Abstract
Background
Long noncoding RNAs (lncRNAs) with dysregulated expression levels have been investigated in numerous types of different cancer. Whether lncRNAs can predict the progression of gastric cancer (GC) still remains largely unclear. The aim of our study was to investigate whether KRT18P55, a novel intergenic lncRNA, can be a predictive biomarker for GC.
Methods
To determine the expression levels of KRT18P55 in GC, we evaluated it in five GC cell lines (SGC-7901, MGC-803, BGC-823, AGS, and HG27) and 97 GC tissue samples in comparison with a normal control by quantitative polymerase chain reaction. In addition, the association with patient clinicopathological characteristics was analyzed to identify the clinical significance of KRT18P55. We also used publicly accessible data from The Cancer Genome Atlas (TCGA) to further verify the expression levels and clinical significance of KRT18P55. Furthermore, a receiver operating characteristic curve was also conducted to evaluate the diagnostic value of KRT18P55 for GC.
Results
A significant upregulation was observed in GC cell lines (P<0.01) and tissue samples (P<0.01). This finding was consistent with the results of 29 pairs of GC tissue samples from TCGA (P<0.01). Additionally, we indicated that the increased expression of KRT18P55 was related to the progression of intestinal type (P=0.032), which was also supported by results of independent GC cohorts from TCGA (P<0.01). However, we did not find significant difference in prognosis between patients with high and low expression of KRT18P55 (P>0.05). Finally, KRT18P55 showed potential diagnostic value for GC with an area under the receiver operating characteristic curve of 0.733 (P<0.01).
Conclusion
Upregulated KRT18P55 was a novel biomarker for the progression of GC, especially for the intestinal type.
Keywords: biomarker, gastric cancer, intestinal type, KRT18P55, long noncoding RNA
Background
Gastric cancer (GC) is the fourth most commonly diagnosed cancer and ranks the second most lethal malignant cancer throughout the world.1 In general, incidence rates are highest in Eastern Asia (particularly in Korea, Mongolia, Japan, and the People’s Republic of China).2 Despite improvements in diagnostic techniques, most patients are typically diagnosed at a late stage with poor prognosis.3 In addition, although an early stage of gastric tumor can be cured by chemotherapy, radiotherapy, and surgical techniques, the 5-year overall survival (OS) rate for patients with GC is still less than 25% because of carcinoma recurrence and metastasis.4,5 Therefore, it is important to identify highly sensitive and specific carcinoma-associated novel biomarkers, which can contribute to improving prevention, detection, diagnosis, and treatment of GC.6
Long noncoding RNAs (lncRNAs) are a significant new class of transcripts longer than 200 nucleotides, which can be classified into five types (sense, antisense, bidirectional, intronic, and intergenic) according to genomic location.7,8 To date, growing evidence has indicated that lncRNAs acting as potential biomarkers are associated with the development of numerous cancer types.9,10 Of note, the dysregulation of lncRNAs has shown clinical significance in patients with GC and also could be emerging biomarkers for the disease.11,12 For example, lncRNA-UCA1 expression was remarkably increased in GC cell lines and tissues, correlating with worse differentiation, tumor size, invasion depth, and TNM stage.13 The expression of lncRNA-BANCR was also upregulated in GC tissues with high expression positively associated with clinical stage, tumor depth, lymph node metastasis, and distant metastasis in GC patients.14 In addition, lncRNA-ABHD11-AS1, RP11-119F7.4, and MALAT2 are potential biomarkers for screening, development, progression, and prognosis of GC.15–17 However, there is still a pressing need to find sensitive and specific dysregulated lncRNAs for further deciphering the mechanisms of GC and to be good biomarkers associated with its progression.
Here, we investigated differentially expressed lncRNAs by the human lncRNA array of six GC tissue samples and their matched normal adjacent tissues (NATs). From the results of the array analysis, we focused on a novel intergenic lncRNA KRT18P55 (NR_028334, 1,950 nucleotides; chromosome 17 (-): 26603012–26634408), which was highly expressed in GC tissue samples when compared with NATs. To determine whether this novel lncRNA can be a potential biomarker for the progression of GC, we first investigated the expression of KRT18P55 in GC cell lines and tissue samples. Then, combined with clinicopathological factors, the potential correlations between KRT18P55 expression levels and patient clinicopathological factors were evaluated. We also reviewed a cohort from The Cancer Genome Atlas (TCGA) to verify the upregulation and clinical utility of KRT18P55. Furthermore, a receiver operating characteristic (ROC) curve was also conducted to explore whether KRT18P55 could be a good candidate to discriminate tumor tissues from nontumorous tissues.
