Abstract
Background & Aims
In fecal samples from patients with chronic constipation, the microbiota differs from that of healthy subjects. However, the profiles of fecal microbiota only partially replicate those of the mucosal microbiota. It is not clear whether these differences are caused by variations in diet or colonic transit, or are associated with methane production (measured by breath tests). We compared the colonic mucosal and fecal microbiota in patients with chronic constipation and in healthy subjects to investigate the relationships between microbiota and other parameters.
Methods
Sigmoid colonic mucosal and fecal microbiota samples were collected from 25 healthy women (controls) and 25 women with chronic constipation and evaluated by 16S ribosomal RNA gene sequencing (average of 49,186 reads/sample). We assessed associations between microbiota (overall composition and operational taxonomic units) and demographic variables, diet, constipation status, colonic transit, and methane production (measured in breath samples after oral lactulose intake).
Results
Fourteen patients with chronic constipation had slow colonic transit. The profile of the colonic mucosal microbiota differed between constipated patients and controls (P<.05). The overall composition of the colonic mucosal microbiota was associated with constipation, independent of colonic transit (P<.05) and discriminated between patients with constipation and controls with 94% accuracy. Genera from Bacteroidetes were more abundant in the colonic mucosal microbiota of patients with constipation. The profile of the fecal microbiota was associated with colonic transit before adjusting for constipation, age, body mass index, and diet; genera from Firmicutes (Faecalibacterium, Lactococcus, and Roseburia) correlated with faster colonic transit. Methane production was associated with the composition of the fecal microbiota, but not with constipation or colonic transit.
Conclusions
After adjusting for diet and colonic transit, the profile of the microbiota in the colonic mucosa could discriminate patients with constipation from healthy individuals. The profile of the fecal microbiota was associated with colonic transit and methane production (measured in breath), but not constipation.
Keywords: microbiome, irritable bowel syndrome
BACKGROUND
Irritable bowel syndrome (IBS) is associated with alterations in colonic mucosal and fecal microbiota1–8 that may conceivably perturb the crosstalk between the gut microbiota, the enteroendocrine system, the immune system, and intestinal permeability.9 However, a review of 22 studies that evaluated the gut microbiome in IBS concluded that “results to date are inconsistent and sometimes contradictory” perhaps partly due to differences in molecular techniques, the use of single samples, and other factors (e.g., diet and phenotypic characterization of patients).10 Most studies were conducted outside the United States6 and assessed the fecal microbiota, which are readily accessible, but do not “fully replicate mucosally associated profiles.”11 This is a significant limitation because the mucosal microbiota might affect epithelial and mucosal function to a greater degree than the fecal bacteria.12–16 It is unclear whether differences between health and IBS are primary or secondary to other factors that affect gut microbiota such as diet, body mass index (BMI), and gastrointestinal (GI) transit.9 Indeed, colonization of germ-free mice with human fecal microbiota suggests that alterations in microbiota affect GI transit and vice versa.17 Alterations in diet can also directly affect the microbiota, both directly and by modifying GI transit.17
Most studies of the gut microbiota have evaluated people with IBS and diarrhea-predominant IBS. Eight studies, involving a total of 98 patients, assessed the gut microbiota in constipation-predominant irritable bowel syndrome (IBS-C).9 The results of 4 studies, 2 each based on bacterial cultures and 16S rRNA pyrosequencing, observed conflicting results in regard to the microbiota in functional constipation.18–21 A comparison of fecal microbiota in 20 healthy people and 22 constipated patients (i.e., 10 patients with IBS-C and 12 with alternating IBS) identified several taxa that were associated with delayed colonic transit. The IBS patients fell into 2 clusters characterized by normal microbiota and increased Firmicutes-associated taxa and a depletion of Bacteroidetes-related taxa.7 Similarly, an increase in several genera of Firmicutes was observed in obese constipated children.20
Breath methane excretion after oral glucose is greater in adults with slow transit constipation than in healthy adults or adults with normal transit constipation.22 Increased breath CH4 production was correlated with slower colonic transit. Methane increased non-propagating small bowel contractile activity and decreased small bowel transit in animal models,23 prompting the hypothesis that alterations in colonic microbiota with increased methane production may delay colonic transit.24
Hence, the objectives of this study were to: (i) compare the colonic mucosal and fecal microbiota in healthy people and patients with chronic constipation; (ii) ascertain if the differences in microbiota between constipated patients and healthy people can be explained by differences in demographic features, diet, and colonic transit between groups; and (iii) evaluate the relationship between microbiota and breath methane production. Our hypotheses were that: (i) there are differences in the fecal and mucosal microbiota between healthy people and constipated patients; (ii) these differences are independent of differences in GI transit and diet between healthy people and constipated patients; and (iii) breath methane production is associated with specific profiles of microbiota.
