Researchers examined the role of a domain within the stromal interaction molecule (STIM) in the mechanism by which Ca2+ release–activated Ca2+ channels can be inactivated as Ca2+ ions enter cells.
Abstract
The inactivation domain of STIM1 (IDSTIM: amino acids 470–491) has been described as necessary for Ca2+-dependent inactivation (CDI) of Ca2+ release–activated Ca2+ (CRAC) channels, but its mechanism of action is unknown. Here we identify acidic residues within IDSTIM that control the extent of CDI and examine functional interactions of IDSTIM with Orai1 pore residues W76 and Y80. Alanine scanning revealed three IDSTIM residues (D476/D478/D479) that are critical for generating full CDI. Disabling IDSTIM by a triple alanine substitution for these three residues (“STIM1 3A”) or by truncation of the entire domain (STIM11–469) reduced CDI to the same residual level observed for the Orai1 pore mutant W76A (approximately one third of the extent seen with full-length STIM1). Results of noise analysis showed that STIM11–469 and Orai1 W76A mutants do not reduce channel open probability or unitary Ca2+ conductance, factors that determine local Ca2+ accumulation, suggesting that they diminish CDI instead by inhibiting the CDI gating mechanism. We tested for functional coupling between IDSTIM and the Orai1 pore by double-mutant cycle analysis. The effects on CDI of mutations disabling IDSTIM or W76 were not additive, demonstrating that IDSTIM and W76 are strongly coupled and act in concert to generate full-strength CDI. Interestingly, disabling IDSTIM and W76 separately gave opposite results in Orai1 Y80A channels: channels with W76 but lacking IDSTIM generated approximately two thirds of the WT extent of CDI but those with IDSTIM but lacking W76 completely failed to inactivate. Together, our results suggest that Y80 alone is sufficient to generate residual CDI, but acts as a barrier to full CDI. Although IDSTIM is not required as a Ca2+ sensor for CDI, it acts in concert with W76 to progress beyond the residual inactivated state and enable CRAC channels to reach the full extent of inactivation.
INTRODUCTION
Ca2+ release–activated Ca2+ (CRAC) channels are prototypic store operated channels that are encoded by the Orai family of plasma membrane proteins and activated by depletion of Ca2+ from the ER (Prakriya and Lewis, 2015). The ER protein STIM1 is best recognized for its function in sensing ER Ca2+ depletion and, in response, activating CRAC channels in the plasma membrane. However, an early clue that STIM1 is more than an activating ligand for Orai1 was the observation that channels activated by an isolated fragment of STIM1 (the CRAC activation domain or CAD; STIM1 aa 342–448), failed to show characteristic fast Ca2+-dependent inactivation (CDI) in response to Ca2+ entry (Park et al., 2009). A series of C-terminal truncations identified a region of STIM1 (aa 470–491) that is required for CDI, termed the inactivation domain of STIM, or IDSTIM (Mullins et al., 2009). Additional evidence for a role of STIM1 in CDI came from observations that in heterologous systems the STIM1/Orai1 expression ratio influences the extent of CDI, with a ratio of ∼2–4:1 being required for CDI to approach the level seen for native CRAC channels (Scrimgeour et al., 2009; Hoover and Lewis, 2011).
A highly negatively charged region within IDSTIM (475DDVDDMDEE483) attracted attention from several groups as a potential Ca2+ binding site for CDI, based on analogy to the highly acidic Ca2+ bowl binding site of the BK channel (Cox, 2011). Consistent with such a role, alanine or glycine substitutions for various combinations of acidic residues within this region reduced CDI, and full neutralization of all acidic residues eliminated CDI (Derler et al., 2009b; Lee et al., 2009; Mullins et al., 2009). However, an attempt to test whether the acidic region is the Ca2+ sensor for CDI yielded equivocal results. 45Ca2+ overlay experiments showed that Ca2+ bound weakly to a peptide containing this region, and mutations that reduced or eliminated CDI reduced 45Ca2+ binding. However, other mutations (STIM1 482/483 EE>AA) paradoxically increased the Ca2+ sensitivity of CDI while reducing 45Ca2+ binding, raising doubts about its function as a simple Ca2+ sensor (Mullins et al., 2009).
Although IDSTIM clearly has an important function in the CDI process, its basis is as yet poorly understood. Because all studies to date have altered multiple rather than individual residues in IDSTIM, the specific acidic residues that are most important for CDI have yet to be identified. Furthermore, it is not known whether IDSTIM affects CDI directly by contributing to the conformational changes involved in inactivation gating or indirectly by changing the CRAC channel’s open probability or unitary current and hence Ca2+ accumulation near the inner pore. If IDSTIM contributes to conformational changes, does this occur via functional coupling to Orai1 pore residues W76 and Y80, which appear to control conformational changes within the pore leading to inactivation (see Mullins et al. in this issue)? Finally, as discussed, it is unknown whether IDSTIM is required for Ca2+ sensing in CDI.
In this paper, we address the four questions introduced above. By identifying new experimental conditions under which CRAC channels inactivate significantly in the absence of IDSTIM, we find that IDSTIM is not absolutely required as a Ca2+ sensor. Alanine scanning of the negatively charged region of IDSTIM defines the contributions of specific acidic residues to CDI, and three residues (D476/D478/D479) emerge as the functional core of the domain. Deletion of IDSTIM does not change channel open probability or unitary current, suggesting that IDSTIM acts directly to promote conformational changes leading to CDI. Finally, mutant cycle analysis of STIM1 and Orai1 mutants reveals a close functional coupling between IDSTIM and Orai1 pore residue W76, but not Y80, defining a functional IDSTIM-W76 module that drives full-strength CDI.
MATERIALS AND METHODS
Abbreviated methods are summarized here. A detailed description of all methods, including equations used in data analysis, can be found in Mullins et al. (2016).
Cells
Experiments used HEK293-H cells (Gibco) grown in Dulbecco’s modified Eagle medium with 10% FBS, 1% penicillin/streptomycin, and 1% l-glutamine (Gemini) in a humidified 5% CO2 incubator at 37°C.
Plasmid cDNA constructs
Experiments used WT or mutant versions of previously described mCherry-STIM1 (Luik et al., 2006) and Orai1-GFP (Xu et al., 2006) constructs. For alanine scanning of IDSTIM, all alanine substitutions were made by QuikChange mutagenesis (Agilent).
Transfection
In the standard experimental design, HEK293-H cells were transfected with mCh-STIM1– and GFP-Orai1–derived constructs at a 4:1 mass ratio (or 12:1 for noise analysis experiments) 16–48 h before recording. Importantly, these ratios are higher than the 1:1 mass ratio used in our previous study of CDI (Mullins et al., 2009). Cells with a high mCh-STIM1/GFP-Orai1 ratio were selected for recording by the relative fluorescence of mCherry and GFP as described in Mullins et al. (2016).