Materials and methods
Cell lines and culture conditions
Approval for the use of human cell lines was obtained from the ATCC Institutional Review Board for Human Subjects Research. A normal gastric epithelium cell line (GES-1, control) and five human GC cell lines (SGC-7901, MGC-803, BGC-823, AGS, and HG27) were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, People’s Republic of China). AGS and GES-1 were cultured in F-12K Medium (Invitrogen, Carlsbad, CA, USA) and Dulbecco’s Modified Eagle’s Medium (Invitrogen), respectively, other cell lines were cultured in Roswell Park Memorial Institute 1640 medium (Invitrogen). Medium was supplemented with 10% fetal bovine serum (Clark Bioscience, Houston, TX, USA). Cells were maintained in a humidified incubator at 37°C in the presence of 5% CO2.
Tissue samples and clinical data collection
A total of 97 GC patients who underwent D2 radical gastrectomy after a definite histopathological diagnosis of GC between May 2009 and July 2010 in our hospital (the First Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China) were enrolled in our study. In addition, there was no radiotherapy or chemotherapy prior to the operations. All the tissue samples were snap-frozen in liquid nitrogen immediately after resection and stored at −80°C until used. Matched NATs were obtained from tissues to the edge of the GC (≥5 cm).
The clinicopathological data were mainly collected from medical records, pathology reports, and personal interviews including baseline of patients, information of surgery, follow-up, and pathological features of the tumor. The histologic grade assessment was based on the World Health Organization histological classification of gastric carcinoma. The clinical staging criteria (pT, pN, pM, and pTNM classification) were assessed according to the seventh edition of the American Joint Committee on Cancer TNM classification. Based on the Lauren criteria, two pathologists reviewed original diagnostic slides and assigned the histological type into intestinal type (tumor with glandula architecture), diffuse type (tumor composed of solitary small clusters of cells, lacking glandular structures), and mixed type (combination of these two features). The designation of tumor growth pattern was in accordance with the third English edition of the Japanese classification of gastric carcinomas.
Follow-up was performed until February 2014 using telephone inquiries or questionnaires. The follow-up time ranged from 1 to 54 months (median 42 months). Except for one patient lost during follow-up, 96 patients were included in the final analysis of the Kaplan–Meier curve (OS: the length of time between surgery and the last follow-up examination or death; cancer-specific survival (CSS): the length of time between surgery and the last follow-up examination or death due to cancer).
Ethical approval of the study protocol
This research was carried out according to the principles expressed in the Declaration of Helsinki. Written informed consent of GC tissue samples collection was obtained from all patients and the study protocol and ethical guidelines were approved by the Research Ethics Committee of China Medical University (Shenyang, Liaoning, People’s Republic of China).
TCGA datasets
Gene expression data from TCGA stomach adenocarcinoma were downloaded from https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm. RNA sequencing (RNA-Seq) experiments of TCGA stomach adenocarcinoma had been performed in a combination of two different Illumina platforms (IlluminaGA RNA-Seq and IlluminaHiSeq RNA-Seq datasets) for a total of 307 patients with GC after removing the duplicates. We used the reads per kilobase of exon per million reads mapped (RPKM) value to represent the expression levels of KRT18P55 for each patient from TCGA datasets.18 In addition, the clinicopathologic and prognostic information of these patients were downloaded from https://tcga-data.nci.nih.gov/tcga/findArchives.htm. We combined the clinicopathologic and prognostic information with these 307 GC patients using structured query language. Finally, based on patient ID, 274 GC patients linked with their clinicopathologic and follow-up data information were available for final analysis. For OS analysis, patients were divided into two groups according to KRT18P55 median expression. Among these 274 GC patients, GC tissue samples and matched NATs were available for 29 GC patients who were used to further verify the differential expression of KRT18P55.
RNA isolation, reverse transcription, and quantitative polymerase chain reaction
Total RNAs were extracted from cultured cells and tissue samples using TRIzol (Invitrogen) following the manufacturer’s protocol. The A260/A280 (1.80–2.00) was evaluated to examine total RNA purity by using a Nanophotometer P-Class (Implen, GmbH, Münich, Germany). RNA was reversely transcribed into 20 μL complementary DNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, Liaoning, People’s Republic of China) by GeneAmp® PCR System 9700 Thermocycler (Applied Biosystems Life Technologies, Foster City, CA, USA).
