidation of the endogenously synthesized lipids, and this cooperation is a key mechanism for cancer cell survival (127). Consequently microRNAs implicated in the regulation of FA synthesis and FAO may be relevant for tumor biology. In this regard, microRNA-378/378*, an intronic microRNA located within the PGC1α genomic sequence, affects important aspects of adipogenesis (128) and microRNA-27 that inhibits PPARγ and C/EBPa during adipocyte differentiation (129), and so, they may contribute to cancer cell metabolism in specific scenarios different to adipocytes.
MicroRNAs in the cross-talk between lipid storage and lipid mobilization (lipolysis and lipophagy)
The majority of research on cancer lipid metabolism has focused on the increase of FA synthesis. This inexorably affects the overall intracellular lipid content and, consequently, complementary mechanisms to regulate lipid homeostasis are needed.
Interestingly, lipolytic remodeling of lipid species have been reported to promote the tumorigenic properties of cancer cells (68). In this sense, the activity of MAGL has been found to be elevated in aggressive cancer cells from multiple tissues and inhibition of MAGL diminishes migration, invasion, and survival of cancer cells in vitro and xenograft tumor growth in mice (68). Moreover, MAGL affects the production of signaling lipids (PA, LPA, and prostaglandin E2) and microRNAs downregulating MAGL, when associated to cancer aggressiveness, may be relevant to block tumor progression.
Investigation on microRNAs that disrupt the equilibrium between lipid storage and lipid mobilization and intra/extracellular lipid trafficking may be crucial for tumor biology. In this sense, microRNA-122 has been reported to stimulate the production of ER-associated LDs and cholesterol-rich membrane domains (caveolae) (123), and inhibition of microRNA-122 with anti-microRNAs may contribute to alter the equilibrium between lipid storage and lipid mobilization (130). MicroRNAs-33a/b are well-known players that affect cholesterol and lipid metabolism and energy homeostasis. They affect the expression of cholesterol export transporters, such as ABCA, as well as regulators of energy homeostasis, such as AMPKa1, SIRT6, and insulin receptor substrate 2 (82, 131, 132).
MicroRNAs could be directed to inhibit LD biogenesis that will have consequences on the accommodation of the increased lipogenesis and the additional regulatory funtions of these organelles. On the other hand, microRNA-mediated inhibition of lipid mobilization can promote cell death due to the decreased availability of lipids for membrane biogenesis, signaling, and/or energy production. In this regard, a better knowledge of the mechanisms that promote lipolysis and lipophagy are required. The discovery of TFEB and the modulation of lysosome-mediated cellular clearance by lipid content opens new therapeutic scenarios that need to be further explored.
STRATEGIES FOR MicroRNA THERAPEUTIC DELIVERY TO TARGET CANCER METABOLISM
In order to use a microRNA-based therapy to target cancer, it is important to define the way by which gain- or loss-of-function of the microRNA is achieved, as well as the delivery strategy.
Currently, when repression of microRNAs is required, antisense oligonucleotides complementary to the mature miRNA are used.
Specific modifications, such as methylated hydroxyl groups (2′-OMe) or locked nucleic acid (133), increase the stability and the affinity to the target microRNA (134). Cholesterol-modified antisense oligonucleotides to repress microRNA-10b successfully suppressed metastasis in a mouse mammary tumor model (135). In addition, peptide nucleic acids have been reported to efficiently target microRNAs in vitro and in vivo (136). Another strategy is the use of “microRNA sponges” that contain multiple binding sites for a specific microRNA to reduce the endogenous levels of the microRNA. This strategy has been effective in inhibiting the proliferation of B-cell lymphoma cells by sequestering the microRNA-17/92 cluster (137).
On the contrary, in order to restore microRNA expression in cells, microRNA mimics, plasmids, or viral constructs have been used.
MicroRNA mimics are double-stranded RNA molecules similar to the endogenous Dicer product. Specific chemical modifications may improve pharmacodynamics and reduce off-target effects (138). An alternative strategy involves the expression of a shRNA or pri-miR mimic from a plasmid or viral construct that provides a more stable expression of the mature microRNA (124). MicroRNA-34a exogenously expressed in cultured breast cancer cells was reported to reduce tumor cell proliferation, migration, and invasion (139), and an adenoviral vector expressing shRNAs against microRNA-221 and -222 has been described in cultured glioblastoma cells (GBM) (140).
