Abstract
Cluster of differentiation 44 (CD44) is a transmembrane glycoprotein that serves as the receptor for the extracellular matrix component hyaluronic acid. CD44 has been reported to play key roles in cell proliferation, motility, and survival, but its role in breast cancer remains controversial. In this study, we conducted a meta-analysis. A total of 23 published Gene Expression Omnibus databases were included to evaluate the association between CD44 mRNA expression and clinicopathological characteristics or prognosis of the patients with breast cancer. Our analysis revealed that CD44 expression was associated with clinicopathological features, including the histological grade, estrogen receptor status, progesterone receptor status, and human epidermal growth factor receptor-2 status. Higher levels of CD44 expression were observed in the basal subtype of breast cancer both at the mRNA and protein levels (odds ratio [OR] =2.08, 95% confidence interval [CI]: 1.72–2.52; OR =2.11, 95% CI: 1.67–2.68). Patients with CD44 overexpression exhibited significantly worse overall survival (hazard ratio =1.27; 95% CI: 1.04–1.55). Whole gene profile analysis revealed that CD44 expression was enriched in basal-type breast cancer and correlated with epithelial–mesenchymal transition and cancer stem cell gene profiles. In summary, our analyses indicated that CD44 potentially might be a prognostic marker for breast cancer and thus can serve as a therapeutic target for basal-type breast cancer.
Keywords: breast cancer, CD44, survival prediction, meta-analysis, biomarker
Introduction
Breast cancer is one of the most common female cancers, accounting for approximately 28% of all female cancers and the second leading cause of cancer-related deaths in women.1 Progress has been made to the earlier diagnosis and better treatment of breast cancer during the past few decades, leading to the 5-year survival rates of breast cancer patients at approximately 85%. However, distant metastasis and recurrence still occur and result in poor prognosis. Therefore, there is an urgent need for identifying novel biomarkers that can be used to screen high-risk patients and help predict the progression and prognosis of breast cancer.2–4
Cluster of differentiation 44 (CD44) is a complex transmembrane glycoprotein that is encoded by the CD44 gene on chromosome 11p13.5 CD44 consists of seven extracellular domains, a transmembrane domain, and a cytoplasmic domain.6 CD44 has several isoforms, including CD44s and CD44v.7,8 Functionally, CD44 was initially identified as the receptor for the extracellular matrix component, hyaluronic acid (HA), and was involved in multiple physiological and pathological processes, such as angiogenesis, cell adhesion, inflammation, and cancer development.9 In addition, CD44 has been reported to play important roles in cell proliferation, motility, and survival.9,10 A recent study indicated that CD44 expression was elevated in tumor-initiating cells in many kinds of cancer.11 Thus, CD44 is thought to be a biomarker for cancer stem cells (CSCs).12 Subsequent functional studies have shown that CD44 is involved in tumorigenesis and metastasis in many cancer types such as colon,13–15 bladder,16 gastric,17 and breast cancers.18–20 Studies on CD44 expression have suggested a correlation between it and clinical outcome in patients with breast cancer. It has been shown that the overexpression of CD44 has a bad impact on survival of breast cancer patients,21 but different results were also reported.22 Currently, the role of CD44 in breast cancer has not been clearly defined. To investigate the role of CD44 in breast cancer, a meta-analysis was performed. Our analysis indicated that CD44 expression was elevated in basal-type breast cancer. Currently, there are no effectively targeted therapies for patients with this subtype of breast cancer and prognosis is poor compared with other subtypes.23 Since CD44 expression is associated with mesenchymal and CSC signature and predicts poor prognosis,24,25 our study indicates that CD44 may represent a potential therapeutic target for basal-type breast cancer.
Materials and methods
Database and literature search
We performed a comprehensive search of relevant Gene Expression Omnibus (GEO) databases for CD44 mRNA expression and literatures for CD44 protein level. First, we searched the ArrayExpress for uploaded databases within the topic of interest, using the search terms “breast cancer” by filtering Homo sapiens, RNA array, array assay, and all arrays. We also searched Oncomine for databases of breast cancer with mRNA information of CD44. Second, PubMed was reviewed to identify potentially relevant literatures using the search terms associated with CD44 (“CD44 antigen”, “hyaluronan-binding protein”, “receptors”, “hyaluronan”) and breast cancer (“breast neoplasm”, “breast tumor”, “breast carcinoma”, “mammary cancer”). The references were also searched to discover additional relevant publications.
Inclusion and exclusion criteria
This meta-analysis collected data aimed at evaluating the role of CD44 expression in breast cancer at both mRNA and protein levels. Databases that met the following criteria were included: 1) the datasets were about breast cancer; 2) CD44 expression was measured in these databases; 3) the sample capacity was >50; and 4) clinical information of patients was showed in these databases. The exclusion criteria were as follows: 1) the datasets were about animals such as mice and rabbits and 2) the datasets were about DNA, rather than RNA. When several databases shared the same patient population, only the latest and most complete datasets were included. Literature that met the following criteria were included: 1) patients recruited in the study were pathologically diagnosed with breast cancer; 2) CD44 expression was measured in breast cancer tissues; and 3) the hazard ratio (HR)/odds ratio (OR) and corresponding 95% confidence interval (CI) were reported or could be statistically extracted from the study. The exclusion criteria were as follows: 1) reviews, case reports, comments, letters, and conference abstract and 2) ineligible samples or those where the required data were not available. When several articles were from the same patient population, the latest or most complete article was included.
Data extraction
Data were abstracted in a standardized collection form, with information recorded as follows: last name of first author, publication year, country, duration, tumor–node–metastasis (TNM) stage, quality score, detection, and cutoff values for CD44. We reviewed ArrayExpress and Oncomine and identified 23 independent human breast cancer microarray datasets with CD44 mRNA expression and clinical data. Overall survival (OS), recurrence-free survival (RFS), and metastasis-free survival (MFS) were evaluated by Cox proportional HRs and 95% CIs using these numerical data. If HRs were not given in an article, we used the methods described by Tierney et al to calculate the statistical variables from published survival curves.26 The quality of observational studies was evaluated according to the Newcastle–Ottawa Quality Assessment Scale. This scale reflects patient selection, study comparability, and outcomes and is based on the identification of eight sources of potential study bias. Two reviewers performed the literature search, study selection, and data abstraction independently, and disagreements between the reviewers were solved by discussion.
Statistical analysis
Statistical analysis was performed based on the requirements of the meta-analysis of observational studies. The STATA software package (Version 12.0; StataCorp LP, College Station, TX, USA) was utilized to perform the meta-analysis. The random-effect model was employed when heterogeneity was present, and the fixed-effect model was used when homogeneity was demonstrated. The heterogeneity of publication was evaluated by means of the chi-square-based Q statistic and inconsistency index (I2) statistic. Begg’s and Egger’s tests were employed to assess the publication bias. HRs were employed to assess the survival outcome of patients with breast cancer who had high CD44 expression, and HR >1 indicated that high expression of CD44 predicted worse survival of patients. The OR and 95% CI were used to evaluate the association between CD44 expression and clinicopathological parameters.