METHODS
Study Design and Participants
From February 2013 through April 2014, 25 female patients with chronic constipation and 25 healthy women, both non-smokers and aged between 18–80 years, consented to participate in this study that had been approved by the Institutional Review Board at Mayo Clinic. Patients had Rome III symptom criteria for a lower functional gastrointestinal disorder with significant constipation.25 Twenty of 25 patients reported having symptoms for at least 4 years before the study. With one exception, 24 of 25 patients lived within 350 miles of Mayo Clinic, Rochester, MN; 21 of these lived within 150 miles. Neither the healthy participants nor the constipated patients had clinical evidence of significant systemic (e.g., cardiovascular) disease that could potentially interfere with the objectives of the study and/or pose safety concerns; prior gastric, intestinal, or colonic resection; inflammatory bowel disease; gastrointestinal cancer; or antibiotic use within 3 months prior to the study.
Study procedures were performed in the following order: collection of a stool specimen, evaluation of breath hydrogen and methane excretion after oral lactulose, assessment of gastric emptying and small intestinal and colonic transit by scintigraphy, and a flexible sigmoidoscopy with colonic mucosal biopsies. Dietary intake and bowel patterns were evaluated as detailed below. Medications that could affect gastrointestinal motility were discontinued for 2 days prior to and for the duration of the study. On average, 15 days (range 2–43 days) elapsed between the collection of stool samples and the flexible sigmoidoscopy.
Assessment of Dietary Intake
Before starting study procedures, a registered dietitian advised participants to maintain a stable diet for 1 week before and throughout the study; and to follow a low fiber diet and avoid dairy products, high fructose corn syrup, fruits, fruit juices, honey, "sugar-free" candies, gums or products containing sorbitol, and live or active cultured-containing foods for 24 hours prior to the lactulose breath test. Participants were instructed to complete a food record for 3 days before the stool collection. Nutrition analysis of the food records was performed using the ESHA Food Processor software (Version 10.14, ESHA Research, Salem, OR).
Lactulose breath test
This test was performed to identify methane producers. After an overnight fast, an antiseptic mouthwash was administered to prevent lactulose fermentation by oropharyngeal bacteria. Breath samples were collected at baseline, at 15-minute intervals for 60 minutes, and at 30-minute intervals for the next 120 minutes after administration of lactulose syrup (10 g) followed by sterile water (250 mL). The duration for which samples were collected was prolonged to 120 minutes because slow transit constipation is associated with delayed small intestinal transit.26 Breath methane and hydrogen concentrations were measured with gas chromatography (Quintron Instrument Company, Milwaukee, WI) and quantified as the area under the curve by the trapezoidal rule.
Scintigraphic Assessment of Gastric Emptying, Small Intestinal, and Colonic Transit
Gastric emptying, small bowel transit, and colonic transit were measured with standard, validated scintigraphic methods.27, 28 Gastric and small bowel transit was measured with 99m-labeled technetium (99mTc), while colonic transit was evaluated with a methacrylate-coated, delayed-release capsule containing indium-111 (111In) adsorbed on activated charcoal particles. Colonic transit was summarized as the colonic geometric center (GC), which is the weighted average of counts in the different colonic regions at 24 (GC24) and 48 (GC48) hours. A higher GC reflects a faster colonic transit.
Stool Collection
Standardized instructions and stool kits were provided to patients for collecting stool samples, which were frozen and stored in a −80°C freezer. One patient required an enema, and one patient required a laxative before providing a stool sample.
Colonic Biopsies
After 1–2 Fleet’s enemas, a flexible sigmoidoscopy was performed. The mucosa was normal in all participants. Five mucosal biopsies were obtained with a 2 mm forceps without a pin from the mid-sigmoid colon, snap frozen, and stored in a −80°C freezer.