Recording solutions
The standard 20 mM Ca2+ Ringer’s solution contained (in mM) 130 NaCl, 4.5 KCl, 20 CaCl2, 1 MgCl2, 10 D-glucose, and 5 HEPES, pH 7.4 with NaOH. The 2 mM Ca2+ Ringer’s solution substituted (in mM) 155 NaCl and 2 CaCl2. The base DVF solution (DVF) contained (in mM) 150 NaCl, 10 HEDTA, 10 HEPES, 10 TEA-Cl, and 1 EDTA, pH 7.4 with NaOH. DVF solutions with micromolar levels of free [Ca2+] (calculated with MaxChelator; Bers et al., 2010) were made by adding CaCl2 from a 1-M stock before titrating the pH.
The standard EGTA internal solution included 150 mM Cs aspartate, 8 mM MgCl2, 10 mM EGTA, and 10 mM HEPES, pH 7.2 with CsOH. The BAPTA internal solution contained 135 mM Cs aspartate, 8 mM MgCl2, 8 mM Cs4BAPTA(Molecular Probes), and 10 mM HEPES, pH 7.2 with CsOH.
Electrophysiology
Currents were recorded with the standard whole-cell patch-clamp technique at 22–25°C, filtering at 2 kHz and sampling at 5 kHz, with all voltages corrected for liquid junction potentials. 2 mM Ca2+ Ringer’s solution with 100 µM LaCl3 was used for leak subtraction.
CDI was evoked by 200-ms hyperpolarizing steps from a holding potential of +30 mV and quantified by dividing the current measured at 195 ms by the peak current at the start of the pulse. Peak current was taken 1–3 ms after the step to minimize the impact of uncompensated capacitance. Peak current was taken at 1–1.5 ms after the step for the most rapidly inactivating channels, using only those cells with the smallest leak-subtracted capacitance artifacts.
Data analysis
The kinetics of Orai1 current inactivation were fit with a biexponential function to generate fast and slow time constants and amplitudes summarized in Figs. S1, S2, and S4. Unitary properties for Na+ currents were derived by applying ensemble variance analysis (Sigworth, 1980; Prakriya and Lewis, 2006) to 200-ms current sweeps collected under continuous holding at −100 mV as in Fig. 3. Estimates of unitary Ca2+ current (iCa) were made by adjusting the unitary Na+ current (iNa) by the relative magnitude of macroscopic ICa and INa as measured using rapid solution exchanges and correcting for the residual level of CDI in 2 mM Ca2+ Ringer’s (Fig. S3; Mullins et al., 2016).
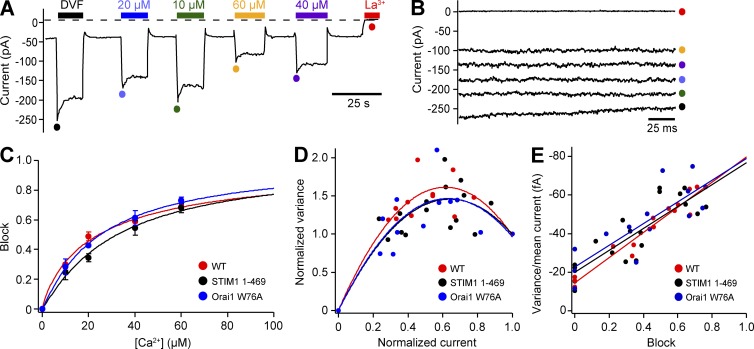
Figure 3.
Ensemble variance analysis of CRAC channels with residual inactivation phenotypes. (A) A thapsigargin-pretreated cell expressing Ch-STIM11-469 and WT Orai1-GFP was held continuously at −100 mV in 2 mM Ca2+o, and as indicated by the bars, DVF solutions containing variable low [Ca2+]o were applied transiently to produce and partially block the Na+-CRAC current. Colored circles indicate peak currents corresponding to the 200-ms sweeps shown in (B), which were divided into 25-ms segments for analysis (see Materials and methods). (C) Ca2+ block of Na+ currents for WT STIM1 + WT Orai1, STIM11–469 + WT Orai1, and WT STIM1 + Orai1 W76A. Na+ current was measured immediately after applying DVF solution containing the indicated [Ca2+]o, and normalized to the value for the pure DVF solution. Each point represents the mean ± SEM for n = 3–4 cells. Lines are fits of the Hill equation, block = 1/[1 + (K1/2/[Ca])nH]. (D) Normalized variance versus current plots for each condition were fitted by a parabolic function to determine open probability (see Materials and methods). (E) Variance/current versus block plots were fitted by a linear function to determine unitary current (see Materials and methods). Unitary channel properties are summarized in Table 1. WT data are reproduced from Mullins et al. (2016).
Data in all summary figures show mean ± SEM. Two-sample t tests were used for statistical comparisons.
Online supplemental material
Fig. S1 shows biexponential time constants and amplitudes of CDI for channels with a residual inactivation phenotype. Fig. S2 shows biexponential time constants and amplitudes of CDI for the alanine scan of IDSTIM. Fig. S3 shows representative data used to estimate iCa from iNa. Fig. S4 compares kinetic parameters of CDI for the STIM11-469 and Orai1 Y80A mutations studied individually and in combination. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201511438/DC1.
RESULTS
CRAC channels inactivate to a limited extent without IDSTIM
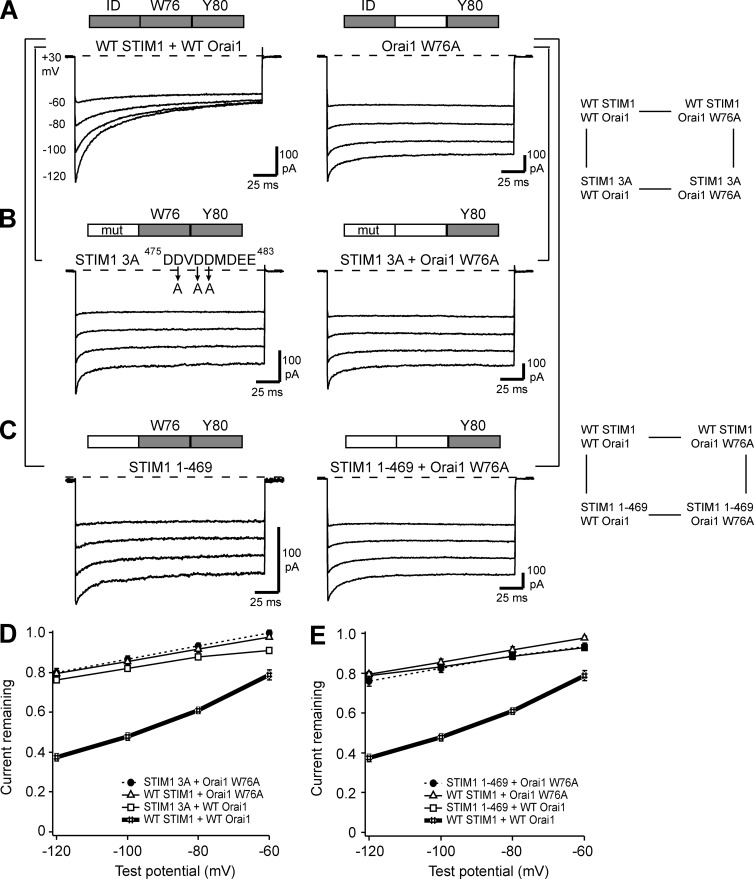
IDSTIM (aa 470–491 of human STIM1) was initially thought to be absolutely required for CDI based on observations that heterologous expression of STIM11–469 with Orai1 produced currents that did not inactivate, whereas STIM11–491 with Orai1 produced currents with normal CDI (Mullins et al., 2009). In this study, we found that expressing STIM11–469 and WT Orai1 at a higher STIM1/Orai1 ratio (see Materials and methods) generated currents with a low but consistent level of inactivation equivalent to approximately one third of the level produced with full-length STIM1 (Fig. 1, A and B). For shorthand, we refer to the inactivation supported by STIM11–469 as “residual inactivation.” We attribute this increased level of inactivation for STIM11–469, like that observed for Orai1 W76A in Mullins et al. (2016), to a higher and more well-controlled STIM1/Orai1 ratio relative to the earlier study.