On the other hand, several approaches are currently being developed to deliver microRNAs into the cell. Lipid- and polymer-based carriers have been tested for siRNA/shRNA delivery in preclinical studies (141). Liposome-mediated delivery of a plasmid encoding microRNA-34a reduced several microRNA-34a target genes in cultured breast cancer cells (139), and liposome-formulated microRNA-34a is being tested in a phase I clinical trial in patients with primary or metastatic liver cancer (http://clinicaltrials.gov/ct2/show/NCT01829971).
CONCLUSIONS
Most of the work done to target cancer metabolism has been focused on the identification of oncogenes and tumor suppressors that mediate the cancer-associated metabolic reprogramming: increased aerobic glycolysis, glutaminolysis, and de novo FA biosynthesis. Moreover, cancer cells present increased flexibility to adapt to substrate and oxygen availability at microenvironment.
Several therapeutic approaches have been proposed for cancer treatment, such as the inhibition of specific glycolytic enzymes (HK, PK, and LDH), reactivation of mitochondrial metabolism by stimulating Pyr oxidation, or inhibition of the expression and/or activity of HIF-1α. Nevertheless, there are exciting scenarios related to lipid metabolism that need to be further investigated: 1) FAO as a complementary partner of the increased lipogenesis in specific situations; and 2) plasticity to regulate the overall lipid content that will support cell proliferation and survival.
In this scenario microRNAs are good candidates for therapeutic approaches to face cancer. MicroRNAs affect nearly all cellular processes based on their mechanism of action. Combination of microRNAs can modulate complete pathways, depending on the specific genetic alteration driving the tumor. Special interest has been shown in targeting aerobic glycolysis by microRNAs. However, in an attempt to explore new therapeutic opportunities, an integrative knowledge of both the fueling substrates and the end products of biosynthetic pathways is required. MicroRNAs may cover the complete and different scenarios to be targeted. In this sense, microRNAs can be directed against biosynthetic pathways, such as lipogenesis and cholesterogenesis, and/or against FA storage versus mobilization.
Defining lipid-related signaling pathways altered in cancer cells will provide rational targets for microRNA-based therapeutic strategies. Future clinical trials should provide new insights into the safety and efficacy of the development of microRNA-based anti-cancer therapies (142).
Footnotes
Abbreviations:
- ACC
- acetyl-CoA carboxylase
- AcCoA
- acetyl-CoA
- ACL
- ATP citrate lyase
- AMACR
- α-methyl-acyl-CoA racemase
- CPT
- carnitine palmitoyltransferase
- DAG
- diacylglycerol
- ER
- endoplasmic reticulum
- FAO
- FA oxidation
- GLS
- glutaminase
- HCC
- hepatocellular carcinoma
- HIF
- hypoxia induction factor
- HK
- hexokinase
- IDH
- isocitrate dehydrogenase
- LAL
- lysosomal acid lipase
- LD
- lipid droplet
- LDH
- lactate dehydrogenase
- LOA
- loss of attachment
- LPA
- lysophosphatidic acid
- MAGL
- monoacylglycerol lipase
- ME
- malic enzyme
- mTOR
- mammalian target of rapamycin
- OAA
- oxaloacetate
- PA
- phosphatidic acid
- PC
- pyruvate carboxylase
- PCa
- prostate cancer
- PEP
- phosphoenolpyruvate
- PFK
- phosphofructokinase
- PK
- pyruvate kinase
- PPP
- pentose phosphate pathway
- pri-miR
- primary-microRNA
- Pyr
- pyruvate
- ROS
- reactive oxygen species
- SIRT
- sirtuin
- SREBP
- sterol regulatory element-binding protein
- TAG
- triacylglycerol
- TCA
- tricarboxylic acid
- TFEB
- transcription factor EB
This work was supported by Ministerio de Economía y Competitividad del Gobierno de España (MINECO, Plan Nacional I+D+i AGL2013-48943-C2), Gobierno regional de la Comunidad de Madrid (P2013/ABI-2728, ALIBIRD-CM), and EU Structural Funds.