Quantitative polymerase chain reaction (qPCR) was performed using SYBR® Premix Ex Taq™ II (TaKaRa) in a Light Cycler® 480 II real-time PCR system (Roche Diagnostics, Basel, Switzerland). Each 25 μL qPCR reaction mixture contained 2 μL complementary DNA, 12.5 μL SYBR, 0.3 μL forward and reverse primers, and 9.9 μL RNase-free water. qPCR was performed using 1 cycle of 95°C for 30 seconds, 45 cycles of 95°C for 5 seconds, and 60°C for 30 seconds. The primers used in qPCR for KRT18P55 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were as follows: KRT18P55 forward primer: 5′-CAGGAATGGGAGTCATCCAG-3′ and reverse primer: 5′-CCAGGCTCCTAACTCTGTCC-3′; GAPDH forward primer: 5′-CGGATTTGGTCGTATTGGG-3′, and reverse primer: 5′-CTGGAAGATGGTGATGGGATT-3′ (Sangon Biotech, Shanghai, People’s Republic of China). The relative expression of KRT18P55 was calculated and normalized by using the comparative cycle threshold (Ct) method relative to an endogenous control GAPDH. qPCR experiments were repeated three times in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5.0 software (GraphPad Software, La Jolla, CA, USA). We calculated RNA expression levels between groups (GC cell lines relative to GES-1; cancer tissue relative to NATs) using the 2−ΔΔCt method (ΔCt = CtlncRNA − CtGAPDH; ΔΔCt = ΔCtGC cell lines/cancer tissue − ΔCtGES-1/NATs). All data were expressed as mean ± standard deviation from at least three separate experiments. The qPCR expression levels of lncRNA between groups were estimated by Student’s t-test, and the expression levels from TCGA datasets between groups were calculated by a nonparametric test (Wilcoxon test). In addition, the correlations between lncRNA expression levels and clinicopathological parameters were analyzed by a nonparametric test (Mann–Whitney U-test for two groups and Kruskal–Wallis test for three or more groups). Furthermore, OS and CSS rates were calculated by the Kaplan–Meier method using the log-rank test applied for comparison. Univariate and multivariate analyses were used to further verify whether the expression of lncRNA was related to OS and CSS. A ROC curve was constructed to evaluate the diagnostic value of lncRNA levels. Differences were considered statistically significant with a two-tailed P<0.05.
Results
KRT18P55 upregulated in human GC
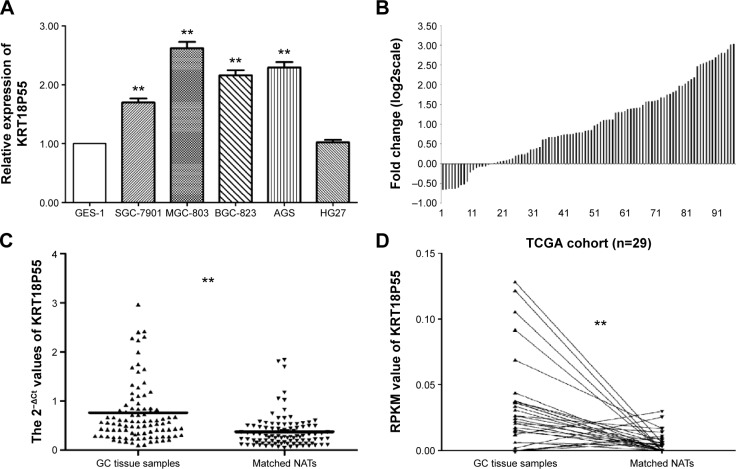
As presented in Figure 1A, we found that four cell lines (SGC-7901, MGC-803, BGC-823, and AGS) expressed significantly higher levels of KRT18P55 when normalized to GES-1. In addition, we further explored the relative expression of KRT18P55 in 97 pairs of GC tissues and matched NATs by qPCR analysis normalized to GAPDH. Figure 1B shows that in 80 of 97 (82.5%) pairs, KRT18P55 was upregulated (2−ΔΔCt fold change >1.0) and the average expression of KRT18P55 was 2.56-fold in 97 pairs of GC. Furthermore, Figure 1C demonstrates that the expression of KRT18P55 was significantly higher in cancer tissues when compared with their matched NATs (2−ΔCtGC, 0.76±0.73; 2−ΔCtNATs, 0.38±0.34; P<0.01). To further confirm our results, the TCGA dataset consistently indicated that KRT18P55 expression was higher in 29 GC tissue samples in comparison with matched NATs (RPKMGC, 0.034±0.035; RPKMNATs, 0.006±0.008; P<0.01; Figure 1D).
Figure 1.
KRT18P55 was upregulated in gastric cancer.
Notes: (A) The expression of KRT18P55 in five GC cell lines: SGC-7901, MGC-803, BGC-823, AGS, and HG27 showed significant higher expression of KRT18P55 in comparison with normal control by qPCR (**P<0.01). (B) In 80 of 97 patients with GC, KRT18P55 was upregulated in GC tissue samples in comparison with matched NATs. (C) KRT18P55 was significantly higher in GC tissues samples when compared with their matched NATs by qPCR (**P<0.01). (D) TCGA datasets indicated that KRT18P55 expression was higher in 29 GC tissue samples in comparison with matched NATs (**P<0.01).