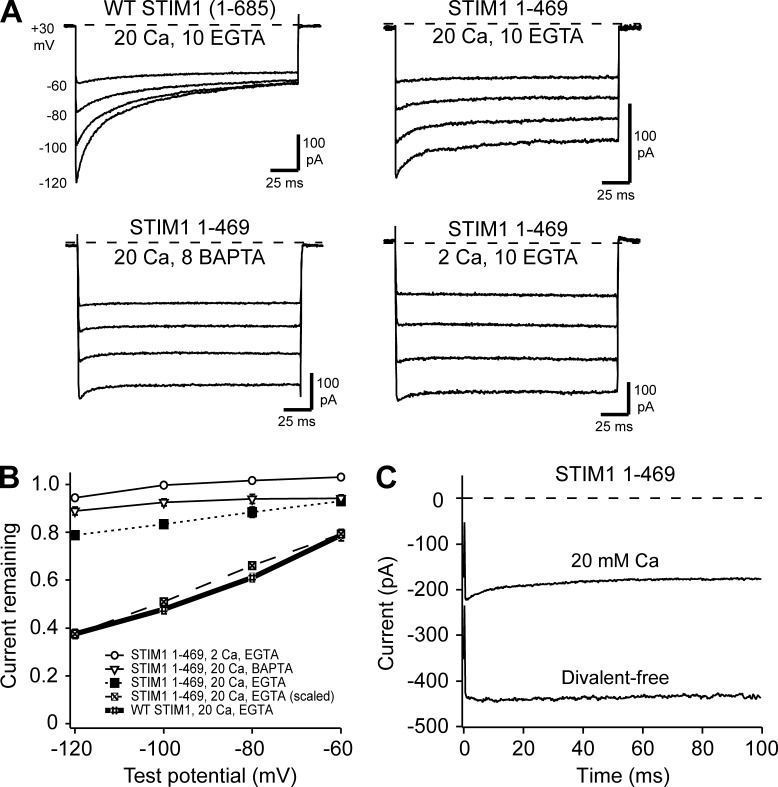
Figure 1.
Residual inactivation in the absence of IDSTIM remains Ca2+-sensitive. All currents were recorded in HEK293H cells cotransfected with WT Orai1-GFP together with either WT mCh-STIM1 (aa 1–685) or mCh-STIM11-469 after induction of ICRAC reached a maximum (∼300 s after break-in). Currents were evoked by 200-ms hyperpolarizations to the voltages indicated. (A) Representative currents are shown with the STIM1 variant and extracellular [Ca2+] and intracellular [EGTA] or [BAPTA] indicated in mM. WT traces are from Fig. 3 A in Mullins et al. (2016). (B) The extent of CRAC channel inactivation for the various conditions, summarized as current remaining at the end of the pulse. Each point represents the mean ± SEM for n = 5–6 cells. (C) Current during 100-ms steps from +30 to −100 mV was recorded from a representative cell expressing STIM11–469 + WT Orai1, first in 20 mM Ca2+o and then in divalent-free (DVF) extracellular solution.
Despite its small amplitude, residual inactivation resembles full CDI in several important ways. Like CDI of WT channels, it follows a double-exponential time course (Fig. S1), and its extent increases with hyperpolarization (Fig. 1 B). Several observations suggest that, like full CDI, the apparent voltage dependence of residual CDI arises from the voltage dependence of Ca2+ entry (Zweifach and Lewis, 1995). The extent of residual inactivation was significantly reduced by lowering extracellular Ca2+ from 20 to 2 mM (P < 10−5 at −120 mV) or by substituting the fast buffer BAPTA for the slower EGTA in the intracellular solution (P < 0.01 at −120 mV; Fig. 1, A and B), as previously reported for CDI of endogenous CRAC channels (Hoth and Penner, 1993; Zweifach and Lewis, 1995). Finally, residual inactivation was completely absent in Na+-ICRAC recordings made under divalent-free conditions (Fig. 1 C). Together, these results demonstrate that residual inactivation observed in the absence of IDSTIM is Ca2+ dependent, similar to native CDI supported by full-length STIM1.
Alanine scan of acidic residues in IDSTIM
Neutralization of combinations of charged residues within IDSTIM has been shown to eliminate, reduce, or enhance CDI, but mutations of individual residues have not been studied (Derler et al., 2009b; Lee et al., 2009; Mullins et al., 2009). To clarify the contributions of individual residues to CDI, we performed a full alanine scan of this negative charge cluster within IDSTIM (475DDVDDMDEE483).
The effects of alanine substitution separated the acidic residues into two groups. In the central group (D476, D478, and D479), individual alanine substitutions reduced the extent of CDI by approximately half (Fig. 2, C–E). A triple alanine substitution for all three residues (Fig. 2 I, “3A”) further reduced CDI to the residual level of approximately one third seen with STIM11–469 (Fig. 2 K; P = 0.28 at −120 mV) and the Orai1 W76A/V/L/I mutants (Fig. 5 G of Mullins et al. [2016]; P = 0.13 for STIM1 3A vs. W76A at −120 mV), indicating that the 3A mutation had eliminated IDSTIM’s ability to boost CDI above the residual level.