REFERENCES
- 1.Hanahan D., and Weinberg R. A.. 2011. Hallmarks of cancer: the next generation. Cell. 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 2.Parks S. K., Chiche J., and Pouyssegur J.. 2011. pH control mechanisms of tumor survival and growth. J. Cell. Physiol. 226: 299–308. [DOI] [PubMed] [Google Scholar]
- 3.Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C. J., Slusher B. S., Zhang H., et al. 2012. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J. Q., and Russo J.. 2012. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim. Biophys. Acta. 1826: 370–384. [DOI] [PubMed] [Google Scholar]
- 5.Kosaka N., Iguchi H., and Ochiya T.. 2010. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 101: 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng Y., Yu S., Li H., Xiang H., Peng J., and Jiang S.. 2014. MicroRNAs: emerging roles in adipogenesis and obesity. Cell. Signal. 26: 1888–1896. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe H., Aida Y., Ishiguro H., Yoshizawa K., Seki N., Miyazaki T., Itagaki M., Sutoh S., Ika M., Kato K., et al. 2013. New proposal for response-guided peg-interferon-plus-ribavirin combination therapy for chronic hepatitis C virus genotype 2 infection. J. Med. Virol. 85: 1523–1533. [DOI] [PubMed] [Google Scholar]
- 9.Kung H. N., Marks J. R., and Chi J. T.. 2011. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 7: e1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeBerardinis R. J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., and Thompson C. B.. 2007. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 104: 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng T., Sudderth J., Yang C., Mullen A. R., Jin E. S., Mates J. M., and DeBerardinis R. J.. 2011. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. USA. 108: 8674–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warburg O., Geissler A. W., and Lorenz S.. 1967. On growth of cancer cells in media in which glucose is replaced by galactose [Article in German]. Hoppe Seylers Z. Physiol. Chem. 348: 1686–1687. [PubMed] [Google Scholar]
- 13.Macheda M. L., Rogers S., and Best J. D.. 2005. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J. Cell. Physiol. 202: 654–662. [DOI] [PubMed] [Google Scholar]
- 14.Vander Heiden M. G., Lunt S. Y., Dayton T. L., Fiske B. P., Israelsen W. J., Mattaini K. R., Vokes N. I., Stephanopoulos G., Cantley L. C., Metallo C. M., et al. 2011. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb. Symp. Quant. Biol. 76: 325–334. [DOI] [PubMed] [Google Scholar]
- 15.Cheung E. C., Athineos D., Lee P., Ridgway R. A., Lambie W., Nixon C., Strathdee D., Blyth K., Sansom O. J., and Vousden K. H.. 2013. TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev. Cell. 25: 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X., Xie W., Li Q., Zhang Y., Zhang J., Zhao X., Liu J., and Huang G.. 2013. TIGAR is correlated with maximal standardized uptake value on FDG-PET and survival in non-small cell lung cancer. PLoS One. 8: e80576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi R., Qin Y., Macara I. G., and Cullen B. R.. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sini G., Colombino M., Lissia A., Maxia S., Gulino M., Paliogiannis P., Palomba G., Palmieri G., Cossu A., and Rubino C.. 2013. Primary dermal melanoma in a patient with a history of multiple malignancies: a case report with molecular characterization. Case Rep. Dermatol. 5: 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deberardinis R. J., Sayed N., Ditsworth D., and Thompson C. B.. 2008. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18: 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBerardinis R. J., and Cheng T.. 2010. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 29: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daye D., and Wellen K. E.. 2012. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin. Cell Dev. Biol. 23: 362–369. [DOI] [PubMed] [Google Scholar]
- 22.Dang C. V. 2010. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 70: 859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baenke F., Peck B., Miess H., and Schulze A.. 2013. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 6: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., and Thompson C. B.. 2008. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7: 11–20. [DOI] [PubMed] [Google Scholar]
- 25.Lunt S. Y., and Vander Heiden M. G.. 2011. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27: 441–464. [DOI] [PubMed] [Google Scholar]
- 26.Weinberg F., Hamanaka R., Wheaton W. W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G. M., Budinger G. R., and Chandel N. S.. 2010. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA. 107: 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mashima T., Sato S., Okabe S., Miyata S., Matsuura M., Sugimoto Y., Tsuruo T., and Seimiya H.. 2009. Acyl-CoA synthetase as a cancer survival factor: its inhibition enhances the efficacy of etoposide. Cancer Sci. 100: 1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos C. R., and Schulze A.. 2012. Lipid metabolism in cancer. FEBS J. 279: 2610–2623. [DOI] [PubMed] [Google Scholar]