Abbreviations: GC, gastric cancer; NATs, normal adjacent tissues; qPCR, quantitative polymerase chain reaction; RPKM, reads per kilobase of exon per million reads mapped; TCGA, The Cancer Genome Atlas.
Expression levels of KRT18P55 and clinicopathologic characteristics of GC patients
To further explore the significance of KRT18P55 overexpression in GC, analyses were carried out to assess the correlation between KRT18P55 expression levels and the clinicopathological features of 97 patients (Table 1). Noticeably, KRT18P55 was associated with Lauren type (intestinal, 2.90 (1.24–5.92) vs diffuse/mixed, 1.69 (1.07–2.68); P=0.032). However, there was no significant correlation between KRT18P55 and other features (age, sex, tumor size, macroscopic type, histologic grade, invasion, lymphatic metastasis, tumor stage, invasion into lymphatic vessels, and growth pattern). Of note, a similar tendency of results from TCGA datasets also identified that higher expression of KRT18P55 was associated with the progression of intestinal type (intestinal, 0.0028 [0.009–0.057] vs diffuse, 0.016 [0.007–0.030]; P=0.009), but not other clinicopathological features (Table 2).
Table 1.
Relationship of KRT18P55 expression in tissue samples with clinicopathological factors of gastric cancer patients
| Characteristics | Number of cases (%) | KRT18P55a | P-value |
|---|---|---|---|
| Age (years) | |||
| ≤63 | 50 (51.5) | 2.16 (1.31–3.21) | 0.307 |
| >63 | 47 (48.5) | 1.67 (1.03–3.35) | |
| Sex | |||
| Male | 70 (72.2) | 1.88 (1.26–3.39) | 0.147 |
| Female | 27 (27.8) | 1.60 (1.02–2.46) | |
| Maximum tumor size (cm) | |||
| ≤5.5 | 54 (55.7) | 1.90 (1.06–3.01) | 0.706 |
| >5.5 | 43 (44.3) | 1.73 (1.18–3.35) | |
| Macroscopic typeb | |||
| Borrmann I + II | 10 (10.3) | 2.58 (1.60–3.94) | 0.272 |
| Borrmann III + IV | 82 (84.5) | 1.79 (1.15–3.25) | |
| Histologic grade | |||
| Well/moderately | 39 (40.2) | 2.63 (1.18–3.94) | 0.195 |
| Poorly | 58 (59.8) | 1.73 (1.07–3.03) | |
| Lauren type | |||
| Intestinal | 28 (28.9) | 2.90 (1.24–5.92) | 0.032 |
| Diffuse/mixed | 69 (71.1) | 1.69 (1.07–2.68) | |
| pT stage | |||
| T1 | 5 (5.2) | 1.04 (0.67–1.84) | 0.108 |
| T2 | 9 (9.3) | 1.31 (0.84–3.01) | |
| T3 | 17 (17.5) | 2.67 (1.19–4.88) | |
| T4 | 66 (68.0) | 1.78 (1.20–3.25) | |
| pN stage | |||
| N0 | 21 (21.7) | 1.59 (1.03–2.92) | 0.564 |
| N1 | 17 (17.5) | 2.48 (1.37–5.00) | |
| N2 | 23 (23.7) | 1.79 (1.05–3.05) | |
| N3 | 36 (37.1) | 1.72 (1.19–3.31) | |
| pTNM stage | |||
| IA, IB | 9 (9.3) | 1.31 (0.87–2.92) | 0.532 |
| IIA, IIB | 19 (19.6) | 2.18 (0.97–4.57) | |
| IIIA, IIIB, IIIC | 69 (71.1) | 1.79 (1.19–3.28) | |
| Invasion into lymphatic vessels | |||
| Absent | 51 (52.6) | 1.68 (1.07–3.01) | 0.337 |
| Present | 46 (47.4) | 2.05 (1.18–3.37) | |
| Growth pattern | |||
| Expanding | 7 (7.2) | 1.68 (1.59–3.42) | 0.476 |
| Intermediate | 37 (38.1) | 2.46 (1.12–3.94) | |
| Infiltrative | 53 (54.7) | 1.69 (1.07–3.13) | |
Notes:
Median of relative expression, with 25th–75th percentile in parenthesis.
Five patients were diagnosed with early gastric cancer. Statistical significance are marked by bold type (P<0.05).
Abbreviations: pT, pathological Tumor; pN, pathological Node; pTNM, pathological Tumor Node Metastasis.
Table 2.