Results
Search results
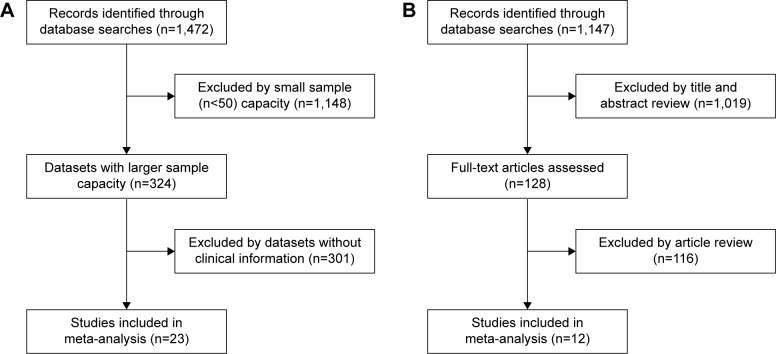
The flow diagram for the identification of relevant studies is shown in Figure 1. A total of 1,472 datasets and 1,147 literatures were initially identified by our search approach. For GEO databases, after the sample capacity and clinical information were checked, 23 datasets21,27–48 met the criteria for this analysis. For 1,147 literatures, after title/abstract scanning and full-text reading, 12 eligible articles22,49–59 were included. Table 1 shows the features of these 23 studies. Four Gene Expression Omnibus series (GSE) datasets were analyzed for finding the difference in CD44 mRNA expression between breast tumors and normal breast tissues. For finding the association between CD44 mRNA expression and TNM stage, tumor grade, estrogen receptor (ER) status, progesterone receptor (PR) status, human epidermal growth factor receptor-2 (HER2) status, and basal-like breast cancer, four, 13, eleven, four, six, and seven GSE datasets, respectively, were analyzed. To estimate the prognostic role of CD44 mRNA expression in OS, RFS, and MFS, eleven, ten, and nine, respectively, GSE datasets were adopted. Three GSE datasets were analyzed for the association between CD44 mRNA expression and the RFS in basal-like breast cancer. Table 2 shows the characteristics of 12 studies. A total of nine, eight, seven, and five articles were assessed for the correlation between CD44 protein abundance and ER status, PR status, HER2 status, and basal-like breast cancer, respectively. Clinical stages I and II were grouped as early-stage disease, whereas stages III and IV were grouped as late-stage disease. Clinical T stages 1 and 2 were identified as early T stage, and 3 and 4 were identified as late T stage. Clinical N stages 1 and 2 were classified into early N stage, and 3 and 4 were classified into late N stage. Histological grades I and II were pooled as low-grade disease, and III and IV were pooled as high-grade disease.
Figure 1.
Flow diagram of article selection.
Notes: (A) Initial dataset search and selection process for CD44 mRNA expression in this meta-analysis. (B) Initial dataset search and selection process for CD44 protein abundance in this meta-analysis.
Abbreviation: CD44, cluster of differentiation 44.
Table 1.
Characteristics of the included studies by CD44 mRNA expression in the meta-analysis
| References | Year | Country or area | Duration (months) | Stage | Quality score | Detection | Cutoff values | Patients with CD44 overexpression (total number) |
|---|---|---|---|---|---|---|---|---|
| Desmedt et al21 | 2007 | Canada | NA | NA | 8 | Microarray | Median expression: 9.34 | 99 (198) |
| Heikkinen et al27 | 2011 | Finland | 36 | NA | 8 | Microarray | Median expression: 11.05 | 92 (183) |
| Terunuma et al28 | 2014 | USA | NA | I–III | 9 | Microarray | Median expression: 11.12 | 31 (61) |
| Sircoulomb et al29 | 2010 | France | 240 | NA | 7 | Microarray | Median expression: 7.60 | 26 (51) |
| Clarke et al30 | 2013 | Canada | 180 | NA | 8 | Microarray | Median expression: NA | 39 (77) |
| Ma et al31 | 2009 | USA | 36 | NA | 7 | Microarray | Median expression: 2.33 | 19 (38) |
| Hatzis et al32 | 2011 | USA | 120 | I–IV | 9 | Microarray | Median expression: 9.63 | 254 (508) |
| Tofigh et al33 | 2014 | Canada | NA | I–IV | 8 | Microarray | Median expression: 11.63 | 161 (321) |
| Kao et al34 | 2011 | Taiwan | 156 | I–IV | 9 | Microarray | Median expression: 8.06 | 164 (327) |
| Minn et al35 | 2005 | USA | NA | NA | 8 | Microarray | Median expression: 1,674.60 | 50 (99) |
| Desmedt et al36 | 2011 | Canada | NA | NA | 8 | Microarray | Median expression: 2.94 | 61 (120) |
| Schmidt et al37 | 2008 | Germany | 120 | NA | 9 | Microarray | Median expression: 1,595 | 100 (200) |
| Nagalla et al38 | 2013 | USA | NA | NA | 8 | Microarray | Median expression: 10.33 | 70 (139) |
| Loi et al39 | 2007 | Canada | NA | NA | 9 | Microarray | Median expression: 6.52 | 164 (327) |
| Dedeurwaerder et al40 | 2011 | Canada | NA | NA | 8 | Microarray | Median expression: 7.18 | 45 (90) |
| Miller et al41 | 2005 | Singapore | NA | NA | 9 | Microarray | Median expression: 8.04 | 126 (251) |
| Symmans et al42 | 2010 | USA | NA | NA | 9 | Microarray | Median expression: 10.78 | 149 (298) |
| Pawitan et al43 | 2005 | Sweden | 24 | NA | 8 | Microarray | Median expression: 7.04 | 80 (159) |
| Wang et al44 | 2005 | USA | 180 | NA | 9 | Microarray | Median expression: 1,285.15 | 143 (286) |
| Hu et al45 | 2009 | USA | NA | NA | 7 | Microarray | Median expression: NA | 40 (80) |
| Hennessy et al46 | 2009 | USA | NA | NA | 9 | Microarray | Median expression: NA | 47 (94) |
| Bild et al47 | 2006 | USA | NA | NA | 9 | Microarray | Median expression: 62.00 | 80 (158) |
| Minn et al48 | 2007 | USA | NA | NA | 7 | Microarray | Median expression: 1,636.50 | 29 (58) |
Abbreviations: CD44, cluster of differentiation 44; NA, not available.
Table 2.
Characteristics of the included studies by CD44 protein abundance in the meta-analysis
| References | Year | Country or area | Duration (months) | Stage | Quality score | Detection | Cutoff values | Patients with CD44 overexpression (total number) |
|---|---|---|---|---|---|---|---|---|
| Dan et al22 | 2014 | USA | 96 | I–II | 8 | IHC | Staining of score | 22 (51) |
| Adamczyk et al49 | 2014 | Poland | 120 | NA | 9 | IHC | Percentage of immunopositive cells >10% | 70 (144) |
| Wu et al50 | 2013 | USA | 144 | NA | 8 | IHC | Staining of H-score | 60 (126) |
| Bernardi et al51 | 2012 | Brazil | 72 | I–IV | 9 | IHC | Staining of score | 55 (78) |
| Uchoa Dde et al52 | 2014 | Brazil | 60 | NA | 8 | IHC | Percentage of immunopositive cells >0.1% | 34 (94) |
| Ahmed et al53 | 2012 | Egypt | 96 | NA | 9 | IHC | Staining of H-score | 501 (786) |
| Wang et al54 | 2011 | People’s Republic of China | NA | NA | 8 | IHC | Percentage of immunopositive cells >10% | 51 (117) |
| Ricardo et al55 | 2011 | Portugal | 168 | NA | 7 | IHC | Percentage of immunopositive cells >10% | 237 (463) |
| Kim et al56 | 2011 | Korea | 60 | NA | 8 | IHC | Percentage of immunopositive cells >10% | 173 (643) |
| Giatromanolaki et al57 | 2011 | Greece | 156 | NA | 9 | IHC | Percentage of immunopositive cells >10% | 72 (139) |
| Looi et al58 | 2006 | Malaysia | NA | NA | 9 | IHC | Percentage of immunopositive cells >10% | 24 (60) |
| Han et al59 | 2015 | People’s Republic of China | 48 | I–IV | 8 | IHC | Staining of score: 0–2 vs 3–12 | 175 (325) |
Abbreviations: CD44, cluster of differentiation 44; IHC, immunohistochemistry; NA, not available.