- 29.Menendez J. A., and Lupu R.. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 7: 763–777. [DOI] [PubMed] [Google Scholar]
- 30.Ramírez de Molina A., Sarmentero-Estrada J., Belda-Iniesta C., Tarón M., Ramírez de Molina V., Cejas P., Skrzypski M., Gallego-Ortega D., de Castro J., Casado E., et al. 2007. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 8: 889–897. [DOI] [PubMed] [Google Scholar]
- 31.Roongta U. V., Pabalan J. G., Wang X., Ryseck R. P., Fargnoli J., Henley B. J., Yang W. P., Zhu J., Madireddi M. T., Lawrence R. M., et al. 2011. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol. Cancer Res. 9: 1551–1561. [DOI] [PubMed] [Google Scholar]
- 32.Abramson H. N. 2011. The lipogenesis pathway as a cancer target. J. Med. Chem. 54: 5615–5638. [DOI] [PubMed] [Google Scholar]
- 33.Zaidi N., Swinnen J. V., and Smans K.. 2012. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 72: 3709–3714. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C., Zhang Z., Zhu Y., and Qin S.. 2014. Glucose-6-phosphate dehydrogenase: a biomarker and potential therapeutic target for cancer. Anticancer. Agents Med. Chem. 14: 280–289. [DOI] [PubMed] [Google Scholar]
- 35.Wang J., Yuan W., Chen Z., Wu S., Chen J., Ge J., Hou F., and Chen Z.. 2012. Overexpression of G6PD is associated with poor clinical outcome in gastric cancer. Tumour Biol. 33: 95–101. [DOI] [PubMed] [Google Scholar]
- 36.Cairns R. A., Harris I. S., and Mak T. W.. 2011. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 11: 85–95. [DOI] [PubMed] [Google Scholar]
- 37.Zheng F. J., Ye H. B., Wu M. S., Lian Y. F., Qian C. N., and Zeng Y. X.. 2012. Repressing malic enzyme 1 redirects glucose metabolism, unbalances the redox state, and attenuates migratory and invasive abilities in nasopharyngeal carcinoma cell lines. Chin. J. Cancer. 31: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wise D. R., Ward P. S., Shay J. E., Cross J. R., Gruber J. J., Sachdeva U. M., Platt J. M., DeMatteo R. G., Simon M. C., and Thompson C. B.. 2011. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA. 108: 19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swinnen J. V., Vanderhoydonc F., Elgamal A. A., Eelen M., Vercaeren I., Joniau S., Van Poppel H., Baert L., Goossens K., Heyns W., et al. 2000. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int. J. Cancer. 88: 176–179. [DOI] [PubMed] [Google Scholar]
- 40.Bauer D. E., Hatzivassiliou G., Zhao F., Andreadis C., and Thompson C. B.. 2005. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 24: 6314–6322. [DOI] [PubMed] [Google Scholar]
- 41.Clendening J. W., Pandyra A., Boutros P. C., El Ghamrasni S., Khosravi F., Trentin G. A., Martirosyan A., Hakem A., Hakem R., Jurisica I., et al. 2010. Dysregulation of the mevalonate pathway promotes transformation. Proc. Natl. Acad. Sci. USA. 107: 15051–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatzivassiliou G., Zhao F., Bauer D. E., Andreadis C., Shaw A. N., Dhanak D., Hingorani S. R., Tuveson D. A., and Thompson C. B.. 2005. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 8: 311–321. [DOI] [PubMed] [Google Scholar]
- 43.Beckers A., Organe S., Timmermans L., Scheys K., Peeters A., Brusselmans K., Verhoeven G., and Swinnen J. V.. 2007. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 67: 8180–8187. [DOI] [PubMed] [Google Scholar]
- 44.Hager M. H., Solomon K. R., and Freeman M. R.. 2006. The role of cholesterol in prostate cancer. Curr. Opin. Clin. Nutr. Metab. Care. 9: 379–385. [DOI] [PubMed] [Google Scholar]
- 45.Rao S., Lowe M., Herliczek T. W., and Keyomarsi K.. 1998. Lovastatin mediated G1 arrest in normal and tumor breast cells is through inhibition of CDK2 activity and redistribution of p21 and p27, independent of p53. Oncogene. 17: 2393–2402. [DOI] [PubMed] [Google Scholar]
- 46.Kollmann K., Mutenda K. E., Balleininger M., Eckermann E., von Figura K., Schmidt B., and Lubke T.. 2005. Identification of novel lysosomal matrix proteins by proteome analysis. Proteomics. 5: 3966–3978. [DOI] [PubMed] [Google Scholar]
- 47.Michalik L., and Wahli W.. 2008. PPARs mediate lipid signaling in inflammation and cancer. PPAR Res. 2008: 134059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dharancy S., Louvet A., Hollebecque A., Desreumaux P., Mathurin P., and Dubuquoy L.. 2008. Nuclear receptor PPAR and hepatology: pathophysiological and therapeutical aspects [Article in French]. Gastroenterol. Clin. Biol. 32: 339–350. [DOI] [PubMed] [Google Scholar]