RPKM value of KRT18P55 linked with clinicopathological factors of gastric cancer patients from cohort of TCGA
| Characteristics | Number of cases (%)a | KRT18P55b | P-value |
|---|---|---|---|
| Sex | |||
| Male | 171 (62.4) | 0.021 (0.009–0.037) | 0.918 |
| Female | 103 (37.6) | 0.023 (0.007–0.043) | |
| Lauren typec | |||
| Intestinal | 75 (59.5) | 0.028 (0.009–0.057) | 0.009 |
| Diffuse | 51 (40.5) | 0.016 (0.007–0.030) | |
| Histologic grade | |||
| G1 | 7 (2.6) | 0.025 (0.012–0.040) | |
| G2 | 87 (32.3) | 0.021 (0.009–0.037) | 0.885 |
| G3 | 175 (65.1) | 0.022 (0.008–0.042) | |
| pT stage | |||
| T1 | 13 (4.9) | 0.017 (0.008–0.030) | |
| T2 | 45 (17.0) | 0.026 (0.009–0.056) | 0.387 |
| T3 | 103 (38.9) | 0.020 (0.007–0.041) | |
| T4 | 104 (39.2) | 0.020 (0.008–0.037) | |
| pN stage | |||
| N0 | 91 (34.7) | 0.025 (0.009–0.406) | |
| N1 | 58 (22.1) | 0.019 (0.007–0.035) | 0.876 |
| N2 | 58 (22.1) | 0.022 (0.008–0.041) | |
| N3 | 55 (21.1) | 0.019 (0.008–0.045) | |
| pM stage | |||
| M0 | 243 (93.1) | 0.022 (0.009–0.041) | 0.941 |
| M1 | 18 (6.9) | 0.023 (0.006–0.040) | |
| pTNM stage | |||
| I | 32 (12.4) | 0.028 (0.008–0.047) | |
| II | 98 (38.0) | 0.022 (0.010–0.040) | 0.679 |
| III | 111 (43.0) | 0.018 (0.007–0.037) | |
| IV | 17 (6.6) | 0.019 (0.005–0.037) | |
Notes:
Information absent patients were excluded in the relevant factors analysis.
Median of RPKM value, with 25th–75th percentile in parenthesis.
Based on TCGA datasets, histologic type were originally divided into seven types: 1) stomach intestinal adenocarcinoma papillary type; 2) stomach intestinal adenocarcinoma mucinous type; 3) stomach adenocarcinoma signet ring; 4) stomach intestinal adenocarcinoma tubular type; 5) stomach intestinal adenocarcinoma not otherwise specified; 6) stomach adenocarcinoma not otherwise specified; 7) stomach adenocarcinoma diffuse type. Intestinal: 1, 2, 4, 5; diffuse: 7. Statistical significance are marked by bold type (P<0.05).
Abbreviations: RPKM, the reads per kilobase of exon per million mapped sequence reads; TCGA, The Cancer Genome Atlas; pT, pathological Tumor; pN, pathological Node; pM, pathological Metastasis; pTNM, pathological Tumor Node Metastasis.
Association of KRT18P55 expression with patients’ survival time
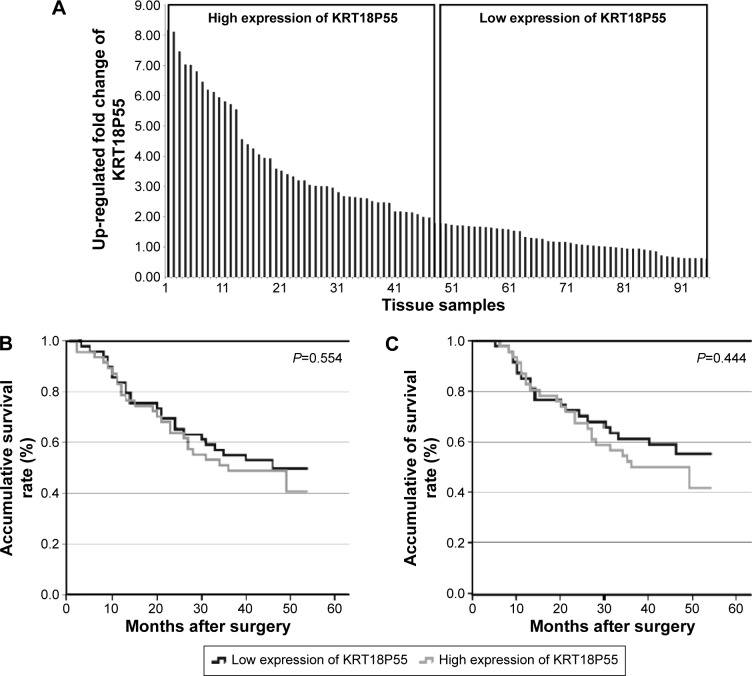
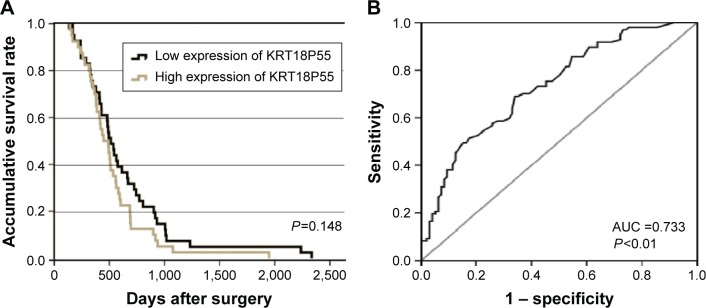
To investigate the correlation of KRT18P55 expression with survival of GC patients, Kaplan–Meier analysis and log-rank test were performed with the cohort of 96 patients (one patient lost in follow-up) on OS and CSS. According to the median expression of KRT18P55 in 96 pairs of tissues, high (n=48) and low (n=48) expression groups were divided (Figure 2A). We did not find significant difference in prognosis between patients with high and low expression of KRT18P55 (OS, P=0.554; CSS, P=0.444; Figure 2B and C). In addition, neither univariate nor multivariate analyses of Cox proportional hazards model demonstrated that KRT18P55 expression was not a significant prognostic factor on OS (univariate, P=0.557; multivariate, P=0.678) and CSS (univariate, P=0.448; multivariate, P=0.570). Furthermore, the results were also verified with TCGA datasets (P=0.148; Figure 3A).