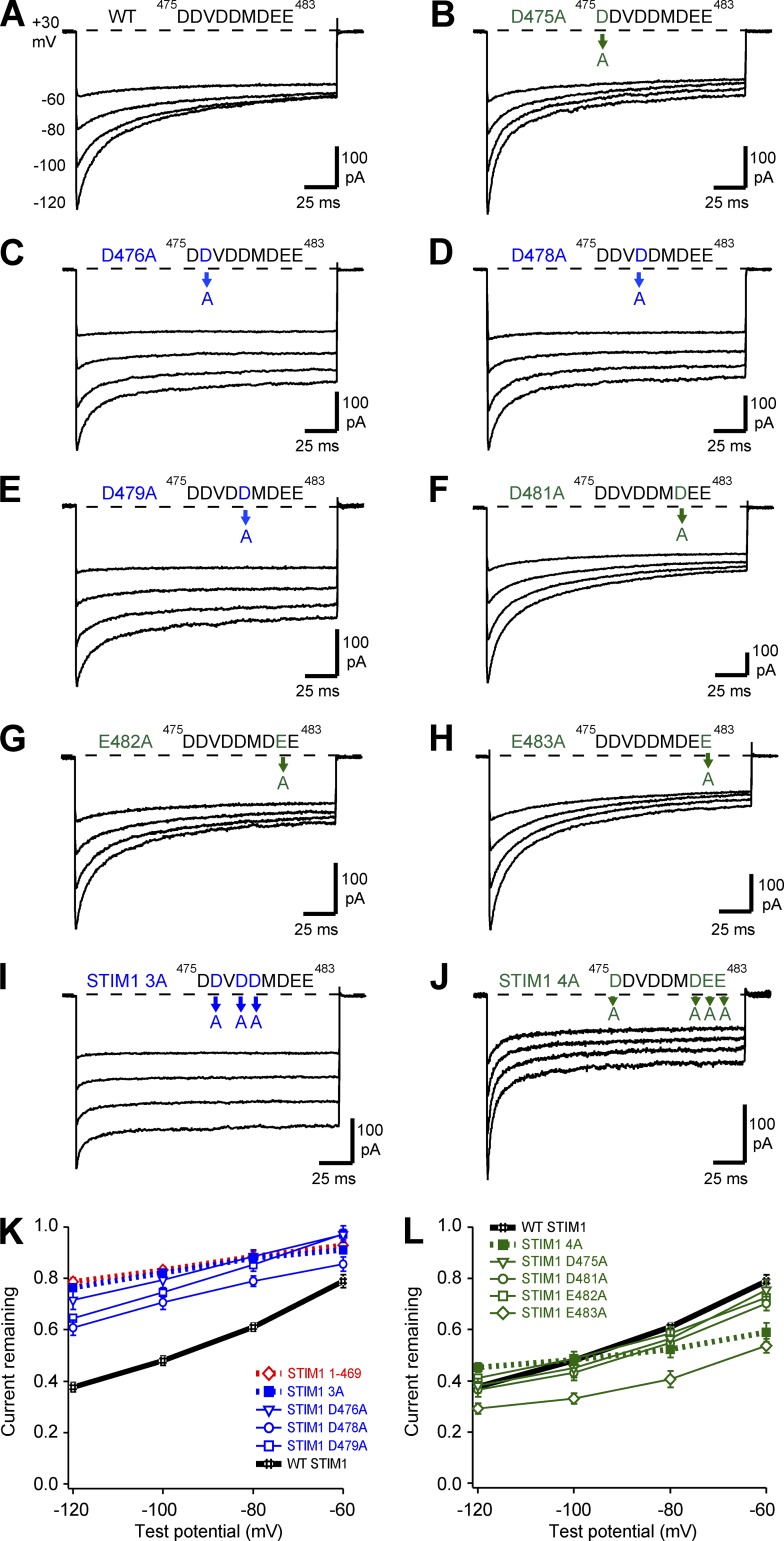
Figure 2.
Alanine scan of IDSTIM reveals two functional groups of acidic residues. All currents were recorded as described in Fig. 1 in cells cotransfected with WT Orai1-GFP together with either WT or mutant Ch-STIM1, with 20 mM Ca2+o and the standard 10 mM EGTA internal solution. Representative currents are shown for WT STIM1 (A, data from Fig. 1 A), D475A (B), D476A (C), D478A (D), D479A (E), D481A (F), E482A (G), and E483A (H). Representative currents for the STIM1 3A triple mutant (I; D476A D478A D479A) and the STIM1 4A quadruple mutant (J; D475A D481A E482A E483A) are also shown. (K) One group of neutralizations, highlighted in blue, strongly reduced the extent of CDI. Summary data for STIM11–469 are shown in red for comparison. (L) A second group of neutralizations, plotted in green, retained strong CDI. Each point on the summary graphs represents the mean ± SEM for n = 5–6 cells.
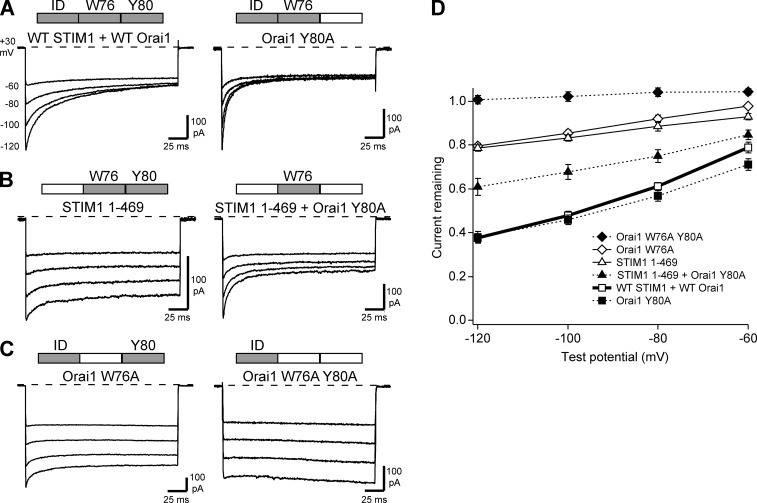
Figure 5.
Orai1 W76A and STIM11–469 have opposite effects on CDI in the Orai1 Y80A background. All currents were recorded as described in Fig. 1 with 20 mM Ca2+o and the standard 10 mM EGTA internal solution. Bars above traces indicate the native elements present in each condition. (A) Representative currents for WT STIM1 + WT Orai1 (from Fig. 1 A) and WT STIM1 + Orai1 Y80A (from Fig. 3 B in Mullins et al., 2016). (B) Representative currents for STIM11–469 + WT Orai1 (from Fig. 1 A) and STIM11-469 + Orai1 Y80A. (C) Representative currents for WT STIM1 + Orai1 W76A (from Fig. 4 A) and WT STIM1 + Orai1 W76A Y80A. (D) Summary of the extent of CDI measured for the different conditions. Each point on the summary graph represents the mean ± SEM for n = 5–6 cells. Although both STIM11–469 and Orai1 W76A have a residual inactivation phenotype when studied on a WT background (B and C, left; D, open symbols), they modulate CDI in opposite directions in the background of Orai1 Y80A (B and C, right; D, closed symbols).
In contrast, individual alanine substitutions in the flanking group of acidic residues either had minimal effects (D475, D481, and E482) or significantly increased (E483; P < 0.02 vs. WT at −120 mV) the extent of CDI (Fig. 2, B and F–H). A quadruple alanine substitution for all four residues (Fig. 2 J, “4A”) inactivated strongly but with an altered apparent voltage dependence and was not studied further (Fig. 2 L, summary). Many individual alanine mutations and the combination mutations in both groups of acidic residues modestly accelerated the kinetics of CDI, with the 4A mutation producing the largest effect (Fig. S2).
Thus, the contributions of individual IDSTIM residues to CDI are complex; although the central group of acidic residues (D476, D478, and D479; Fig. 2, blue) is required to support the full extent of CDI, reducing side chain size and/or charge in either the central or the flanking (green) group of residues appears to accelerate the process (Fig. S2 A).