- 49.Li G., and Guo G. L.. 2011. Role of class II nuclear receptors in liver carcinogenesis. Anticancer. Agents Med. Chem. 11: 529–542. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y. X. 2010. PPARs: diverse regulators in energy metabolism and metabolic diseases. Cell Res. 20: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y. 2006. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 9: 230–234. [DOI] [PubMed] [Google Scholar]
- 52.Ludtmann M. H., Angelova P. R., Zhang Y., Abramov A. Y., and Dinkova-Kostova A. T.. 2014. Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem. J. 457: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khasawneh J., Schulz M. D., Walch A., Rozman J., Hrabe de Angelis M., Klingenspor M., Buck A., Schwaiger M., Saur D., Schmid R. M., et al. 2009. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc. Natl. Acad. Sci. USA. 106: 3354–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee Y. E., He H. L., Lee S. W., Chen T. J., Chang K. Y., Hsing C. H., and Li C. F.. 2014. AMACR overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biol. 35: 7983–7991. [DOI] [PubMed] [Google Scholar]
- 55.Festuccia C., Gravina G. L., Mancini A., Muzi P., Cesare E. D., Kirk R., Smith M., Hughes S., Gibson R., Lian L. Y., et al. 2014. Trifluoroibuprofen inhibits α-methylacyl coenzyme A racemase (AMACR/P504S), reduces cancer cell proliferation and inhibits in vivo tumor growth in aggressive prostate cancer models. Anticancer Agents Med. Chem. 14: 1031–1041. [DOI] [PubMed] [Google Scholar]
- 56.Lin H., Lu J. P., Laflamme P., Qiao S., Shayegan B., Bryskin I., Monardo L., Wilson B. C., Singh G., and Pinthus J. H.. 2010. Inter-related in vitro effects of androgens, fatty acids and oxidative stress in prostate cancer: a mechanistic model supporting prevention strategies. Int. J. Oncol. 37: 761–766. [DOI] [PubMed] [Google Scholar]
- 57.Li S., Zhou T., Li C., Dai Z., Che D., Yao Y., Li L., Ma J., Yang X., and Gao G.. 2014. High metastaticgastric and breast cancer cells consume oleic acid in an AMPK dependent manner. PLoS One. 9: e97330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeon S. M., Chandel N. S., and Hay N.. 2012. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 485: 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schafer Z. T., Grassian A. R., Song L., Jiang Z., Gerhart-Hines Z., Irie H. Y., Gao S., Puigserver P., and Brugge J. S.. 2009. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 461: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samudio I., Harmancey R., Fiegl M., Kantarjian H., Konopleva M., Korchin B., Kaluarachchi K., Bornmann W., Duvvuri S., Taegtmeyer H., et al. 2010. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Invest. 120: 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pike L. S., Smift A. L., Croteau N. J., Ferrick D. A., and Wu M.. 2011. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim. Biophys. Acta. 1807: 726–734. [DOI] [PubMed] [Google Scholar]
- 62.Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D. E., Schafer Z. T., Evans R. M., Suda T., Lee C. H., et al. 2012. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 18: 1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carracedo A., Weiss D., Leliaert A. K., Bhasin M., de Boer V. C., Laurent G., Adams A. C., Sundvall M., Song S. J., Ito K., et al. 2012. A metabolic prosurvival role for PML in breast cancer. J. Clin. Invest. 122: 3088–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Przybytkowski E., Joly E., Nolan C. J., Hardy S., Francoeur A. M., Langelier Y., and Prentki M.. 2007. Upregulation of cellular triacylglycerol - free fatty acid cycling by oleate is associated with long-term serum-free survival of human breast cancer cells. Biochem. Cell Biol. 85: 301–310. [DOI] [PubMed] [Google Scholar]
- 65.Pol A., Gross S. P., and Parton R. G.. 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332: 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kao C., Chao A., Tsai C. L., Chuang W. C., Huang W. P., Chen G. C., Lin C. Y., Wang T. H., Wang H. S., and Lai C. H.. 2014. Bortezomib enhances cancer cell death by blocking the autophagic flux through stimulating ERK phosphorylation. Cell Death Dis. 5: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nomura D. K., Long J. Z., Niessen S., Hoover H. S., Ng S. W., and Cravatt B. F.. 2010. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 140: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., and Czaja M. J.. 2009. Autophagy regulates lipid metabolism. Nature. 458: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johansen T., and Lamark T.. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7: 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lübke T., Lobel P., and Sleat D. E.. 2009. Proteomics of the lysosome. Biochim. Biophys. Acta. 1793: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amir M., and Czaja M. J.. 2011. Autophagy in nonalcoholic steatohepatitis. Expert Rev. Gastroenterol. Hepatol. 5: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., and Ballabio A.. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20: 3852–3866. [DOI] [PubMed] [Google Scholar]