CD44 expression correlates with clinicopathological features of breast cancer
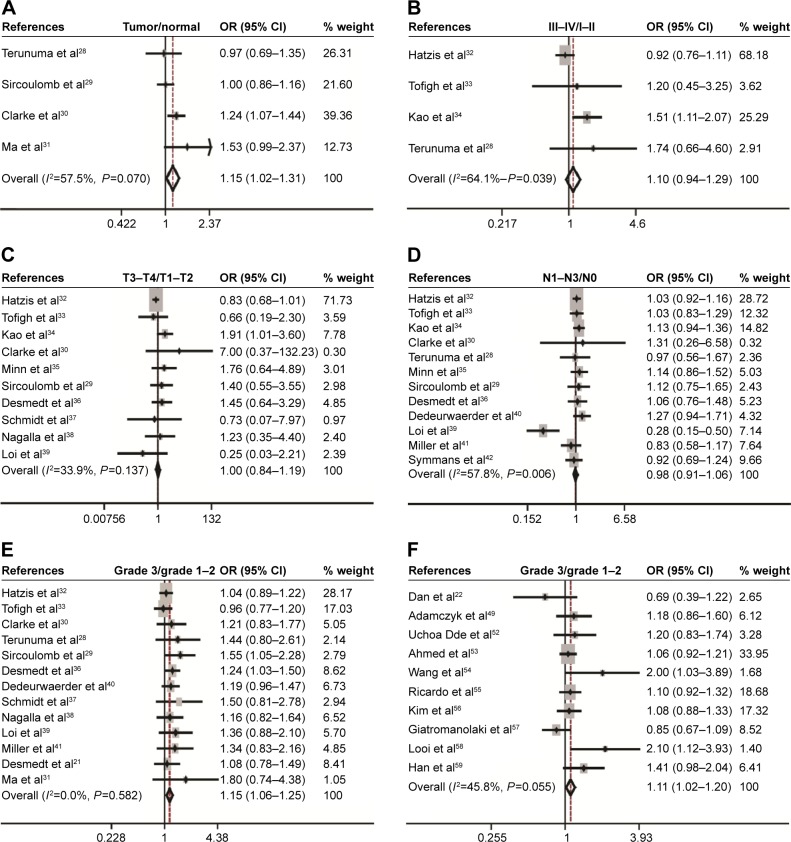
Eighteen studies assessed the association between CD44 mRNA expression and tumor clinicopathological features. Our meta-analysis indicated that CD44 expression in breast cancer tissues was increased when compared with that in normal breast tissues (pooled OR =1.15, 95% CI: 1.02–1.31, Cochran’s Q test P=0.070, and I2=57.5%; Figure 2A). However, there was no statistically significant correlation between CD44 expression and tumor TNM stage (pooled OR =1.10, 95% CI: 0.94–1.29, Cochran’s Q test P=0.039, and I2=64.1%; Figure 2B), T stage (pooled OR =1.00, 95% CI: 0.84–1.19, Cochran’s Q test P=0.137, and I2=33.9%; Figure 2C), and N status (pooled OR =0.98, 95% CI: 0.91–1.06, Cochran’s Q test P=0.006, and I2=57.8%; Figure 2D). Patients with breast cancer with higher histological grade were likely to have a higher content of CD44 at both mRNA (pooled OR =1.15, 95% CI: 1.06–1.25, Cochran’s Q test P=0.582, and I2=0.0%; Figure 2E) and protein levels (pooled OR =1.11, 95% CI: 1.02–1.20, Cochran’s Q test P=0.055, and I2=45.8%; Figure 2F).
Figure 2.
Correlation between CD44 mRNA expression or CD44 protein and breast cancer development and progression as evaluated by the OR.
Notes: Association between CD44 mRNA expression with breast cancer risk compared with normal breast tissue (A), clinical TNM stage (B), T stage (C), N status (D), and histological grade (E). Association between CD44 protein and tumor grade (F).
Abbreviations: CD44, cluster of differentiation 44; CI, confidence interval; OR, odds ratio; TNM, tumor–node–metastasis.
CD44 expression correlates with molecular subtypes of breast cancer
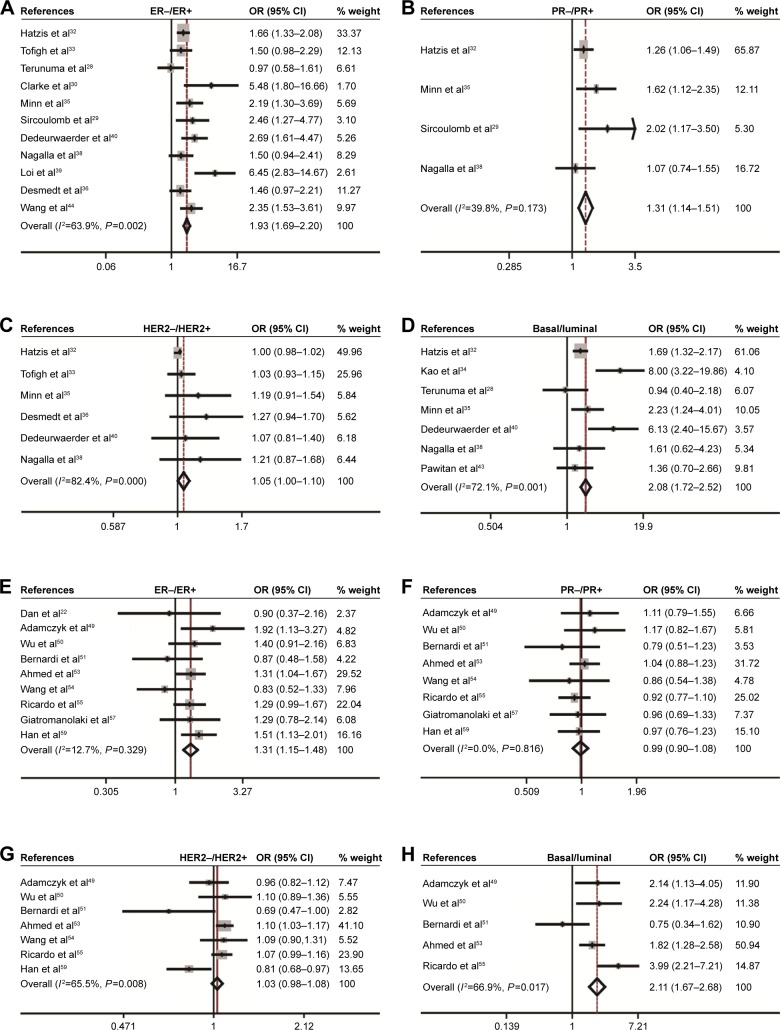
The association of CD44 expression with ER, PR, HER2 status, and basal-like breast cancer was also analyzed. At the mRNA level, CD44 was inversely correlated with ER status (pooled OR =1.93, 95% CI: 1.69–2.20, Cochran’s Q test P=0.002, and I2=63.9%; Figure 3A), PR status (pooled OR =1.31, 95% CI: 1.14–1.51, Cochran’s Q test P=0.173, and I2=39.8%; Figure 3B), and HER2 status (pooled OR =1.05, 95% CI: 1.00–1.10, Cochran’s Q test P=0.000, and I2=82.4%; Figure 3C). Interestingly, CD44 mRNA expression was higher in basal-like tumors than in the luminal subtype of breast cancer (pooled OR =2.08, 95% CI: 1.72–2.52, Cochran’s Q test P=0.001, and I2=72.1%; Figure 3D). At the protein level, CD44 expression was conversely linked to ER status (pooled OR =1.31, 95% CI: 1.15–1.48, Cochran’s Q test P=0.329, and I2=12.7%; Figure 3E). However, there is no statistical significance in terms of an association between CD44 expression and PR status (pooled OR =0.99, 95% CI: 0.90–1.08, Cochran’s Q test P=0.816, and I2=0.0%; Figure 3F) or HER2 status (pooled OR =1.03, 95% CI: 0.98–1.08, Cochran’s Q test P=0.008, and I2=65.5%; Figure 3G) at protein level. Moreover, CD44 protein abundance in basal-like tumors was much higher than in the luminal subtype of breast cancer (pooled OR =2.11, 95% CI: 1.67–2.68, Cochran’s Q test P=0.017, and I2=66.9%; Figure 3H).