Sequencing and Analytical Methods
DNA was extracted from stool with a commercial kit (MoBio DNA extraction kit, Carlsbad, CA) following standard Human Microbiome Project guidelines.29 After extraction, total DNA was quantified using Qubit assay kit (Life Technologies Corporation, NY, USA); in all cases, this was >100 ng/μL, with an average yield of 3799 (range 134 – 40,800) ng/μL. 16S-based sequencing was performed with an Illumina MiSeq sequencer (Illumina Inc, San Diego, CA). Phylotype profiles of the microbiota from healthy and constipated populations were generated using deep rDNA hypervariable tag sequencing of the hypervariable V3-V5 region of the small subunit (SSU) rRNA gene, which has been validated for use with human microbiota and is the preferred technique in the Human Microbiome Project. With the longer reads from the MiSeq (300x300 paired end reads), sequencing included the V3-V5 regions, thereby optimizing the phylogenetic analysis30; the 300 base pair reads ensured optimal phylogenetic identification. Barcoding of samples prior to sequencing yielded an average of 49,186 reads per sample (stool: range 9,438 – 161,117; mucosal biopsy: range 3,116 – 425,701), ensuring detection of both dominant (core microbiota) and poorly-represented taxa (variable microbiota). Paired end reads were stitched, aligned, and classified using a custom pipeline (TORNADO v2.0).31 Briefly, low base quality reads were either trimmed or discarded,32 and these reads were not classified as a bacteria kingdom33 or matched to the bacteria 16S rRNA secondary structure.34 To evaluate the microbial diversity and abundance, UPARSE was used for Operational Taxonomical Units (OTU) clustering,35 and FastTree was used for phylogeny.36 The 16S data were clustered into OTUs at 97% sequence similarity, and the taxonomy was assigned using the Ribosomal Database Project classifier.
Statistical analysis
To assess the differences between microbiome profiles, i.e., β diversity, we used both unweighted and weighted Unifrac distances (‘GUniFrac’ function in the R package ‘GUniFrac’). These metrics capture different parameters, with unweighted UniFrac reflecting differences in community membership (i.e., presence or absence of OTU), while the weighted UniFrac better reflects differences in OTU abundance.37 To prevent artifacts in the analysis that can arise from unequal sequencing depth, rarefaction was performed on the OTU table before calculating the unweighted UniFrac distance. In order to identify associations between microbial community profiles and variables of interest (e.g., constipation status, colonic transit), we used a permutational multivariate analysis of variance (PERMANOVA) approach (‘adonis’ function in the R package ‘vegan’) that adjusts for potential confounding covariates. For all variables, statistical significance of the PERMANOVA results was assessed using 1,000 random microbial community/variable permutations.
In addition to performing an association analysis with the OTU-based microbiome profiles, we also uncovered associations between specific taxa and variables of interest. This was done by comparing relative taxa abundance and variables of interest using a linear model with adjustments for covariates where appropriate. Because the taxa data is not normally distributed, assessment of statistical significance considered 1,000 permutations with the F-statistic as the test statistic. Associations were evaluated at the phylum, family, and genus level. The false discovery rate (FDR) control was performed based on the Benjamini-Hochberg procedure to correct for multiple testing, i.e., ‘p.adjust’ in R. Analysis was confined to taxa with a prevalence greater than 10% and a maximum proportion (relative abundance) greater than 0.002. An FDR-adjusted P value (or Q-value) less than 5% was considered to be significant. A machine learning (“random forest [RF]”) algorithm,38 due to its non-parametric assumptions, was used to evaluate both linear and nonlinear relationships between gut microbiota (i.e., genus or OTU-level relative abundance data) and specific phenotypes (i.e., constipation versus health, colonic transit, and methane production). This process employed bootstrapping to evaluate the accuracy of prediction. The RF-based prediction was compared to random guess, where the prediction was based on the majority class (binary phenotype) or outcome mean (continuous phenotype) in the training set. The Friedman Rank Sum test was used to test the significance of different prediction models. The Boruta algorithm was used to identify the most predictive taxa based on the importance values produced by RF.39 The importance value of a taxon was calculated by RF based on the loss of accuracy by random permutation of the abundance profile of the taxon. To assess whether the importance was significant, the Boruta algorithm compared the observed importance to those produced by the spiked-in ‘shadow’ taxa, which were randomized versions of real taxa. Hence, this algorithm generally provides more power to identify taxa that jointly predict a phenotype.
Receiver Operating Characteristic (ROC) curves were used to estimate the extent to which the differences in clinical parameters (e.g., constipation status and colonic transit) could be discriminated by microbiota profiles, and the incremental utility of the microbiota over other clinical parameters (e.g., age, diet) for evaluating the same. The extent to which differences between the microbiota profiles of the two groups can be explained by the relative abundances of those select taxa that were found to be significantly different was assessed by a principal component analysis.
A post-hoc power assessment40 demonstrated that for a comparison of microbiota between two groups, a sample size of 20 participants per group provides approximately 80% power to detect a medium to strong difference in the microbiota profile between groups (Ω2=0.07), and 20% power to detect a weak difference (Ω2=0.02) (See Appendix for details). Therefore, the positive results for major associations of interest indicate the variable of interest (e.g., constipation status) has a strong effect on the gut microbiota while the negative results probably indicate a weaker effect, if it exists. All statistical analyses were performed in R-3.0.2 (R Development Core Teams, Vienna, Austria).