- 74.Settembre C., De Cegli R., Mansueto G., Saha P. K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T. J., et al. 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez-Lopez N., Athonvarangkul D., Mishall P., Sahu S., and Singh R.. 2013. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 4: 2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodriguez-Navarro J. A., Kaushik S., Koga H., Dall’Armi C., Shui G., Wenk M. R., Di Paolo G., and Cuervo A. M.. 2012. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 109: E705–E714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whitaker-Menezes D., Martinez-Outschoorn U. E., Flomenberg N., Birbe R. C., Witkiewicz A. K., Howell A., Pavlides S., Tsirigos A., Ertel A., Pestell R. G., et al. 2011. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle. 10: 4047–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mullen A. R., Hu Z., Shi X., Jiang L., Boroughs L. K., Kovacs Z., Boriack R., Rakheja D., Sullivan L. B., Linehan W. M., et al. 2014. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Reports. 7: 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martinez-Outschoorn U. E., Pestell R. G., Howell A., Tykocinski M. L., Nagajyothi F., Machado F. S., Tanowitz H. B., Sotgia F., and Lisanti M. P.. 2011. Energy transfer in “parasitic” cancer metabolism: mitochondria are the powerhouse and Achilles’ heel of tumor cells. Cell Cycle. 10: 4208–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nieman K. M., Kenny H. A., Penicka C. V., Ladanyi A., Buell-Gutbrod R., Zillhardt M. R., Romero I. L., Carey M. S., Mills G. B., Hotamisligil G. S., et al. 2011. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17: 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruby J. G., Jan C. H., and Bartel D. P.. 2007. Intronic microRNA precursors that bypass Drosha processing. Nature. 448: 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rottiers V., Najafi-Shoushtari S. H., Kristo F., Gurumurthy S., Zhong L., Li Y., Cohen D. E., Gerszten R. E., Bardeesy N., Mostoslavsky R., et al. 2011. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb. Symp. Quant. Biol. 76: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen B., Li H., Zeng X., Yang P., Liu X., Zhao X., and Liang S.. 2012. Roles of microRNA on cancer cell metabolism. J. Transl. Med. 10: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X., Chen J., Hu X., Huang Y., Li Z., Zhou L., Tian Z., Ma H., Wu Z., Chen M., et al. 2011. Comparative mRNA and microRNA expression profiling of three genitourinary cancers reveals common hallmarks and cancer-specific molecular events. PLoS One. 6: e22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gao P., Sun L., He X., Cao Y., and Zhang H.. 2012. MicroRNAs and the Warburg Effect: new players in an old arena. Curr. Gene Ther. 12: 285–291. [DOI] [PubMed] [Google Scholar]
- 86.Dang C. V., Le A., and Gao P.. 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15: 6479–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fei X., Qi M., Wu B., Song Y., Wang Y., and Li T.. 2012. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 586: 392–397. [DOI] [PubMed] [Google Scholar]
- 88.Laios A., O’Toole S., Flavin R., Martin C., Kelly L., Ring M., Finn S. P., Barrett C., Loda M., Gleeson N., et al. 2008. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol. Cancer. 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang R., Xiao T., Fang Z., Sun Y., Li F., Gao Y., Feng Y., Li L., Wang Y., Liu X., et al. 2012. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 287: 23227–23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Calin G. A., Cimmino A., Fabbri M., Ferracin M., Wojcik S. E., Shimizu M., Taccioli C., Zanesi N., Garzon R., Aqeilan R. I., et al. 2008. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA. 105: 5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu A. M., Xu Z., Shek F. H., Wong K. F., Lee N. P., Poon R. T., Chen J., and Luk J. M.. 2014. miR-122 targets pyruvate kinase M2 and affects metabolism of hepatocellular carcinoma. PLoS One. 9: e86872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He J., Xie G., Tong J., Peng Y., Huang H., Li J., Wang N., and Liang H.. 2014. Overexpression of microRNA-122 re-sensitizes 5-FU-resistant colon cancer cells to 5-FU through the inhibition of PKM2 in vitro and in vivo. Cell Biochem. Biophys. 70: 1343–1350. [DOI] [PubMed] [Google Scholar]
- 93.Kefas B., Comeau L., Erdle N., Montgomery E., Amos S., and Purow B.. 2010. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro-oncol. 12: 1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun Y., Zhao X., Zhou Y., and Hu Y.. 2012. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 28: 1346–1352. [DOI] [PubMed] [Google Scholar]