Figure 2.
Association between expression of KRT18P55 and patient survival.
Notes: (A) According to the median expression of KRT18P55 in 96 paired tissue samples, high (n=48) and low (n=48) expression groups were divided. (B) Overall survival of patients in the high and low expression groups (P=0.554). (C) Cancer-specific survival of patients in the high and low expression groups (P=0.444).
Figure 3.
(A) Overall survival of patients from TCGA datasets (P=0.148). (B) ROC curve of patients based on KRT18P55 expression in GC tissue samples and matched NATs.
Abbreviations: AUC, area under the ROC curve; GC, gastric cancer; NATs, normal adjacent tissues; ROC, receiver operating characteristic; TCGA, The Cancer Genome Atlas.
KRT18P55 is a potential diagnosis biomarker for GC
We carried out ROC curve analysis to explore whether KRT18P55 could be a good candidate to discriminate tumor tissues from nontumorous tissues. The area under the ROC curve (AUC) was 0.733 (95% confidence interval 0.664–0.802; P<0.01; sensitivity, 0.691; specificity, 0.660; the Youden index, 0.351; Figure 3B).
Discussion
The progression of GC is complex, heterogeneous, and includes multiple processing steps, involving numerous genetic and epigenetic alterations.19 Although cellular phenomena in parallel with various molecular events promoting GC initiation, progression, and metastasis have been verified,20 it is still necessary to identify diagnosis and progression-specific biomarkers, especially novel biomarkers with significant clinical diagnosis value, for exploring gastric carcinogenesis.
To date, the tissue-specific expression patterns of lnc-RNAs in various diseases are causing a paradigmatic change in our understanding of lncRNAs in cancer development.7 In addition, accumulating numbers of investigations have focused on dysregulated lncRNAs in GC.21 Among these investigations, Yang et al22 elucidated the contributions of H19 to human GC, which highlighted the connection between lncRNAs and GC. After this research, the dysregulation of many specific lncRNAs showed biological and clinical relevance in GC, including HOTAIR, GAS5, ANRIL, CCAT1, LINC00152.23–27 It is worth mentioning that the dysregulated HOTAIR and its genetic variations have been recognized as a potential biomarker for risk assessment, early detection, and the therapeutic target of GC in Chinese populations.28 In fact, to the best of our knowledge, our research is first to investigate a novel lncRNA KRT18P55 in GC, which is significantly upregulated in GC cell lines and tissues samples. Results from TCGA datasets consistently verified our findings. With all of the above taken into consideration, we believed that dysregulated KRT18P55 was a biomarker associated with the progression of GC.
Based on combination of the expression levels of KRT18P55 with clinicopathological characteristics from the cohorts of our hospital and TCGA datasets, we found that increases in KRT18P55 expression showed significant relevance with the occurrence of intestinal type. The conception of classification was first proposed by Lauren in 1965, GC could be divided into intestinal, diffuse, and mixed types.29 Recently, several studies have also demonstrated the relationship between molecular biomarkers and the Lauren classification of GC.30–32 Interestingly, although diffuse types of GC carried a worse prognosis comparing with intestinal types in Chinese populations,33 we found patients with the intestinal type expressed relatively higher levels of KRT18P55 in comparison with the diffuse type. This phenomenon showed a similar tendency with claudin 1, which is verified to be overexpressed in GC in Chinese populations34 and at a higher level in intestinal than in diffuse types from studies in the People’s Republic of China, Finland, and the USA.34–36 Hence, KRT18P55 shows a clinical significance in acting as a biomarker, especially in intestinal types of GC, in a similar role to claudin 1.