IDSTIM and Orai1 residue W76 affect the channel’s response to local [Ca2+]
The reduced level of inactivation observed for the STIM11–469 and Orai1 W76A/V/L/I mutants could occur in two ways. The mutants could directly affect the ability of the channel to respond to local Ca2+ accumulation, or they could diminish CDI indirectly by reducing the single-channel Ca2+ current or open probability (Po). Unfortunately, the extremely small unitary conductance of CRAC channels precludes single-channel recording. Therefore, to address this point we applied ensemble current noise analysis (Prakriya and Lewis, 2006) to estimate the unitary current and Po in cells expressing STIM11–469 + WT Orai1 or WT-STIM1 + Orai1 W76A. Monovalent currents were measured under DVF conditions, and current noise and Po were varied by adding micromolar amounts of Ca2+ to produce flickery block. At a constant holding potential of −100 mV, switching from 2 mM Ca2+ to DVF produced a step increase in current due to Na+ influx, followed by a slow decline resulting from Orai1 depotentiation, and returning to 2 mM Ca2+ allowed the channels to repotentiate (Zweifach and Lewis, 1996; Fig. 3 A). The sequence of solution changes was then repeated using DVF containing varying levels of Ca2+, and for each application a single 200-ms trace collected at the peak of the Na+ current was analyzed (Fig. 3, A and B, colored dots). The Ca2+ blocking affinity was similar for WT, Orai1 W76A, and STIM11–469 channels, being well described by the Hill equation with a K1/2 of 24–32 µM and Hill coefficient (nH) of 0.9–1.1 (Fig. 3 C and Table 1). Po values obtained from parabolic fits to the variance versus mean current (Fig. 3 D; see Materials and methods) were similar for the three channels (0.76–0.81; Table 1; P > 0.3 for both STIM11–469 vs. WT and Orai1 W76A vs. WT). Finally, the unitary Na+ current amplitude (iNa) was estimated by extrapolating the variance/current ratio to the limit of complete block (Po = 0; Fig. 3 E); again, all three channels had similar values for iNa of −80 to −83 fA at −100 mV (Table 1; P > 0.7 for both STIM11–469 vs. WT and Orai1 W76A vs. WT). Thus, the Ca2+ affinity, unitary Na+ current, and open probability of channels incorporating STIM11–469 or Orai1 W76A were indistinguishable from that of WT channels.
Table 1.
Unitary channel properties for WT CRAC, STIM11–469, and Orai1 W76A
| Channel | Po | iNa | iCa | Ca2+ affinity (K1/2) | nH | n |
| fA | fA | µM | ||||
| WT CRACa | 0.81 ± 0.02 | 79.6 ± 2.4 | 9.6 ± 0.4 | 24.2 ± 2.0 | 0.87 ± 0.10 | 3 |
| STIM11–469b | 0.78 ± 0.02 | 79.9 ± 6.6 | 14.0 ± 1.5 | 32.0 ± 4.5 | 1.13 ± 0.11 | 4 |
| Orai1 W76Ac | 0.76 ± 0.04 | 83.4 ± 9.2 | 12.4 ± 2.3 | 26.4 ± 4.4 | 1.11 ± 0.08 | 3 |
All channel properties were measured at −100 mV.
WT STIM1 + WT Orai1; data reproduced from Mullins et al. (2016).
STIM11–469 + WT Orai1.
WT STIM1 + Orai1 W76A.
To determine whether the STIM11–469 or Orai1 W76A mutations affected the Ca2+ conductance of single CRAC channels, we made estimates based on our values for iNa. When the extracellular solution is changed from 2 mM Ca2+ to DVF, the whole-cell current increases as the current carrier is switched from Ca2+ to Na+ (Fig. S3 A). Therefore, iCa can be estimated by dividing iNa by the ratio of INa/ICa, after adjusting ICa for the resting level of inactivation in 2 mM Ca2+ (Fig. S3 B; see Materials and methods). Using this approach, we estimated iCa for WT, STIM1 1–469, and Orai1 W76A channels (Table 1). Although iCa values estimated for STIM11–469 and Orai1 W76A are somewhat higher than for WT channels, the differences are not statistically significant (P = 0.063 for STIM11-469 vs. WT; P = 0.363 for Orai1 W76A vs. WT). Thus, we would expect local [Ca2+] near the inner pore to be similar for WT, STIM11–469, and Orai1 W76A channels. Based on these results, we conclude that IDSTIM and Orai1 residue W76 promote CDI by directly affecting the channel’s ability to respond to local [Ca2+]i rather than by altering the local [Ca2+]i itself.
IDSTIM and Orai1 W76 act in concert to drive full-strength CDI
The Orai1 W76A mutation and mutations that abrogate IDSTIM function (STIM1 3A, STIM11–469) all support a similar residual level of CDI. To test for functional coupling between IDSTIM and W76, we applied a double-mutant cycle strategy based on coexpressing IDSTIM-deficient STIM1 mutants (STIM1 3A or STIM11–469) with WT Orai1, WT STIM1 with Orai1 W76A, or the two mutant proteins together (Fig. 4, A–C). STIM1 3A and STIM11–469 produced similar levels of residual inactivation whether they were coexpressed with WT or W76A Orai1 (Fig. 4, D and E). The fact that the residual CDI of the Orai1 W76A mutant was not further reduced when activated by IDSTIM-deficient mutants indicates that IDSTIM and Orai1 W76 are strongly coupled and comprise a functional module that is required to boost CDI from the residual to the full level.
Figure 4.
IDSTIM and Orai1 W76 act in concert to enable full-strength CDI. All currents were recorded as described in Fig. 1 with 20 mM Ca2+o and the standard 10 mM EGTA internal solution. The brackets indicate overlapping mutant cycles. Cells coexpressed the indicated STIM1 and Orai1 variants, and where a label is absent the WT STIM1 or Orai1 protein was expressed. Bars above traces indicate the native CDI elements present in each condition. (A) Representative currents for WT STIM1 + WT Orai1 (from Fig. 1 A) and WT STIM1 + Orai1 W76A (from Fig. 5 B in Mullins et al., 2016). (B) Representative currents for STIM1 3A + WT Orai1 (from Fig. 2 I) and STIM1 3A + Orai1 W76A. (C) Representative currents for STIM11–469 + WT Orai1 (from Fig. 1 A) and STIM11–469 + Orai1 W76A. (D and E) Summary of the extent of CDI measured for the mutant cycles involving STIM1 3A and STIM11–469, respectively. Each point on the summary graphs represents the mean ± SEM for n = 5–6 cells. The residual inactivation persists even when mutant STIM1 and mutant Orai1 are expressed together.
Different functions of IDSTIM and W76 are revealed in the Y80A mutant
Although IDSTIM and W76 appear to act in concert to promote CDI, they are likely to play different roles. We explored these by examining the effects of IDSTIM or W76A mutants on an Orai1 Y80A mutant background. As described, loss of IDSTIM limits CDI to the residual level (STIM11–469; Fig. 5 B, left), whereas Y80A accelerates the kinetics of CDI without altering its extent (Fig. 5 A, right). However, coexpression of Orai1 Y80A with STIM11–469 increased CDI to an intermediate level between that of STIM11–469 and WT STIM1 (Fig. 5 B, right; Fig. 5 D, summary). Importantly, these results clearly show that Orai1 can inactivate to a significant degree (two thirds of the WT extent) without IDSTIM function.