- 95.Guo C., Sah J. F., Beard L., Willson J. K., Markowitz S. D., and Guda K.. 2008. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 47: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagaraja A. K., Creighton C. J., Yu Z., Zhu H., Gunaratne P. H., Reid J. G., Olokpa E., Itamochi H., Ueno N. T., Hawkins S. M., et al. 2010. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol. Endocrinol. 24: 447–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fornari F., Milazzo M., Chieco P., Negrini M., Calin G. A., Grazi G. L., Pollutri D., Croce C. M., Bolondi L., and Gramantieri L.. 2010. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 70: 5184–5193. [DOI] [PubMed] [Google Scholar]
- 98.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., and Patel T.. 2007. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 133: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Godlewski J., Nowicki M. O., Bronisz A., Nuovo G., Palatini J., De Lay M., Van Brocklyn J., Ostrowski M. C., Chiocca E. A., and Lawler S. E.. 2010. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell. 37: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kikuchi H., Pino M. S., Zeng M., Shirasawa S., and Chung D. C.. 2009. Oncogenic KRAS and BRAF differentially regulate hypoxia-inducible factor-1alpha and -2alpha in colon cancer. Cancer Res. 69: 8499–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y., et al. 2009. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 28: 1385–1392. [DOI] [PubMed] [Google Scholar]
- 102.Tsang W. P., and Kwok T. T.. 2009. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 30: 953–959. [DOI] [PubMed] [Google Scholar]
- 103.Zhao W. G., Yu S. N., Lu Z. H., Ma Y. H., Gu Y. M., and Chen J.. 2010. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 31: 1726–1733. [DOI] [PubMed] [Google Scholar]
- 104.Sun T., Wang C., Xing J., and Wu D.. 2011. miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur. J. Cancer. 47: 2552–2559. [DOI] [PubMed] [Google Scholar]
- 105.Wu S., Lin Y., Xu D., Chen J., Shu M., Zhou Y., Zhu W., Su X., Qiu P., and Yan G.. 2012. MiR-135a functions as a selective killer of malignant glioma. Oncogene. 31: 3866–3874. [DOI] [PubMed] [Google Scholar]
- 106.Takwi A. A., Li Y., Becker Buscaglia L. E., Zhang J., Choudhury S., Park A. K., Liu M., Young K. H., Park W. Y., and Martin R. C.. 2012. A statin-regulated microRNA represses human c-Myc expression and function. EMBO Mol. Med. 4: 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gao P., Tchernyshyov I., Chang T. C., Lee Y. S., Kita K., Ochi T., Zeller K. I., De Marzo A. M., Van Eyk J. E., Mendell J. T., et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 458: 762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He X. Y., Chen J. X., Zhang Z., Li C. L., Peng Q. L., and Peng H. M.. 2010. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J. Cancer Res. Clin. Oncol. 136: 1023–1028. [DOI] [PubMed] [Google Scholar]
- 109.Le M. T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H. F., and Lim B.. 2009. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23: 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu W., Chan C. S., Wu R., Zhang C., Sun Y., Song J. S., Tang L. H., Levine A. J., and Feng Z.. 2010. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell. 38: 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamakuchi M., Ferlito M., and Lowenstein C. J.. 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA. 105: 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen H., Untiveros G. M., McKee L. A., Perez J., Li J., Antin P. B., and Konhilas J. P.. 2012. Micro-RNA-195 and -451 regulate the LKB1/AMPK signaling axis by targeting MO25. PLoS One. 7: e41574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Godlewski J., Bronisz A., Nowicki M. O., Chiocca E. A., and Lawler S.. 2010. microRNA-451: A conditional switch controlling glioma cell proliferation and migration. Cell Cycle. 9: 2742–2748. [PubMed] [Google Scholar]
- 114.Hardie D. G. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8: 774–785. [DOI] [PubMed] [Google Scholar]
- 115.Semenza G. L. 2010. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim J. W., Tchernyshyov I., Semenza G. L., and Dang C. V.. 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3: 177–185. [DOI] [PubMed] [Google Scholar]