GC is frequently not diagnosed until a relatively advanced stage,3 so it is urgently necessary to find plasma or tissue biomarkers with high sensitivity and specificity. CEA, CA19-9, and CA72-4 are the most frequently used serological biomarkers with respective advantages in the diagnosis of GC.37 However, there is currently still no excellent tumor marker for GC.37 Recently, lncRNAs acting as biomarkers for GC has attracted the attention of researchers. Zhou et al38 identified that H19 in plasma can be a novel biomarker for the diagnosis of GC (AUC =0.877).38 Shao et al39 found lncRNA-AA174084 in gastric juice can be used as a tumor marker for screening GC (AUC =0.848). Chen et al40 demonstrated lncRNA HIF1A–AS2 in tissues was a diagnosis biomarker for GC (AUC =0.673). Based on the aforementioned studies, lncRNAs can act as emerging biomarkers in plasma, gastric juice, and tissue for the diagnosis of GC. Notably, the dysregulated lncRNAs in plasma and gastric juice exhibit potential ability in the early diagnosis of GC. A recent study indicated that a combination of three circulating lncRNAs (CUDR, LSINCT-5, and PTENP1) showed a significant higher diagnostic value than CEA and CA19-9 for GC detection.41 However, because of the limitation of our samples, we could not provide a direct conclusion whether KRT18P55 in plasma or gastric juice can be a better biomarker than CEA, CA19-9, and CA72-4. In the future, we will focus on the expression level of KRT18P55 in plasma or gastric juice to explore its value in early diagnosis.
Long intergenic noncoding RNAs (lincRNAs) are derived from the regions between two genes.10 Through the University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/), we identified the location of KRT18P55 to be between the LGALS9 and RAB34 genes. Of note, a recent study confirmed that RAB34 expression was correlated with glioma patient survival and glioma grade, and RAB34 could be used as a progression and prognostic biomarker for poor outcome.42 In addition, increasing evidence also suggests that many lincRNAs function through specific interactions (scaffolds, chromatin-modification, directly DNA interaction, RNA base pairing) with other cellular factors, namely proteins, DNA, and other RNA molecules.43 Unfortunately, the association and effecting patterns between KRT18P55 and its downstream gene still remain largely unknown and need further exploration. At the same time, the roles of lincRNAs have surely laid a good foundation and have provided promising directions for our further research of KRT18P55.
On the whole, our study showed the advantage of enrolling a large number of GC patients, which promoted our strongly reliable results. In addition, thorough pathological examination, long-term follow-up assessment, and scientific statistical methods were also performed to ensure the clinical significance of KRT18P55 with the lowest bias. Most importantly, TCGA datasets further highlighted the ability of KRT18P55 to be a predictive progression biomarker for GC.
Conclusion
To the best of our knowledge, we are the first to present a novel intergenic lncRNA KRT18P55, which increased expression in GC cell lines and tissue samples. In addition, KRT18P55 could be a novel biomarker for the progression of GC, especially the intestinal type.
Acknowledgments
The authors would like to thank the Department of Surgical Oncology of the First Hospital of China Medical University for providing human gastric tissue samples. They would also like to thank the College of China Medical University for technical assistance in experiments. This work was supported by National Science Foundation of China (No 81201888, 81372549, and 81172370), the Special Prophase Program for National Key Basic Research Program of China (No 2014CB560712), Program of Education Department of Liaoning Province (L2014307).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.de Martel C, Forman D, Plummer M. Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42(2):219–240. doi: 10.1016/j.gtc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71(2):127–164. doi: 10.1016/j.critrevonc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 5.Shen L, Shan YS, Hu HM, et al. Management of gastric cancer in Asia: resource-stratified guidelines. Lancet Oncol. 2013;14(12):e535–e547. doi: 10.1016/S1470-2045(13)70436-4. [DOI] [PubMed] [Google Scholar]
- 6.Duraes C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Arch. 2014;464(3):367–378. doi: 10.1007/s00428-013-1533-y. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5(4):e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2015 Jul 4; doi: 10.1016/j.bbagrm.2015.06.015. Epub. [DOI] [PubMed] [Google Scholar]
- 10.Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015;3:5. doi: 10.1186/s40591-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356(2 Pt B):357–366. doi: 10.1016/j.canlet.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Sun M, Nie FQ, Wang ZX, De W. Involvement of lncRNA dysregulation in gastric cancer. Histol Histopathol. 2015;31:33–39. doi: 10.14670/HH-11-655. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Q, Wu F, Dai WY, et al. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17(8):640–646. doi: 10.1007/s12094-015-1290-2. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Zhang L, Zhang Y, Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109–112. doi: 10.1016/j.biopha.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Shao Y, Zhu M, et al. Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2015 Aug 18; doi: 10.1007/s13277-015-3903-3. Epub. [DOI] [PubMed] [Google Scholar]