RESULTS
Patient Characteristics and Dietary Assessment
Of the 25 patients, 13 had symptoms of functional constipation, 6 had IBS-C, and 6 had mixed IBS. Patients with mixed IBS had predominant symptoms of constipation, which they rated as moderate (2 patients), severe (3 patients), or very severe (1 patient). These patients had several (3 in 2 patients, 4 in 1 patient, 5 in 2 patients, and 6 in 1 patient) of 6 Rome III symptom criteria for constipation. They were categorized as mixed IBS because they also had a history of loose stools. The constipated patients were older (P=.02) than healthy participants but had a similar BMI (Table 1). The total caloric intake was greater (P=.005) in healthy participants. Compared to patients, healthy participants also consumed more carbohydrate (P=.054), protein (P=.002), fat (P=.03), and fiber (P=.01) when expressed as an absolute amount, but not as a proportion of the total calorie intake (Table 1). Breath methane production (AUC) was not different (P=.20) in patients than controls. Gastric emptying at 2 hours, but not at 4 hours, was lower (P<.01) in constipated patients than in healthy participants. There was no difference in small intestinal transit between the two groups. Fourteen patients (9 with functional constipation, 3 with IBS-C, and 2 with mixed IBS) but only 2 controls (P<.005), had delayed colonic transit. Six healthy participants but no patients had rapid colonic transit. Colonic transit (GC24) was directly correlated with total calorie intake (P<.05) and total fiber intake (P<.05), and inversely correlated with age (P<.05).
Table 1.
Summary of Patient Characteristics a
| Variable | Healthy (n=25) | Constipated (n=25) | P value |
|---|---|---|---|
| Age, y | 39±10 | 48±15 | .02 |
|
| |||
| BMI, kg/m2 | 26±4 | 25±4 | .13 |
|
| |||
| Total caloric intake, kcal | 1597±402 | 1265±350 | .005 |
|
| |||
| Carbohydrate, g | 188±54 | 155±62 | .054 |
|
| |||
| Protein, g | 75±19 | 60±26 | .002 |
|
| |||
| Fat, g | 60±26 | 46±15 | .03 |
|
| |||
| Fiber, g | 17±13 | 12±4 | .01 |
|
| |||
| Carbohydrate (% of total calories) | 47±9 | 49±12 | .24 |
|
| |||
| Protein (% of total calories) | 20±6 | 20±7 | .88 |
|
| |||
| Fat (% of total calories) | 33±6 | 32±6 | .85 |
|
| |||
| Breath methane (AUC, ppm*min) | 1488±2895 | 4100±6656 | .20 |
|
| |||
| Gastric emptying, % | |||
| 2 hours | 59±18 | 47±11 | .005 |
| 4 hours | 93±10 | 90±12 | .11 |
|
| |||
| Small intestinal transit (Colonic filling [%] at 6 hours) | 45±27 | 48±26 | .74 |
|
| |||
| Colonic transit, GC24 | 2.6±1.1 | 1.6±0.8 | \.0006 |
|
| |||
| Colonic transit, GC48 | 3.9±0.9 | 2.8±1.0 | .001 |
Abbreviations: GC24, geometric center of colonic transit at 24 hours; GC48, geometric center of colonic transit at 48 hours
All data presented as mean (SD)
Univariate Associations Between Overall Composition of Microbiota and Other Variables
For each univariate association, two P values reflecting the results based on unweighted and weighted UniFrac distance matrices are provided (Table 2). The overall fecal microbial composition was significantly associated with colonic transit, constipation status, age, and total calorie intake. Associations with carbohydrate intake, and fat intake were of borderline significance.
Table 2.
Associations Between Microbiota and Other Variables of Interest
| Variable | Stool | Mucosal | ||
|---|---|---|---|---|
|
| ||||
| Unweighted UniFrac a | Weighted UniFrac a | Unweighted UniFrac a | Weighted UniFrac a | |
| Univariate analyses | ||||
| Age, y | .03 | .02 | .09 | .13 |
| BMI | .62 | .15 | .73 | .72 |
| Calories | .04 | .14 | .92 | .12 |
| Carbohydrate | .26 | .09 | .93 | .86 |
| Fat | .31 | .07 | .73 | .59 |
| Constipation status | .049 | .09 | <.001 | .005 |
| Colonic transit | .22 | .008 | .07 | .07 |
| Breath methane | <.001 | .02 | .01 | .97 |
| Multiple variable analyses | ||||
| Constipation status (adjusted for age, BMI, diet, and colonic transit) | .50 | .39 | <.001 | .048 |
| Colonic transit (adjusted for age, BMI, diet, and constipation status) | .92 | .08 | .55 | .55 |
| Breath methane (age, BMI, constipation status, colonic transit, diet) | <.001 | .03 | .005 | .99 |
P values
Abbreviations: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); UniFrac, ‘GUniFrac’ function in R package
The overall colonic mucosal microbiota composition was significantly associated with constipation status. However, associations with age and colonic transit were of borderline significance (Table 2).