- 117.Kong W., He L., Richards E. J., Challa S., Xu C. X., Permuth-Wey J., Lancaster J. M., Coppola D., Sellers T. A., Djeu J. Y., et al. 2014. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 33: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Babar I. A., Czochor J., Steinmetz A., Weidhaas J. B., Glazer P. M., and Slack F. J.. 2011. Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol. Ther. 12: 908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wan G., Xie W., Liu Z., Xu W., Lao Y., Huang N., Cui K., Liao M., He J., Jiang Y., et al. 2014. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 10: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Favaro E., Ramachandran A., McCormick R., Gee H., Blancher C., Crosby M., Devlin C., Blick C., Buffa F., Li J. L., et al. 2010. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 5: e10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li X., Chen Y. T., Josson S., Mukhopadhyay N. K., Kim J., Freeman M. R., and Huang W. C.. 2013. MicroRNA-185 and 342 inhibit tumorigenicity and induce apoptosis through blockade of the SREBP metabolic pathway in prostate cancer cells. PLoS One. 8: e70987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhong D., Zhang Y., Zeng Y. J., Gao M., Wu G. Z., Hu C. J., Huang G., and He F. T.. 2013. MicroRNA-613 represses lipogenesis in HepG2 cells by downregulating LXRalpha. Lipids Health Dis. 12: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 124.Esau C. C., and Monia B. P.. 2007. Therapeutic potential for microRNAs. Adv. Drug Deliv. Rev. 59: 101–114. [DOI] [PubMed] [Google Scholar]
- 125.Iliopoulos D., Drosatos K., Hiyama Y., Goldberg I. J., and Zannis V. I.. 2010. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J. Lipid Res. 51: 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Erdmann K., Kaulke K., Thomae C., Huebner D., Sergon M., Froehner M., Wirth M. P., and Fuessel S.. 2014. Elevated expression of prostate cancer-associated genes is linked to down-regulation of microRNAs. BMC Cancer. 14: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Casado P., Bilanges B., Rajeeve V., Vanhaesebroeck B., and Cutillas P. R.. 2014. Environmental stress affects the activity of metabolic and growth factor signaling networks and induces autophagy markers in MCF7 breast cancer cells. Mol. Cell. Proteomics. 13: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gerin I., Bommer G. T., McCoin C. S., Sousa K. M., Krishnan V., and MacDougald O. A.. 2010. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 299: E198–E206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lin Q., Gao Z., Alarcon R. M., Ye J., and Yun Z.. 2009. A role of miR-27 in the regulation of adipogenesis. FEBS J. 276: 2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsai W. C., Hsu S. D., Hsu C. S., Lai T. C., Chen S. J., Shen R., Huang Y., Chen H. C., Lee C. H., Tsai T. F., et al. 2012. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 122: 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gerin I., Clerbaux L. A., Haumont O., Lanthier N., Das A. K., Burant C. F., Leclercq I. A., MacDougald O. A., and Bommer G. T.. 2010. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 285: 33652–33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dávalos A., Goedeke L., Smibert P., Ramírez C. M., Warrier N. P., Andreo U., Cirera-Salinas D., Rayner K., Suresh U., Pastor-Pareja J. C., et al. 2011. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA. 108: 9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., and Stoffel M.. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 438: 685–689. [DOI] [PubMed] [Google Scholar]
- 134.Ørom U. A., Kauppinen S., and Lund A. H.. 2006. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 372: 137–141. [DOI] [PubMed] [Google Scholar]
- 135.Ma L., Reinhardt F., Pan E., Soutschek J., Bhat B., Marcusson E. G., Teruya-Feldstein J., Bell G. W., and Weinberg R. A.. 2010. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 28: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Babar I. A., Cheng C. J., Booth C. J., Liang X., Weidhaas J. B., Saltzman W. M., and Slack F. J.. 2012. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. USA. 109: E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kluiver J., Gibcus J. H., Hettinga C., Adema A., Richter M. K., Halsema N., Slezak-Prochazka I., Ding Y., Kroesen B. J., and van den Berg A.. 2012. Rapid generation of microRNA sponges for microRNA inhibition. PLoS One. 7: e29275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Behlke M. A. 2008. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 18: 305–319. [DOI] [PubMed] [Google Scholar]
- 139.Li L., Xie X., Luo J., Liu M., Xi S., Guo J., Kong Y., Wu M., Gao J., Xie Z., et al. 2012. Targeted expression of miR-34a using the T-VISA system suppresses breast cancer cell growth and invasion. Mol. Ther. 20: 2326–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang X., Han L., Zhang A., Wang G., Jia Z., Yang Y., Yue X., Pu P., Shen C., and Kang C.. 2011. Adenovirus-mediated shRNAs for co-repression of miR-221 and miR-222 expression and function in glioblastoma cells. Oncol. Rep. 25: 97–105. [PubMed] [Google Scholar]
- 141.Lu P. Y., Xie F., and Woodle M. C.. 2005. In vivo application of RNA interference: from functional genomics to therapeutics. Adv. Genet. 54: 117–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Costa P. M., and Pedroso de Lima M. C.. 2013. MicroRNAs as molecular targets for cancer therapy: on the modulation of microRNA expression. Pharmaceuticals (Basel). 6: 1195–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]