Figure 3.
Association between CD44 expression and molecular subtype.
Notes: Association between CD44 mRNA with ER status (A), PR (B), HER2 (C), and basal–luminal (D). Association between CD44 protein with ER status (E), PR (F), HER2 (G), and basal–luminal (H).
Abbreviations: CD44, cluster of differentiation 44; CI, confidence interval; ER, estrogen receptor; HER2, human epidermal growth factor receptor-2; OR, odds ratio; PR, progesterone receptor.
CD44 mRNAexpression correlates with breast cancer survival
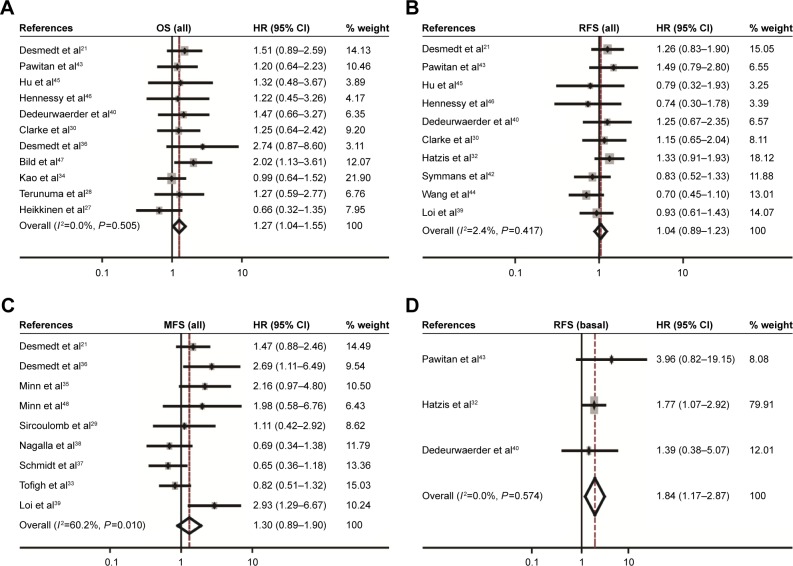
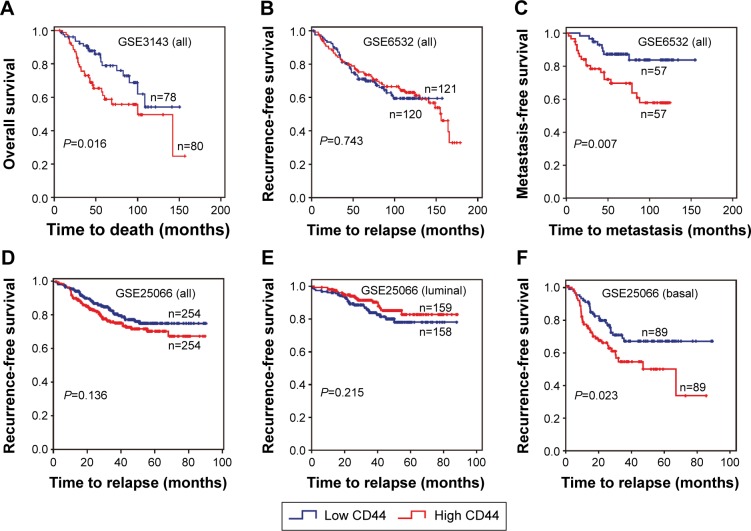
The association between CD44 expression level and breast cancer patient survival was analyzed. Our analysis indicated that there was a significant correlation between CD44 overexpression and the poor OS rate (pooled OR =1.27, 95% CI: 1.04–1.55, Cochran’s Q test P=0.505, and I2=0.0%; Figure 4A). However, CD44 expression was not statistically significant in terms of an association between the RFS rate (pooled OR =1.04, 95% CI: 0.89–1.23, Cochran’s Q test P=0.417, and I2=2.4%; Figure 4B) and the MFS rate (pooled OR =1.30, 95% CI: 0.89–1.90, Cochran’s Q test P=0.010, and I2=60.2%; Figure 4C). Subcategory analyses according to the molecular classification of breast cancer were also performed. We found that higher CD44 mRNA expression correlated with worse RFS in patients with basal-like breast cancer (pooled OR =1.84, 95% CI: 1.17–2.87, Cochran’s Q test P=0.574, and I2=0.0%; Figure 4D). However, there was no statistically significant correlation between CD44 mRNA expression and the survival performance of patients with luminal subtype of breast cancer. The latter included the OS rate (pooled OR =1.14, 95% CI: 0.73–1.79, Cochran’s Q test P=0.296, and I2=17.8%), the RFS rate (pooled OR =0.99, 95% CI: 0.75–1.31, Cochran’s Q test P=0.258, and I2=23.5%), and the MFS rate (pooled OR =1.25, 95% CI: 0.65–2.38, Cochran’s Q test P=0.010, and I2=69.7%). Kaplan–Meier survival analysis of GSE3143 demonstrated that there was a significant effect of CD44 on OS (P=0.016; Figure 5A). Kaplan–Meier survival analysis of GSE6532 showed that there were no significant effects of CD44 on the RFS in all population of breast cancer (P=0.743; Figure 5B), but it was inversely associated with the MFS rate (P=0.007; Figure 5C). Kaplan–Meier survival analysis of GSE25066 demonstrated that there was a significant effect of CD44 on the RFS in basal-like breast cancer (P=0.023; Figure 5F) but no significant association between CD44 mRNA expression and the RFS in all molecular subtypes (P=0.136; Figure 5D) or in luminal breast cancer (P=0.215; Figure 5E). In all, the results from the CD44 mRNA profile indicated that higher CD44 expression predicted a poorer prognosis in patients with breast cancer subtype.
Figure 4.