Multiple Variable Associations Between Overall Composition of Microbiota and Other Variables
After adjusting for age, BMI, diet, and colonic transit, the fecal microbiota was not associated with constipation status (Table 2). In contrast, after adjusting for age, BMI, and colonic transit, the mucosal microbial composition remained strongly associated with constipation status. Because diet was not univariately associated with mucosal microbiota, the analysis for association between mucosal microbiota and constipation did not adjust for the same.
After adjusting for age, BMI, diet, and constipation status, the fecal microbiota was borderline (P=0.9, 0.08) associated with colonic transit with the weighted UniFrac distance only. The colonic mucosal microbiota composition was not associated with colonic transit, after adjusting for age, BMI, and constipation status.
Associations between Microbiota and Breath Methane Production
Breath methane production was associated with fecal and with mucosal microbiota before and after adjusting for age, BMI, diet, constipation status, and colonic transit (Table 2).
Associations Between Taxa Assessed with Single Taxon-based Testing and Other Variables
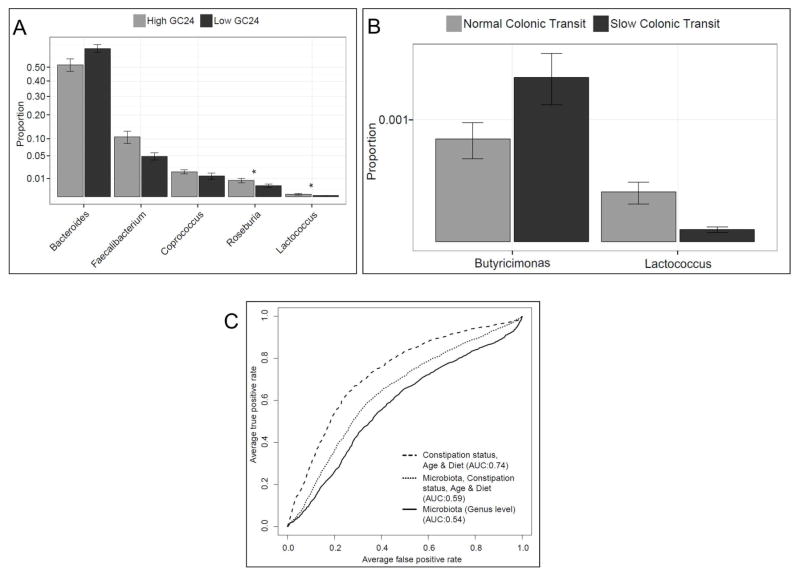
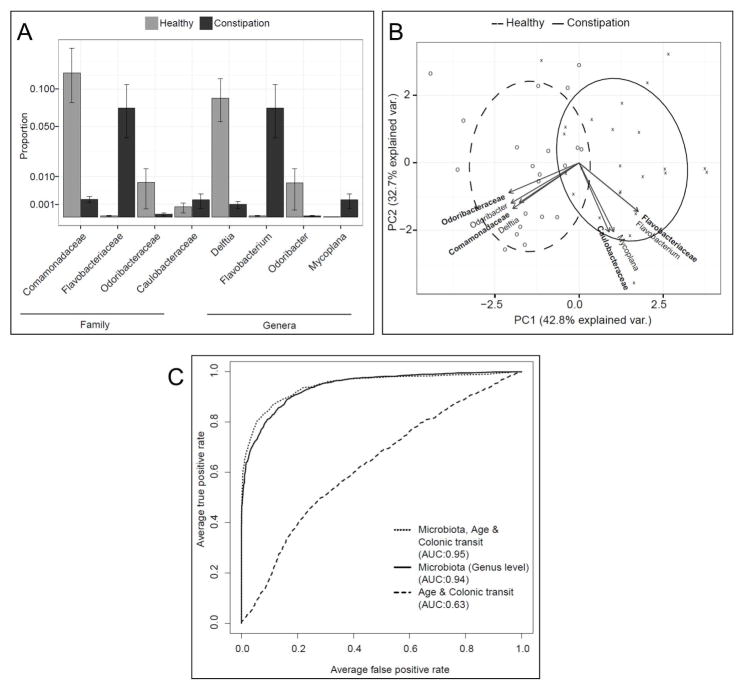
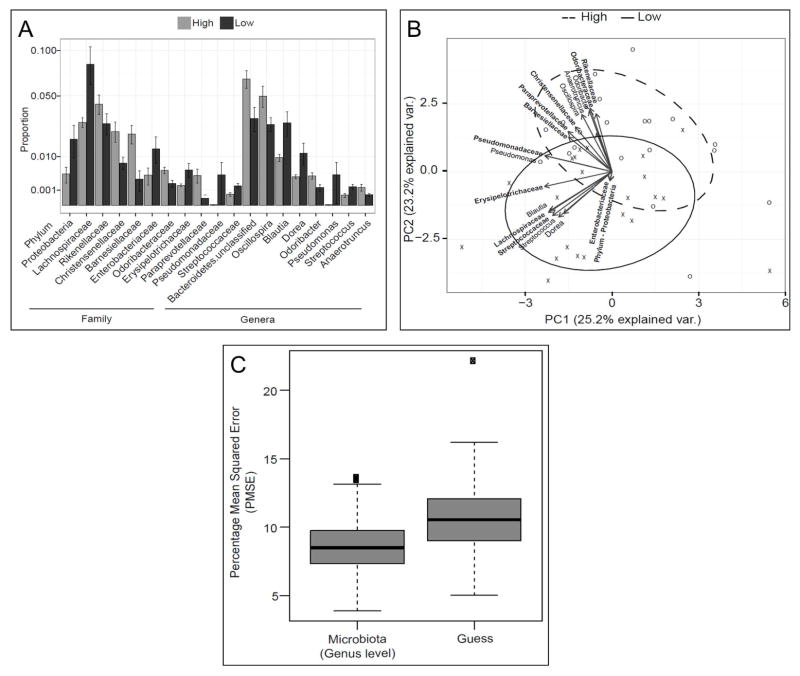
These analyses were guided by the results of the multivariate analyses for overall microbiota composition. They focused on the relationships between the fecal microbiota and colonic transit (Figure 1), the colonic mucosal microbiota and constipation status (Figure 2), and the fecal microbiota and breath methane production (Figure 3).
Figure 1. Relationship between stool microbiota and colonic transit.
The analysis shows microbiota evaluated with the RF algorithm that was associated with colonic transit expressed as continuous (A) or dichotomous variables (B). In A, the bar graph represents the relative abundance of all genera in participants with GC24 less than (Low GC24) and ≥ (High GC24) the median value (1.8) in all participants. The asterisk represents genera that were also significant by single taxon analysis. In B, colonic transit is categorized as normal (GC24 ≥1.4 [10th percentile value in healthy women]) and slow (GC24 <1.4) colonic transit. (C) ROC curves showing the utility of i) stool microbiota at the genus level; ii) constipation status, age, and diet; and iii) both i) and ii) for discriminating between normal and slow colonic transit.