- 16.Sun J, Song Y, Chen X, et al. Novel long non-coding RNA RP11-119F7.4 as a potential biomarker for the development and progression of gastric cancer. Oncol Lett. 2015;10(1):115–120. doi: 10.3892/ol.2015.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F, Tian Y, Pang EJ, Wang Y, Li L. MALAT2-activated long noncoding RNA indicates a biomarker of poor prognosis in gastric cancer. Cancer Gene Ther. 2015 Feb 27; doi: 10.1038/cgt.2015.6. Epub. [DOI] [PubMed] [Google Scholar]
- 18.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 19.Stock M, Otto F. Gene deregulation in gastric cancer. Gene. 2005;360(1):1–19. doi: 10.1016/j.gene.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 20.Li PF, Chen SC, Xia T, et al. Non-coding RNAs and gastric cancer. World J Gastroenterol. 2014;20(18):5411–5419. doi: 10.3748/wjg.v20.i18.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gan L, Xu M, Zhang Y, Zhang X, Guo W. Focusing on long noncoding RNA dysregulation in gastric cancer. Tumour Biol. 2015;36(1):129–141. doi: 10.1007/s13277-014-2894-9. [DOI] [PubMed] [Google Scholar]
- 22.Yang F, Bi J, Xue X, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279(17):3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 23.Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464(3):807–813. doi: 10.1016/j.bbrc.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 24.Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5(8):2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizrahi I, Mazeh H, Grinbaum R, et al. Colon cancer associated transcript-1 (CCAT1) expression in adenocarcinoma of the stomach. J Cancer. 2015;6(2):105–110. doi: 10.7150/jca.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J, Liu Y, Zhang W, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14(19):1–12. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du M, Wang W, Jin H, et al. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6:31255–31262. doi: 10.18632/oncotarget.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 30.Kroepil F, Dulian A, Vallbohmer D, et al. High EpCAM expression is linked to proliferation and Lauren classification in gastric cancer. BMC Res Notes. 2013;6:253. doi: 10.1186/1756-0500-6-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Susman S, Barnoud R, Bibeau F, et al. The Lauren classification highlights the role of epithelial-to-mesenchymal transition in gastric carcinogenesis: an immunohistochemistry study of the STAT3 and adhesion molecules expression. J Gastrointestin Liver Dis. 2015;24(1):77–83. doi: 10.15403/jgld.2014.1121.sus. [DOI] [PubMed] [Google Scholar]
- 32.Qiu M, Zhou Y, Zhang X, et al. Lauren classification combined with HER2 status is a better prognostic factor in Chinese gastric cancer patients. BMC Cancer. 2014;14:823. doi: 10.1186/1471-2407-14-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu MZ, Cai MY, Zhang DS, et al. Clinicopathological characteristics and prognostic analysis of Lauren classification in gastric adenocarcinoma in China. J Transl Med. 2013;11:58. doi: 10.1186/1479-5876-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J, Li J, Qu Y, et al. The expression of claudin 1 correlates with beta-catenin and is a prognostic factor of poor outcome in gastric cancer. Int J Oncol. 2014;44(4):1293–1301. doi: 10.3892/ijo.2014.2298. [DOI] [PubMed] [Google Scholar]
- 35.Soini Y, Tommola S, Helin H, Martikainen P. Claudins 1, 3, 4 and 5 in gastric carcinoma, loss of claudin expression associates with the diffuse subtype. Virchows Arch. 2006;448(1):52–58. doi: 10.1007/s00428-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 36.Resnick MB, Gavilanez M, Newton E, et al. Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol. 2005;36(8):886–892. doi: 10.1016/j.humpath.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 37.Tsai MM, Wang CS, Tsai CY, Chi HC, Tseng YH, Lin KH. Potential prognostic, diagnostic and therapeutic markers for human gastric cancer. World J Gastroenterol. 2014;20(38):13791–13803. doi: 10.3748/wjg.v20.i38.13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao Y, Ye M, Jiang X, et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120(21):3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 40.Chen WM, Huang MD, Kong R, et al. Antisense long noncoding RNA HIF1A-AS2 Is upregulated in gastric cancer and associated with poor prognosis. Dig Dis Sci. 2015;60(6):1655–1662. doi: 10.1007/s10620-015-3524-0. [DOI] [PubMed] [Google Scholar]
- 41.Dong L, Qi P, Xu MD, et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137(5):1128–1135. doi: 10.1002/ijc.29484. [DOI] [PubMed] [Google Scholar]
- 42.Wang HJ, Gao Y, Chen L, Li YL, Jiang CL. RAB34 was a progression-and prognosis-associated biomarker in gliomas. Tumour Biol. 2015;36(3):1573–1578. doi: 10.1007/s13277-014-2732-0. [DOI] [PubMed] [Google Scholar]
- 43.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]