Forest plot for the correlation of CD44 mRNA expression with breast cancer survival.
Notes: Associations between CD44 mRNA expression with breast cancer OS (A), RFS (B), MFS (C) in all population of breast cancer, and RFS (D) in patients with basal-like breast cancer.
Abbreviations: CD44, cluster of differentiation 44; CI, confidence interval; HR, hazard ratio; MFS, metastasis-free survival; OS, overall survival; RFS, recurrence-free survival.
Figure 5.
Kaplan–Meier survival curves for the correlation of CD44 mRNA expression with breast cancer.
Notes: (A) CD44 mRNA expression with respect to OS in patients with breast cancer (GSE3143). CD44 mRNA expression with respect to RFS (B) and MFS (C) in all patients with breast cancer (GSE6532). CD44 mRNA expressions with respect to RFS in all patients (D), those with luminal subtype (E), and those with basal-like breast cancer (F) in database GSE25066.
Abbreviations: CD44, cluster of differentiation 44; MFS, metastasis-free survival; OS, overall survival; RFS, recurrence-free survival.
CD44 correlates with epithelial–mesenchymal transition and CSC markers
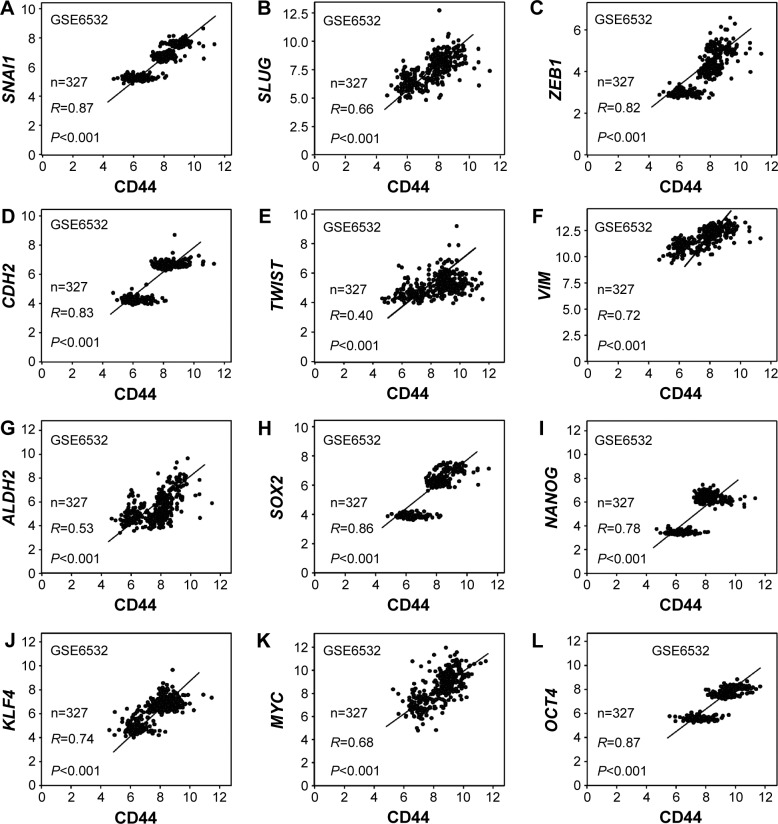
The association between CD44 and epithelial–mesenchymal transition (EMT) or CSC-related genes was also assessed. The results indicated that there was a positive relation between CD44 and SNAI1 (R=0.87, P<0.001; Figure 6A), SLUG (R=0.66, P<0.001; Figure 6B), ZEB1 (R=0.82, P<0.001; Figure 6C), CDH2 (R=0.83, P<0.001; Figure 6D), TWIST (R=0.40, P<0.001; Figure 6E), and VIM (R=0.72, P<0.001; Figure 6F). The association between CD44 and CSC markers was also evaluated. It was shown that CD44 was positively associated with ALDH1 (R=0.53, P<0.001; Figure 6G), SOX2 (R=0.86, P<0.001; Figure 6H), NANOG (R=0.78, P<0.001; Figure 6I), KLF4 (R=0.74, P<0.001; Figure 6J), MYC (R=0.68, P<0.001; Figure 6K), and OCT4 (R=0.87, P<0.001; Figure 6L).
Figure 6.
CD44 expression was associated with stem cell and EMT markers.
Notes: Association between mRNA expressions of CD44 with EMT-related genes SNAI1 (A), SLUG (B), ZEB1 (C), CDH2 (D), TWIST (E), and VIM (F). Association between mRNA expression of CD44 and stem cell factors ALDH1 (G), SOX2 (H), NANOG (I), KLF4 (J), MYC (K), and OCT4 (L).
Abbreviations: CD44, cluster of differentiation 44; EMT, epithelial–mesenchymal transition.
Publication bias
Publication bias statistics were obtained using Begg’s and Egger’s tests, and did not indicate any significant publication bias; CD44 mRNA expression: breast cancer: Begg’s test P=0.734, Egger’s test P=0.905; TNM stage: Begg’s test P=1, Egger’s test P=0.796; tumor size: Begg’s test P=0.466, Egger’s test P=0.362; lymph node metastasis: Begg’s test P=0.945, Egger’s test P=0.097; histological grade: Begg’s test P=0.246, Egger’s test P=0.948; expression of ER: Begg’s test P=0.640, Egger’s test P=0.313; expression of PR: Begg’s test P=1, Egger’s test P=0.809; expression of Her2: Begg’s test P=0.260, Egger’s test P=0.494; basal-like breast cancer: Begg’s test P=1, Egger’s test P=0.77; OS: Begg’s test P=0.436, Egger’s test P=0.436; RFS: Begg’s test P=0.592, Egger’s test P=0.612; MFS: Begg’s test P=0.251, Egger’s test P=0.146; OS of luminal breast cancer: Begg’s test P=1, Egger’s test P=0.642; RFS of luminal breast cancer: Begg’s test P=0.260, Egger’s test P=0.436; MFS of luminal breast cancer: Begg’s test P=0.806, Egger’s test P=0.528; RFS of basal-like breast cancer: Begg’s test P=1, Egger’s test P=0.698. Protein level: histological grade: Begg’s test P=0.917, Egger’s test P=0.911; expression of ER: Begg’s test P=0.917, Egger’s test P=0.009; expression of PR: Begg’s test P=0.266, Egger’s test P=0.743; expression of HER2: Begg’s test P=1, Egger’s test P=0.434; and basal-like breast cancer: Begg’s test P=0.462, Egger’s test P=0.065.
Discussion
Molecular characterization contributes to the discovery of biomarkers and potential targets for anticancer therapy, which is the basis of precise medicine.60 Accumulating evidence suggests that CD44 is a marker of tumor-initiating cells, plays a role in tumorigenesis, and linked to the progression of breast cancer.15,61–63 CD44 was also reported to have an impact on the prognosis of breast cancer including recurrence64 and chemoresistance.65 Uchino et al found that the upregulation of CD44 represented an aggressive subtype in noninvasive breast cancer cell.19 The blockade of CD44 intracellular domain (CD44ICD) cleavage and nuclear translocation have been shown in cancer cells. The activation of CD44 by HA promoted the chemoresistance in breast cancer cells.66 CD44/cellular prion protein interaction has an effect on the responses to neoadjuvant chemotherapy in patients with breast cancer and exhibits aggressive behaviors of breast cancer cells.67 CD44–STAT3 interaction plays an important role in breast cancer invasion.64 Moreover, Cox regression analysis showed that ezrin and CD44 co-expression were independent prognostic factors of breast cancer.68
In our meta-analysis, the role of CD44 in breast cancer, at both mRNA and protein levels, was investigated. We found that the mRNA level of CD44 was higher in breast tumor tissues than in normal breast tissues, indicating that CD44 might participate in the tumorigenesis of specific subtypes of breast cancer. Moreover, our meta-analysis suggests a positive association between histological grade and the CD44 levels. This would indicate that patients with high expression of CD44 mRNA might have poor prognosis, because high-grade tumor tends to be more aggressive and tends toward early recurrence. It has been shown that CD44 was activated in breast cancer cells but inactivated in normal cells in vitro and in vivo.69 However, the association between CD44 mRNA expression and TNM stage, T stage, and N stage was not statistically significant.