Figure 2. Relationship between colonic mucosal microbiota and constipation.
(A) Differential abundance of bacterial taxa in healthy participants and constipated patients. All bacterial taxa so identified by single taxon based analysis with FDR control are shown. (B) Colonic mucosal microbiota discriminates between healthy participants and constipated patients. The relative abundance of only those taxa that were found to be significantly different between the two groups was used in the principal components analysis. Taxa at the family level are shown in bold. Individual colonic mucosal samples of healthy controls are presented as ‘O’ and constipated patients are presented as ‘X’. (C) ROC curves showing the utility of i) colonic mucosal microbiota at the genus level; ii) colonic transit, and age; and iii) both i) and ii) for discriminating between healthy participants and constipated patients.
Figure 3. Relationship between stool microbiota and breath methane production.
(A) Differential abundance of bacterial taxa in participants with breath methane excretion above (high) and below (low) the median value of breath methane excretion in all participants (Area Under Curve). All bacterial taxa so identified by single taxon-based analysis with FDR control are shown. (B) Stool microbiota discriminates between high and low breath methane producers. The relative abundances of only those taxa that were found to be significantly different between the two groups were used in the principal components analysis. Taxa at the family level are shown in bold. Individual stool samples of high methane producers are presented as ‘O’ and low methane producers are presented as ‘X’. (C) Stool microbiota at the genus level predicts breath methane production. The boxes represent the distribution of predictive mean squared error (PMSE) over 1,000 permutations for breath methane as a continuous variable, using overall microbiota composition at the genus level. The PMSE was lower compared to “guess,” signifying that the microbiota predicted breath methane excretion expressed as a continuous variable (by Friedman Rank Sum test).