Given that W76 and IDSTIM show strong functional coupling, the simplest expectation was that W76A would produce the same intermediate CDI phenotype as STIM11–469 when combined with the Orai1 Y80A mutation. However, the W76A/Y80A double mutant completely failed to inactivate (Fig. 5, C [right] and D). Thus, alanine substitution for Y80 reveals functional differences in the components of the IDSTIM–W76 module. Whereas W76 alone is capable of responding to Ca2+ and promoting an intermediate level of CDI (Fig. 5 B, right), IDSTIM alone is not (Fig. 5 C, right). Moreover, Y80 appears to limit the inactivating ability of W76 alone (Fig. 5 B), whereas it enables a residual amount of CDI in the absence of W76 (Fig. 5 C, left), IDSTIM (Fig. 5 B, left), or both (Fig. 4 C, right). Results are summarized in Fig. 5 D.
DISCUSSION
In this study, we examined the role of IDSTIM in Ca2+-dependent inactivation of CRAC channels. We identified a core of acidic residues that are most critical for CDI and showed that IDSTIM operates in concert with the Orai1 pore residue W76 to produce full-strength CDI. The IDSTIM–W76 module appears to function by controlling inactivation gating rather than increasing Ca2+ accumulation near the pore or serving as a Ca2+ sensor, and it acts to overcome Y80-dependent residual inactivation to produce the full extent of CDI.
Individual acidic residues in IDSTIM make distinct contributions to CDI
In prior studies, the importance of IDSTIM for CDI was established by the effects of neutralizing combinations of acidic residues, which did not permit the functional contributions of individual residues to be distinguished. The results of alanine scanning show here that three residues (D476, D478, and D479) are most important for enabling full CDI and that the 3A mutant completely eliminates IDSTIM function, as it reduces CDI to the same residual level observed in the complete absence of IDSTIM (STIM11–469; Fig. 2 K). Alanine substitutions of the flanking residues D475, D481, and E482 had little effect on CDI, whereas E483A significantly increased the extent of CDI (Fig. 2 L), likely because of an increased Ca2+ sensitivity of CDI (Mullins et al., 2009). These results help explain the effects of neutralizing combinations of residues in previous studies. Neutralization of D476 and D478 can account for the reduced CDI observed for 475/476 DD→AA and 478/481 DD→GG mutations (Mullins et al., 2009). Similarly, neutralization of D478 and D479 may be responsible for reduced CDI in the 5xA mutation (D478, D479, D481, E482, and E483 mutated to alanine; Derler et al., 2009b). Finally, preserved CDI for the 3xA mutation (D481/E482/E483>AAA; Derler et al., 2009b) and enhanced CDI for the E482/E483>AA mutation (Mullins et al., 2009) are consistent with the preserved or enhanced CDI we observe for the individual D481A, E482A, and E483A mutations (Fig. 2 L). Overall, our results suggest a functional organization of IDSTIM in which a central core of three neighboring aspartate residues (D476, D478, and D479) promote full-strength CDI and a flanking residue (E483) tempers the Ca2+ sensitivity of the process.
Is IDSTIM a Ca2+ sensor for CDI?
Previous evidence supporting a role of IDSTIM as a Ca2+ sensor for CDI has been equivocal. Neutralization of residues that reduced CDI caused a parallel reduction in binding Ca2+ of IDSTIM-containing peptides in a 45Ca2+ overlay assay; however, the E482/E483→AA mutation increased the apparent Ca2+ sensitivity of CDI while reducing 45Ca2+ binding (Mullins et al., 2009). Our current results provide direct evidence against a required role of IDSTIM as Ca2+ sensor by demonstrating that the Orai1 Y80A mutant channels can inactivate to a significant degree (two thirds of the WT level) in the complete absence of IDSTIM (stimulation with STIM11–469; Fig. 5, B and D). Thus, although a limited contribution to Ca2+ sensing cannot be ruled out, the most parsimonious interpretation is that IDSTIM instead shapes the Ca2+ sensitivity of CDI by coupling Ca2+ binding to CDI gating. Viewed in this manner, the E483A and 482/483 EE→AA mutations may increase the apparent Ca2+ sensitivity of CDI by enhancing coupling rather than by directly affecting the Ca2+ binding site.
Full-strength CDI requires the concerted action of IDSTIM and W76
A striking pattern emerging from our data collected at high STIM1/Orai1 ratio was the occurrence of residual inactivation (approximately one third of the full WT extent) under several conditions in which the function of IDSTIM or Orai1 W76 was lost (STIM11–469, STIM1 3A, or W76A; Fig. 4). In previous studies (Derler et al., 2009b; Mullins et al., 2009), residual CDI was masked by potentiation, presumably because of low STIM1/Orai1 ratio (Scrimgeour et al., 2009; Hoover and Lewis, 2011). Our current results further underscore the importance of a high STIM1/Orai1 ratio for determining mutant CDI phenotypes.