Based on the status of ER, PR, and HER2, breast cancer could be divided into five molecular subtypes, including normal-like, luminal A, luminal B, HER2-overexpressing, and basal-like breast cancer.70,71 Each subtype exhibits distinctive expression patterns of specific molecules, clinical outcomes, and responses to adjuvant chemotherapy.33,72,73 Some studies indicated that CD44 expression was negatively associated with the status of ER,49,53,74,75 PR,74 and HER2.53,74 At the mRNA level, our meta-analysis showed that CD44 expression was significantly inversely associated with the status of ER, PR, and HER2. Consistently, significant correlation between CD44 expression and ER status was found at its protein level. Among the five molecular subtypes, basal-like breast cancer tends to be more aggressive and there is a lack of effective therapy, resulting in poorer outcomes.76 Jang et al showed that CD44(+)/CD24(−) subpopulation was much higher in basal-like breast cancer than that in non-basal-like cancer,24 and that CD44(+)/CD24(−) cells had a high capacity of proliferation, migration, invasion, and tumorigenesis.25 By providing a highly hydrated environment favoring cellular invasion, HA–CD44 interaction contributed to the progression of basal-like breast cancer.77 Consistently, our results showed that CD44 expression was higher in basal-like breast cancers than in luminal breast cancer or all other subtypes.
CD44 is critical for regulating EMT.19 CD44 activation can lead to the expression of epithelial growth factor receptor and the activation of phosphoinositide-3 kinase/Akt. CD44 also upregulates N-cadherin, α-actin, vimentin, fibronectin, and other EMT markers. The latter is involved in cell invasion and migration.78 By knocking down CD44 expression in human hepatoma cell line HLE, the levels of snail and vimentin were decreased, which was correlated with a less-mesenchymal-like phenotype.79 Consistently, our analysis indicated that CD44 expression was significantly associated with mesenchymal gene SNAI1, SLUG, ZEB1, CDH2, and TWIST.
CD44 is a well-known breast CSC marker that plays a role in promoting tumorigenesis of breast cancer through interaction with its intracellular domain and stemness factors such as NANOG, OCT4, and SOX2.20 Analysis of gene expression profiles revealed that CD44 is closely associated with key stem cell genes ALDH1, SOX2, NANOG, KLF4, OCT4, and MYC. Since CSC is thought to be a major cause for cancer progression and therapeutic resistance,80,81 the role of CD44 in breast cancer might be attributable to those stem cell factors.
Studies identified several genes that might have prognostic values for breast cancer, including urokinase plasminogen activator and its inhibitor82 and the genes in the DACH–EYA–SIX pathway.83–85 Interestingly, insulin-like growth factor 1 receptor expression showed different prognostic values for patients with different subtypes of breast cancer.86 Ubiquitin protein D and KLF4 have been reported to predict the response to chemotherapy.87,88 Accumulating evidence indicates that CD44 could be a prognostic biomarker for breast cancer.12 Our meta-analysis suggested that CD44 high expression could be a prognostic marker for OS. Although there was no association between CD44 expression and RFS in the whole population of breast cancer, a significant association between CD44 mRNA expression and RFS in patients with basal-like breast cancer was identified. This agrees with a previous study showing that patients with CSC markers CD44(+)/CD24(−) had a lower survival rate, while patients without this subpopulation had a higher survival rate in basal-like breast cancer.89 Some studies showed that CD44 expression was positively correlated with the metastasis of breast carcinoma,75 but others reported opposite results.90 Martin and Jiang found that CD44 was markedly reduced in patients with ductal breast cancer with metastasis.91 Our meta-analysis showed that CD44 expression has no significant effect on the MFS (Figure 4C), but some GSE data did demonstrate that CD44 was correlated with the MFS (Figure 5C). Breast cancer metastasis is a complicated process which is involved in the alteration of a number of proteins, including epithelial growth factor receptor and transforming growth factor-β.92 Considering the complex regulation of the metastasis process of breast cancer, the effects of CD44 on the MFS might be covered by other factors.
Heterogeneity tests are essential to a meta-analysis. In this study, the evidence of minor heterogeneities was observed with respect to TNM stage, ER status, molecular subtypes, and the MFS. However, there was substantial heterogeneity with respect to HER2 status. This result might be due to the following aspects: 1) The sample size is limited, indicating that multicenter prospective studies are needed. 2) The variations in assessing CD44 mRNA expression might also contribute to heterogeneity. The cutoff value was estimated in 23 studies using the median CD44 level measured by gene microarray. 3) Publication bias is worth considering in meta-analyses. This study was a meta-analysis based on GEO datasets and published studies. Thus, our analysis has the following limitations: 1) we cannot exclude the publication bias; 2) the relevant papers were limited; and 3) methods and cutoff values used to assess CD44 expression were different.