With the single taxon approach, the abundance of several organisms in the feces was significantly correlated with colonic transit before, but not after, adjusting for FDR (Table 3). In contrast, the abundance of several taxa in the colonic mucosa was associated with constipation status even after adjusting for FDR (Table 3). The correlations between breath methane production and taxa in stool, but not colonic mucosa, remained significant even after adjusting for FDR (Table 3).
Table 3.
Differences in Abundance of Taxa at Different Levels
| Mucosal Microbiota in Health and Constipation | ||||
|---|---|---|---|---|
| Taxon | Mean Abundance in Healthy Controls | Mean Abundance in Constipated Patients | P value | FDR- adjusted P value |
| Phyla | ||||
| Bacteroidetes | 0.39 | 0.60 | .04 | .20 |
| Family | ||||
| Bacteroidetes;Flavobacteriaceae | 0.00001 | 0.07 | .001 | .01 |
| Bacteroidetes;Odoribacteraceae | 0.01 | 0.0001 | .001 | .01 |
| Proteobacteria;Caulobacteraceae | 0.001 | 0.002 | .005 | .04 |
| Proteobacteria;Comamonadaceae | 0.13 | 0.002 | .001 | .01 |
| Genus | ||||
| Bacteroidetes;Flavobacterium | 0.00001 | 0.07 | .001 | .01 |
| Bacteroidetes;Odoribacter | 0.01 | 0.00001 | .002 | .02 |
| Proteobacteria;Comamonas | 0.002 | 0.000002 | .008 | .06 |
| Proteobacteria;Delftia | 0.09 | 0.001 | .001 | .01 |
| Proteobacteria;Mycoplana | 0.000001 | 0.002 | .001 | .01 |
| By Boruta feature selection | ||||
| Firmicutes;Faecalibacterium | 0.08 | 0.09 | NA | NA |
| Proteobacteria;Agrobacterium | 0.05 | 0.02 | NA | NA |
| Proteobacteria;Pseudomonas | 0.005 | 0.006 | NA | NA |
| Correlation Between Stool Microbiota and Colonic Transit | ||||
| Taxon | Spearman’s correlation coefficient | P value | FDR- adjusted P value | |
| Phylum | ||||
| Actinobacteria | 0.23 | .02 | .12 | |
| Family | ||||
| Actinobacteria;Coriobacteriaceae | 0.28 | .04 | .32 | |
| Firmicutes;Veillonellaceae | 0.18 | .01 | .30 | |
| Genus | ||||
| Firmicutes;Lactococcus | 0.44 | .01 | .33 | |
| Firmicutes;Roseburia | 0.34 | .01 | .33 | |
| By Boruta feature selection | ||||
| Bacteroidetes;Bacteroides | −0.20 | NA | NA | |
| Firmicutes;Coprococcus | 0.41 | NA | NA | |
| Firmicutes;Faecalibacterium | 0.36 | NA | NA | |
| Bacteroidetes;Butyricimonas | −0.12 | NA | NA | |
| Correlation Between Stool Microbiota and Breath Methane Concentration | ||||
| Taxon | Spearman’s correlation coefficient | P value | FDR- adjusted P value | |
| Phylum | ||||
| Proteobacteria | −0.21 | .006 | .03 | |
| Family | ||||
| Bacteroidetes;Barnesiellaceae | 0.37 | .003 | .02 | |
| Bacteroidetes;Odoribacteraceae | 0.36 | .003 | .02 | |
| Bacteroidetes;Paraprevotellaceae | 0.44 | .03 | .06 | |
| Bacteroidetes;Rikenellaceae | 0.30 | .03 | .06 | |
| Firmicutes;Christensenellaceae | 0.45 | .001 | .02 | |
| Firmicutes;Erysipelotrichaceae | −0.16 | .02 | .046 | |
| Firmicutes;Lachnospiraceae | −0.33 | .003 | .02 | |
| Firmicutes;Peptostreptococcaceae | −0.16 | .04 | .07 | |
| Firmicutes;Streptococcaceae | −0.33 | .005 | .02 | |
| Proteobacteria;Enterobacteriaceae | −0.24 | .008 | .03 | |
| Proteobacteria;Pseudomonadaceae | −0.30 | .006 | .02 | |
| Genus | ||||
| Bacteroidetes;Butyricimonas | 0.30 | .02 | .05 | |
| Bacteroidetes;Odoribacter | 0.38 | .01 | .05 | |
| Bacteroidetes;Paraprevotella | 0.39 | .048 | .12 | |
| Bacteroidetes;unclassified | 0.43 | .001 | .02 | |
| Firmicutes;Anaerotruncus | 0.37 | .005 | .04 | |
| Firmicutes;Blautia | −0.27 | .008 | .04 | |
| Firmicutes;Dorea | −0.43 | .002 | .02 | |
| Firmicutes;Eubacterium | −0.04 | .01 | .04 | |
| Firmicutes;Oscillospira | 0.44 | .002 | .02 | |
| Firmicutes;SMB53 | −0.30 | .03 | .08 | |
| Firmicutes;Streptococcus | −0.45 | .001 | .02 | |
| Firmicutes;unclassified | 0.39 | .02 | .05 | |
| Proteobacteria;Erwinia | −0.19 | .03 | .09 | |
| Proteobacteria;Klebsiella | −0.41 | .01 | .04 | |
| Proteobacteria;Pseudomonas | −0.30 | .008 | .04 | |
| Correlation Between Mucosal Microbiota and Breath Methane Concentration | ||||