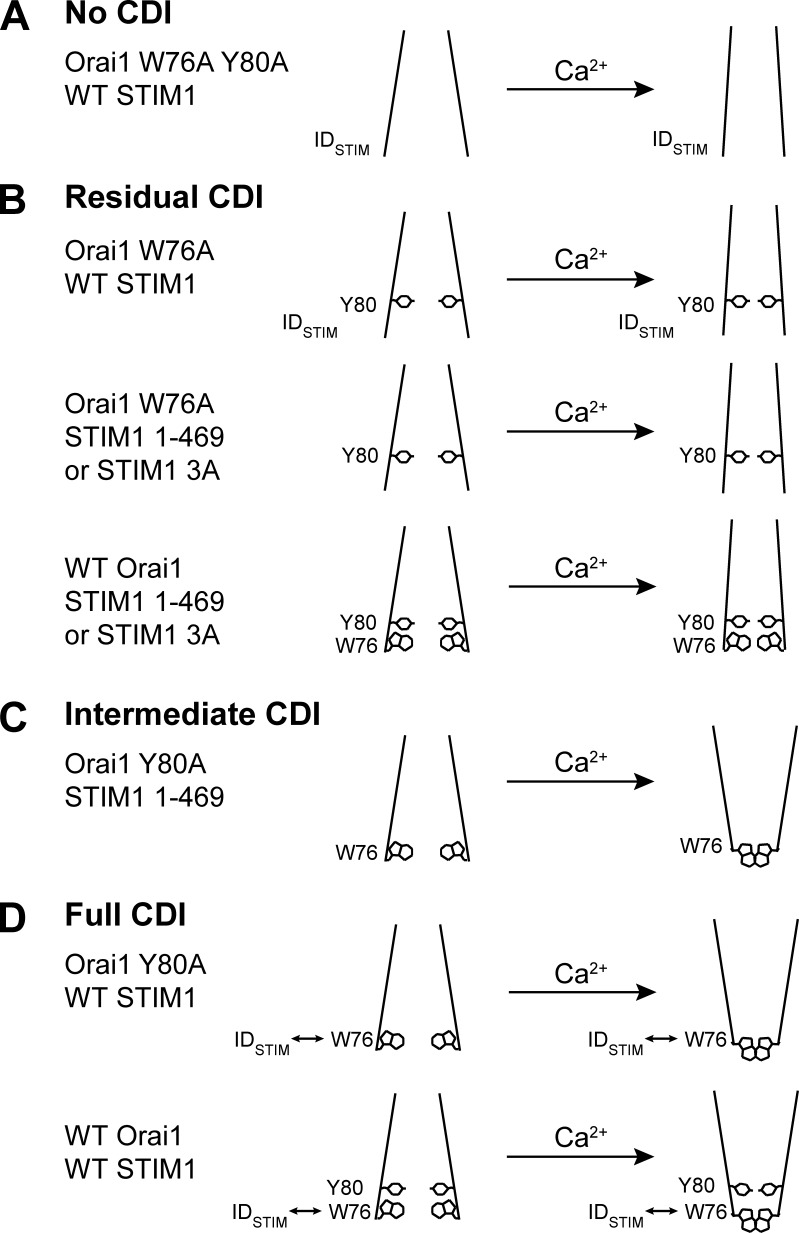
Residual inactivation is Ca2+ dependent with an apparent voltage sensitivity similar to that of full-strength CDI (Fig. 1 B, scaled curve). Because the voltage dependence of CRAC channel CDI can be ascribed to the voltage dependence of Ca2+ entry (Zweifach and Lewis, 1995), this result suggests that the two forms of CDI share a similar Ca2+ sensitivity and thus may use the same Ca2+ sensor. The relation of the residual to full CDI states is not clear, but it could represent an intermediate between open and fully inactivated states, limited occupancy of the fully inactivated state (e.g., through reduced stabilization of the CDI state), or a conformation that is not normally visited when IDSTIM and W76 are functional. Interestingly, the residual inactivated state is distinguished by its dependence on Y80. Of all the mutant combinations we have studied, residual inactivation only occurred when the W76-IDSTIM module was disabled and Y80 was present (Figs. 4 and 5).
In contrast, the full extent of CDI intrinsic to the CRAC channel is independent of Y80 but requires the presence of both W76 and IDSTIM (Fig. 5). A critical finding is that W76 and IDSTIM are strongly coupled and act in concert to generate CDI, based on our observations that loss of W76 and IDSTIM together produces the same level of residual CDI as the loss of each element individually (Fig. 4). Importantly, disruption of the IDSTIM-W76 module does not reduce channel open probability or unitary current (Fig. 3 and Table 1), which suggests that rather than affecting the accumulation of Ca2+, IDSTIM and W76 act together to foster full CDI by enabling the channel’s response to a given local [Ca2+]. Given the evidence discussed against IDSTIM as a Ca2+ sensor itself, a more likely function for IDSTIM is to help couple the Ca2+ sensor to movements of W76 leading to full strength inactivation.
Strong coupling between IDSTIM and W76 does not necessarily imply that the functions of the two elements are identical, and in fact, distinct roles of IDSTIM and W76 are indicated by the effects of disrupting the IDSTIM-W76 module in Orai1 Y80A mutant channels. In this channel background, loss of IDSTIM produced an intermediate level of CDI, whereas loss of W76 eliminated it entirely (Fig. 5). The complete absence of CDI in W76A/Y80A channels underscores the critical function of W76 in enabling CDI. That W76 alone can support a substantial amount of CDI indicates that even in the absence of IDSTIM, W76 can respond to Ca2+ binding and enable a conformational change or blocking particle binding to close the channel.
Possible models for CRAC channel CDI
Functional insights from this study regarding the roles of IDSTIM, W76 and Y80 in CRAC channel CDI are summarized in Fig. 6. Bulky hydrophobic side chains in the inner pore appear to be required for CDI, because CDI is completely eliminated in the absence of W76 and Y80 (Fig. 6 A). Y80 alone is sufficient to support a residual level of CDI, which occurs independently of IDSTIM or W76 (Fig. 6 B). That the removal of Y80 allows W76 alone to support an increased level of CDI (Fig. 6 C) implies that Y80 impedes W76-dependent full CDI. When IDSTIM and W76 act together to boost CDI to its full extent, Y80 slows the kinetics, suggesting that the large hydrophobic Y80 side chain imposes an energy barrier along the pathway to the inactivated state (Fig. 6 D). These results lead us to conclude that a critical function of the IDSTIM-W76 module is to overcome the barriers imposed by Y80 in order to reach the fully inactivated state.
Figure 6.
A summary of functional roles for W76, Y80, and IDSTIM. Open-channel states are on the left, and inactivated states supported by the various combinations of functional elements tested are on the right. For convenience, we depict inactivation schematically as a conformational change in the inner pore, but other mechanisms are also possible (see Discussion). (A) Bulky hydrophobic side chains of W76 and Y80 in the inner pore are required for CDI. (B) The Orai1 Y80 side chain by itself is sufficient to support residual CDI (approximately one third of WT CDI) in the absence of a functional IDSTIM–W76 module. (C) In the absence of Y80 and IDSTIM, Orai1 W76 by itself can support an intermediate level of CDI (approximately two thirds of WT CDI), revealing an inhibitory role for Y80. (D) The IDSTIM–W76 module overcomes the Y80-dependent residual CDI depicted in B to enable full CDI. Y80 slows the kinetics of CDI, suggesting that it imposes an energy barrier on the path to full inactivation.
The ability of W76 alone to support a significant degree of CDI provides new clues regarding its mechanistic role. In principle, W76 could form the binding site for a proteinaceous blocking particle, form a hydrophobic gate, or enable conformational changes leading to closure at another site. The polyanionic character of IDSTIM initially suggested the possibility that it might interact with the three rings of positively charged residues in the pore (R83, K87, and R91) to act as a blocking particle, thus explaining why introduction of negative charges at these pore sites inhibit CDI (Mullins et al., 2016). However, if a blocking particle is involved, it cannot be IDSTIM, as inactivation clearly occurs in its absence in Y80A Orai1 channels (Fig. 5 B). Interestingly, a short sequence within the intracellular Orai1 II-III linker (153NVHNL157) has been proposed to function as a pore-blocking particle during CDI (Srikanth et al., 2010). A mechanism in which the linker sequence binds via hydrophobic interactions with a ring of W76 side chains would be consistent with our results showing that significant levels of inactivation require bulky hydrophobic residues at position 76 (W76F/Y; Fig. 5 in Mullins et al., 2016). Measuring the effects of combined mutations in the linker, W76, and IDSTIM may provide a means of testing for functional interactions of the linker with the IDSTIM-W76 module.
As an alternative gating mechanism a ring of W76 side chains could be brought together during CDI to function as a hydrophobic gate, by analogy to the block of conduction by the Orai1 R91W pore mutation (Feske et al., 2006; Derler et al., 2009a). W76 could also stabilize pore conformations in which other residues form the gate, such as V102 and the adjacent hydrophobic region (McNally et al., 2012; Gudlur et al., 2014). Further studies will be needed to define the structural basis of the inactivated state and its relation to the closed state, and to determine whether residual inactivation represents incomplete stabilization of the full inactivation state or a different conformation with a “leaky” gate.