Conclusion
In conclusion, our meta-analysis suggests that CD44 might be a prognostic factor for patients with breast cancer, particularly for the basal-like breast cancer. Since CD44 expression was elevated in basal-type breast cancer and its expression levels were correlated with EMT and CSC signatures, these considerations might partially explain why patients with basal-type breast cancer have a high risk of metastasis and relapse. Moreover, our meta-analysis might help identify subpopulation of patients with breast cancer for CD44-based therapy in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos 81572608, 81172422, and 81072169).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Radenkovic S, Milosevic Z, Konjevic G, et al. Lactate dehydrogenase, catalase, and superoxide dismutase in tumor tissue of breast cancer patients in respect to mammographic findings. Cell Biochem Biophys. 2013;66(2):287–295. doi: 10.1007/s12013-012-9482-7. [DOI] [PubMed] [Google Scholar]
- 3.Radenkovic S, Konjevic G, Jurisic V, et al. Values of MMP-2 and MMP-9 in tumor tissue of basal-like breast cancer patients. Cell Biochem Biophys. 2014;68(1):143–152. doi: 10.1007/s12013-013-9701-x. [DOI] [PubMed] [Google Scholar]
- 4.Stankovic S, Konjevic G, Gopcevic K, et al. Activity of MMP-2 and MMP-9 in sera of breast cancer patients. Pathol Res Pract. 2010;206(4):241–247. doi: 10.1016/j.prp.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Screaton GR, Caceres JF, Mayeda A, et al. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14(17):4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Windt GJ, Schouten M, Zeerleder S, et al. CD44 is protective during hyperoxia-induced lung injury. Am J Respir Cell Mol Biol. 2011;44(3):377–383. doi: 10.1165/rcmb.2010-0158OC. [DOI] [PubMed] [Google Scholar]
- 7.Xu H, Tian Y, Yuan X, et al. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco Targets Ther. 2015;8:3783–3792. doi: 10.2147/OTT.S95470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erb U, Megaptche AP, Gu X, et al. CD44 standard and CD44v10 isoform expression on leukemia cells distinctly influences niche embedding of hematopoietic stem cells. J Hematol Oncol. 2014;7:29. doi: 10.1186/1756-8722-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 10.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 11.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 12.Basakran NS. CD44 as a potential diagnostic tumor marker. Saudi Med J. 2015;36(3):273–279. doi: 10.15537/smj.2015.3.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du L, Wang H, He L, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14(21):6751–6760. doi: 10.1158/1078-0432.CCR-08-1034. [DOI] [PubMed] [Google Scholar]
- 15.Santoyo-Ramos P, Likhatcheva M, Garcia-Zepeda EA, et al. Hypoxia-inducible factors modulate the stemness and malignancy of colon cancer cells by playing opposite roles in canonical Wnt signaling. PLoS One. 2014;9(11):e112580. doi: 10.1371/journal.pone.0112580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu K, Ning Z, Zeng J, et al. Silibinin inhibits beta-catenin/ZEB1 signaling and suppresses bladder cancer metastasis via dual-blocking epithelial-mesenchymal transition and stemness. Cell Signal. 2013;25(12):2625–2633. doi: 10.1016/j.cellsig.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 17.Yu D, Shin HS, Lee YS, et al. miR-106b modulates cancer stem cell characteristics through TGF-beta/Smad signaling in CD44-positive gastric cancer cells. Lab Invest. 2014;94(12):1370–1381. doi: 10.1038/labinvest.2014.125. [DOI] [PubMed] [Google Scholar]
- 18.Xie G, Yao Q, Liu Y, et al. IL-6-induced epithelial-mesenchymal transition promotes the generation of breast cancer stem-like cells analogous to mammosphere cultures. Int J Oncol. 2012;40(4):1171–1179. doi: 10.3892/ijo.2011.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uchino M, Kojima H, Wada K, et al. Nuclear beta-catenin and CD44 upregulation characterize invasive cell populations in non-aggressive MCF-7 breast cancer cells. BMC Cancer. 2010;10:414. doi: 10.1186/1471-2407-10-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho Y, Lee HW, Kang HG, et al. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget. 2015;6(11):8709–8721. doi: 10.18632/oncotarget.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 22.Dan T, Hewitt SM, Ohri N, et al. CD44 is prognostic for overall survival in the NCI randomized trial on breast conservation with 25 year follow-up. Breast Cancer Res Treat. 2014;143(1):11–18. doi: 10.1007/s10549-013-2758-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomao F, Papa A, Zaccarelli E, et al. Triple-negative breast cancer: new perspectives for targeted therapies. Onco Targets Ther. 2015;8:177–193. doi: 10.2147/OTT.S67673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang MH, Kim HJ, Kim EJ, et al. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum Pathol. 2015;46(9):1267–1274. doi: 10.1016/j.humpath.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Ma F, Li H, Wang H, et al. Enriched CD44(+)/CD24(−) population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC) Cancer Lett. 2014;353(2):153–159. doi: 10.1016/j.canlet.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 26.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heikkinen T, Greco D, Pelttari LM, et al. Variants on the promoter region of PTEN affect breast cancer progression and patient survival. Breast Cancer Res. 2011;13(6):R130. doi: 10.1186/bcr3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terunuma A, Putluri N, Mishra P, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124(1):398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sircoulomb F, Bekhouche I, Finetti P, et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer. 2010;10:539. doi: 10.1186/1471-2407-10-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke C, Madden SF, Doolan P, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34(10):2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 31.Ma XJ, Dahiya S, Richardson E, et al. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11(1):R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tofigh A, Suderman M, Paquet ER, et al. The prognostic ease and difficulty of invasive breast carcinoma. Cell Rep. 2014;9(1):129–142. doi: 10.1016/j.celrep.2014.08.073. [DOI] [PubMed] [Google Scholar]
- 34.Kao KJ, Chang KM, Hsu HC, et al. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desmedt C, Di Leo A, de Azambuja E, et al. Multifactorial approach to predicting resistance to anthracyclines. J Clin Oncol. 2011;29(12):1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M, Bohm D, von Torne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 38.Nagalla S, Chou JW, Willingham MC, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14(4):R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 40.Dedeurwaerder S, Desmedt C, Calonne E, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011;3(12):726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28(27):4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 45.Hu Z, Fan C, Livasy C, et al. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med. 2009;7:9. doi: 10.1186/1741-7015-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69(10):4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 48.Minn AJ, Gupta GP, Padua D, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A. 2007;104(16):6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamczyk A, Niemiec JA, Ambicka A, et al. CD44/CD24 as potential prognostic markers in node-positive invasive ductal breast cancer patients treated with adjuvant chemotherapy. J Mol Histol. 2014;45(1):35–45. doi: 10.1007/s10735-013-9523-6. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Sarkissyan M, Elshimali Y, et al. Triple negative breast tumors in African–American and Hispanic/Latina women are high in CD44+, low in CD24+, and have loss of PTEN. PLoS One. 2013;8(10):e78259. doi: 10.1371/journal.pone.0078259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernardi MA, Logullo AF, Pasini FS, et al. Prognostic significance of CD24 and claudin-7 immunoexpression in ductal invasive breast cancer. Oncol Rep. 2012;27(1):28–38. doi: 10.3892/or.2011.1477. [DOI] [PubMed] [Google Scholar]