Our results defining the functional core of the IDSTIM domain (D476/D478/D479) and its close functional interaction with W76 raise the question of how coupling is achieved. Although coupling could occur through direct contact, in light of steric constraints, it seems more likely to involve allosteric interactions between IDSTIM and W76. The identification of additional residues that mediate the functional connection between W76 and the core of IDSTIM may offer a means to more completely define the pathway leading to inactivation.
The physiological role of CRAC channel CDI is currently unknown. Reduced fast CDI in a human gain-of-function mutation (STIM1 p.R304W) associated with a Stormorken syndrome presentation has been reported, but the mutation also yielded CRAC channels with constitutive activity, confounding clear interpretation with respect to CDI (Nesin et al., 2014). Testing for a physiological role of CRAC channel CDI will require precise ablation or enhancement of CDI, best accomplished by genetic introduction of mutations that alter the amount of CDI without perturbing local Ca2+ accumulation. Based on our results, specific knockin mutations at Orai1 W76, Y80, or IDSTIM that reduce or eliminate inactivation offer a promising new approach to unraveling the physiological roles of CRAC channel CDI.
Supplementary Material
Acknowledgments
The authors thank Luda Lokteva for technical support, members of the Lewis laboratory for their helpful comments throughout the work, Rick Aldrich for critical reading of the manuscript, and Steve Galli for support of physician-scientist development.
This work was supported by a mentored career development award from the National Institute of General Medical Sciences (NIGMS; 5K08GM098880 to F.M. Mullins) and by a MERIT award from NIGMS (R37GM045374 to R.S. Lewis).
The authors declare no competing financial interests.
Kenton J. Swartz served as editor.
Footnotes
Abbreviations used in this paper:
- CDI
- Ca2+-dependent inactivation
- CRAC
- Ca2+ release–activated Ca2+
- DVF
- divalent-free
- IDSTIM
- inactivation domain of STIM
References
- Bers D.M., Patton C.W., and Nuccitelli R.. 2010. A practical guide to the preparation of Ca2+ buffers. Methods Cell Biol. 99:1–26. [DOI] [PubMed] [Google Scholar]
- Cox D.H. 2011. Ca2+-regulated ion channels. BMB Rep. 44:635–646. 10.5483/BMBRep.2011.44.10.635 [DOI] [PubMed] [Google Scholar]
- Derler I., Fahrner M., Carugo O., Muik M., Bergsmann J., Schindl R., Frischauf I., Eshaghi S., and Romanin C.. 2009a. Increased hydrophobicity at the N terminus/membrane interface impairs gating of the severe combined immunodeficiency-related ORAI1 mutant. J. Biol. Chem. 284:15903–15915. 10.1074/jbc.M808312200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derler I., Fahrner M., Muik M., Lackner B., Schindl R., Groschner K., and Romanin C.. 2009b. A Ca2+ release-activated Ca2+ (CRAC) modulatory domain CMD) within STIM1 mediates fast Ca2+-dependent inactivation of ORAI1 channels. J. Biol. Chem. 284:24933–24938. 10.1074/jbc.C109.024083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.H., Tanasa B., Hogan P.G., Lewis R.S., Daly M., and Rao A.. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 441:179–185. 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Gudlur A., Quintana A., Zhou Y., Hirve N., Mahapatra S., and Hogan P.G.. 2014. STIM1 triggers a gating rearrangement at the extracellular mouth of the ORAI1 channel. Nat. Commun. 5:5164 10.1038/ncomms6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover P.J., and Lewis R.S.. 2011. Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. USA. 108:13299–13304. 10.1073/pnas.1101664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth M., and Penner R.. 1993. Calcium release-activated calcium current in rat mast cells. J. Physiol. 465:359–386. 10.1113/jphysiol.1993.sp019681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.P., Yuan J.P., Zeng W., So I., Worley P.F., and Muallem S.. 2009. Molecular determinants of fast Ca2+-dependent inactivation and gating of the Orai channels. Proc. Natl. Acad. Sci. USA. 106:14687–14692. 10.1073/pnas.0904664106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik R.M., Wu M.M., Buchanan J., and Lewis R.S.. 2006. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J. Cell Biol. 174:815–825. 10.1083/jcb.200604015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally B.A., Somasundaram A., Yamashita M., and Prakriya M.. 2012. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 482:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins F.M., Park C.Y., Dolmetsch R.E., and Lewis R.S.. 2009. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. USA. 106:15495–15500. 10.1073/pnas.0906781106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins F.M., Yen M., and Lewis R.S.. 2016. Orai1 pore residues control CRAC channel inactivation independently of calmodulin. J. Gen. Physiol. 10.1085/jgp.201511437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesin V., Wiley G., Kousi M., Ong E.C., Lehmann T., Nicholl D.J., Suri M., Shahrizaila N., Katsanis N., Gaffney P.M., et al. 2014. Activating mutations in STIM1 and ORAI1 cause overlapping syndromes of tubular myopathy and congenital miosis. Proc. Natl. Acad. Sci. USA. 111:4197–4202. 10.1073/pnas.1312520111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Hoover P.J., Mullins F.M., Bachhawat P., Covington E.D., Raunser S., Walz T., Garcia K.C., Dolmetsch R.E., and Lewis R.S.. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 136:876–890. 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M., and Lewis R.S.. 2006. Regulation of CRAC channel activity by recruitment of silent channels to a high open-probability gating mode. J. Gen. Physiol. 128:373–386. 10.1085/jgp.200609588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M., and Lewis R.S.. 2015. Store-operated calcium channels. Physiol. Rev. 95:1383–1436. 10.1152/physrev.00020.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrimgeour N., Litjens T., Ma L., Barritt G.J., and Rychkov G.Y.. 2009. Properties of Orai1 mediated store-operated current depend on the expression levels of STIM1 and Orai1 proteins. J. Physiol. 587:2903–2918. 10.1113/jphysiol.2009.170662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth F.J. 1980. The variance of sodium current fluctuations at the node of Ranvier. J. Physiol. 307:97–129. 10.1113/jphysiol.1980.sp013426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Jung H.J., Ribalet B., and Gwack Y.. 2010. The intracellular loop of Orai1 plays a central role in fast inactivation of Ca2+ release-activated Ca2+ channels. J. Biol. Chem. 285:5066–5075. 10.1074/jbc.M109.072736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Lu J., Li Z., Yu X., Chen L., and Xu T.. 2006. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem. Biophys. Res. Commun. 350:969–976. 10.1016/j.bbrc.2006.09.134 [DOI] [PubMed] [Google Scholar]
- Zweifach A., and Lewis R.S.. 1995. Rapid inactivation of depletion-activated calcium current (ICRAC) due to local calcium feedback. J. Gen. Physiol. 105:209–226. 10.1085/jgp.105.2.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifach A., and Lewis R.S.. 1996. Calcium-dependent potentiation of store-operated calcium channels in T lymphocytes. J. Gen. Physiol. 107:597–610. 10.1085/jgp.107.5.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.