- 52.Uchoa Dde M, Graudenz MS, Callegari-Jacques SM, et al. Expression of cancer stem cell markers in basal and penta-negative breast carcinomas – a study of a series of triple-negative tumors. Pathol Res Pract. 2014;210(7):432–439. doi: 10.1016/j.prp.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 53.Ahmed MA, Aleskandarany MA, Rakha EA, et al. A CD44(−)/CD24(+) phenotype is a poor prognostic marker in early invasive breast cancer. Breast Cancer Res Treat. 2012;133(3):979–995. doi: 10.1007/s10549-011-1865-8. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Shi Q, Wang Z, et al. Clinicopathologic correlation of cancer stem cell markers CD44, CD24, VEGF and HIF-1alpha in ductal carcinoma in situ and invasive ductal carcinoma of breast: an immunohistochemistry-based pilot study. Pathol Res Pract. 2011;207(8):505–513. doi: 10.1016/j.prp.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Ricardo S, Vieira AF, Gerhard R, et al. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011;64(11):937–946. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 56.Kim HJ, Kim MJ, Ahn SH, et al. Different prognostic significance of CD24 and CD44 expression in breast cancer according to hormone receptor status. Breast. 2011;20(1):78–85. doi: 10.1016/j.breast.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Giatromanolaki A, Sivridis E, Fiska A, et al. The CD44+/CD24− phenotype relates to ‘triple-negative’ state and unfavorable prognosis in breast cancer patients. Med Oncol. 2011;28(3):745–752. doi: 10.1007/s12032-010-9530-3. [DOI] [PubMed] [Google Scholar]
- 58.Looi LM, Cheah PL, Zhao W, et al. CD44 expression and axillary lymph node metastasis in infiltrating ductal carcinoma of the breast. Malays J Pathol. 2006;28(2):83–86. [PubMed] [Google Scholar]
- 59.Han Z, Chen Z, Zheng R, et al. Clinicopathological significance of CD133 and CD44 expression in infiltrating ductal carcinoma and their relationship to angiogenesis. World J Surg Oncol. 2015;13:56. doi: 10.1186/s12957-015-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith AD, Roda D, Yap TA. Strategies for modern biomarker and drug development in oncology. J Hematol Oncol. 2014;7:70. doi: 10.1186/s13045-014-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montgomery N, Hill A, McFarlane S, et al. CD44 enhances invasion of basal-like breast cancer cells by upregulating serine protease and collagen-degrading enzymatic expression and activity. Breast Cancer Res. 2012;14(3):R84. doi: 10.1186/bcr3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park J, Schlederer M, Schreiber M, et al. AF1q is a novel TCF7 co-factor which activates CD44 and promotes breast cancer metastasis. Oncotarget. 2015;6(24):20697–20710. doi: 10.18632/oncotarget.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nam K, Oh S, Lee KM, et al. CD44 regulates cell proliferation, migration, and invasion via modulation of c-Src transcription in human breast cancer cells. Cell Signal. 2015;27(9):1882–1894. doi: 10.1016/j.cellsig.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 64.So JY, Smolarek AK, Salerno DM, et al. Targeting CD44-STAT3 signaling by Gemini vitamin D analog leads to inhibition of invasion in basal-like breast cancer. PLoS One. 2013;8(1):e54020. doi: 10.1371/journal.pone.0054020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boulbes DR, Chauhan GB, Jin Q, et al. CD44 expression contributes to trastuzumab resistance in HER2-positive breast cancer cells. Breast Cancer Res Treat. 2015;151(3):501–513. doi: 10.1007/s10549-015-3414-3. [DOI] [PubMed] [Google Scholar]
- 66.Wei X, Xu M, Wei Y, et al. The addition of rituximab to CHOP therapy alters the prognostic significance of CD44 expression. J Hematol Oncol. 2014;7:34. doi: 10.1186/1756-8722-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng Y, Tao L, Xu J, et al. CD44/cellular prion protein interact in multidrug resistant breast cancer cells and correlate with responses to neoadjuvant chemotherapy in breast cancer patients. Mol Carcinog. 2014;53(9):686–697. doi: 10.1002/mc.22021. [DOI] [PubMed] [Google Scholar]
- 68.Ma L, Jiang T. Clinical implications of Ezrin and CD44 coexpression in breast cancer. Oncol Rep. 2013;30(4):1899–1905. doi: 10.3892/or.2013.2641. [DOI] [PubMed] [Google Scholar]
- 69.Yang C, He Y, Zhang H, et al. Selective killing of breast cancer cells expressing activated CD44 using CD44 ligand-coated nanoparticles in vitro and in vivo. Oncotarget. 2015;6(17):15283–15296. doi: 10.18632/oncotarget.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 71.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Camerini A, Donati S, Viacava P, et al. Evaluation of HER2 and p53 expression in predicting response to docetaxel-based first-line chemotherapy in advanced breast cancer. J Exp Clin Canc Res. 2011;30:38. doi: 10.1186/1756-9966-30-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McFarlane S, Coulter JA, Tibbits P, et al. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget. 2015;6(13):11465–11476. doi: 10.18632/oncotarget.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolodeoku J, Yoshida K, Sugino T, et al. CD44 expression in human breast cancer cell lines is related to oestrogen receptor (ER) status and confluency in vitro. Biochem Soc Trans. 1997;25(2):356S. doi: 10.1042/bst025356s. [DOI] [PubMed] [Google Scholar]
- 76.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heldin P, Basu K, Kozlova I, et al. HAS2 and CD44 in breast tumorigenesis. Adv Cancer Res. 2014;123:211–229. doi: 10.1016/B978-0-12-800092-2.00008-3. [DOI] [PubMed] [Google Scholar]
- 78.Cho SH, Park YS, Kim HJ, et al. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int J Oncol. 2012;41(1):211–218. doi: 10.3892/ijo.2012.1453. [DOI] [PubMed] [Google Scholar]
- 79.Fernando J, Malfettone A, Cepeda EB, et al. A mesenchymal-like phenotype and expression of CD44 predict lack of apoptotic response to sorafenib in liver tumor cells. Int J Cancer. 2015;136(4):E161–E172. doi: 10.1002/ijc.29097. [DOI] [PubMed] [Google Scholar]
- 80.Schulenburg A, Blatt K, Cerny-Reiterer S, et al. Cancer stem cells in basic science and in translational oncology: can we translate into clinical application? J Hematol Oncol. 2015;8(1):16. doi: 10.1186/s13045-015-0113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yin X, Zhang BH, Zheng SS, et al. Coexpression of gene Oct4 and Nanog initiates stem cell characteristics in hepatocellular carcinoma and promotes epithelial-mesenchymal transition through activation of Stat3/Snail signaling. J Hematol Oncol. 2015;8:23. doi: 10.1186/s13045-015-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Witzel I, Milde-Langosch K, Schmidt M, et al. Role of urokinase plasminogen activator and plasminogen activator inhibitor mRNA expression as prognostic factors in molecular subtypes of breast cancer. Onco Targets Ther. 2014;7:2205–2213. doi: 10.2147/OTT.S65344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu K, Chen K, Wang C, et al. Cell fate factor DACH1 represses YB-1- mediated oncogenic transcription and translation. Cancer Res. 2014;74(3):829–839. doi: 10.1158/0008-5472.CAN-13-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu K, Li Z, Cai S, et al. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 2013;73(14):4488–4499. doi: 10.1158/0008-5472.CAN-12-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Y, Han N, Zhou S, et al. The DACH/EYA/SIX gene network and its role in tumor initiation and progression. Int J Cancer. 2015 Apr 9; doi: 10.1002/ijc.29560. Epub. [DOI] [PubMed] [Google Scholar]
- 86.Yan S, Jiao X, Li K, et al. The impact of IGF-1R expression on the outcomes of patients with breast cancer: a meta-analysis. Onco Targets Ther. 2015;8:279–287. doi: 10.2147/OTT.S74774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han T, Liu Z, Li H, et al. High expression of UBD correlates with epirubicin resistance and indicates poor prognosis in triple-negative breast cancer. Onco Targets Ther. 2015;8:1643–1649. doi: 10.2147/OTT.S81214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dong MJ, Wang LB, Jiang ZN, et al. The transcription factor KLF4 as an independent predictive marker for pathologic complete remission in breast cancer neoadjuvant chemotherapy: a case-control study. Onco Targets Ther. 2014;7:1963–1969. doi: 10.2147/OTT.S68340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chekhun SV, Zadvorny TV, Tymovska YO, et al. CyrillicD44+/CD24− markers of cancer stem cells in patients with breast cancer of different molecular subtypes. Exp Oncol. 2015;37(1):58–63. [PubMed] [Google Scholar]
- 90.Loh TJ, Moon H, Cho S, et al. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol Rep. 2015;34(3):1231–1238. doi: 10.3892/or.2015.4110. [DOI] [PubMed] [Google Scholar]
- 91.Martin TA, Jiang WG. Evaluation of the expression of stem cell markers in human breast cancer reveals a correlation with clinical progression and metastatic disease in ductal carcinoma. Oncol Rep. 2014;31(1):262–272. doi: 10.3892/or.2013.2813. [DOI] [PubMed] [Google Scholar]
- 92.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]