| Taxon | Spearman’s correlation coefficient | P value | FDR- adjusted P value | |
| Family | ||||
| Firmicutes;Veillonellaceae | 0.47 | .03 | .67 | |
| Genus | ||||
| Bacteroidetes;Cloacibacterium | 0.24 | .03 | .38 | |
| Firmicutes;Dorea | −0.21 | .01 | .38 | |
| Firmicutes;Phascolarctobacterium | 0.44 | .02 | .38 | |
| By Boruta feature selection | ||||
| Bacteroidetes;Bacteroides | −0.01 | NA | NA | |
NA: Not applicable
Associations Between Taxa Assessed with the Random Forest Algorithm and Other Variables
As evidenced by a decreased predictive mean square error compared to random guess, a predictive model utilizing the entire microbiota suggested that the fecal microbiota composition (at the genus level) weakly predicted colonic transit (i.e., GC24; P<.001) and strongly predicted breath methane production (P<.001) as shown in Figure 3c. Based on feature selection by Boruta, the genera Bacteroidetes-Bacteroides, Firmicutes-Coprococcus, Firmicutes-Faecalibacterium, Firmicutes-Lactococcus, and Firmicutes-Roseburia were independently significantly useful for predicting colonic transit (Figure 1a). When participants were categorized as having normal or rapid versus slow colonic transit, the genera Bacteroidetes-Butyricimonas, (which was increased in slow transit) and Firmicutes-Lactococcus (which was decreased in slow transit) were considered important (Figure 1b).
The fecal microbiota at the genus level was not particularly useful for discriminating between normal and slow colonic transit. Indeed, the area under the ROC curve for this analysis was only 0.54 (P<.001) for fecal microbiota compared to 0.74 (P<.001) for the combination of age, diet, and constipation status (P<.001) (Figure 1c). In contrast, the colonic mucosal microbiota at the genus level was very useful for distinguishing between constipated and healthy participants; the area under the ROC curve for this analysis was 0.94 (P<.001), compared to 0.63 (P<.001) for the combination of age and colonic transit (P<.001) (Figure 2c). By Boruta feature selection, the genera Bacteroidetes-Flavobacterium, Firmicutes-Faecalibacterium, Proteobacteria-Agrobacterium, Proteobacteria-Delftia, Proteobacteria-Mycoplana, and Proteobacteria-Pseudomonas, along with age and colonic transit, were significantly important.
For prediction of breath methane production, the Boruta feature selection algorithm identified the genera Bacteroidetes-Odoribacter, Bacteroidetes-unclassified, Firmicutes-Anaerotruncus, Firmicutes-Oscillospira in fecal microbiota, and the genera Bacteroidetes-Bacteroides, Firmicutes-Dorea, and Firmicutes-Phascolarctobacterium in mucosal microbiota as important.
DISCUSSION
This is the first study that compared fecal and colonic mucosal microbiota in healthy participants and constipated patients, employed state-of-the-art analyses to characterize the overall microbiota profile and taxonomic differences, and assessed the contribution of other factors (i.e., dietary intake, colonic transit, and breath methane production after carbohydrate ingestion) to colonic microbiota with multivariate analyses. The overall profile of mucosal microbiota evaluated by 2 approaches (i.e., unweighted and weighted UniFrac) was different between constipated patients and healthy participants, even after adjusting for demographic variables, diet, and colonic transit. In contrast to previous studies, which used a limited set of bacterial species,41 we used the entire microbiota to discriminate between healthy participants and patients. Indeed, this overall profile had 94% accuracy for discriminating between health and constipation. Moreover, the mucosal microbiota profile was not associated with colonic transit and was significantly different between healthy participants and constipated patients even after adjusting for colonic transit. Taken together, these findings strongly suggest that the observed differences in the colonic mucosal profile between healthy participants and constipated patients cannot be explained by slow colonic transit.
Similar to a previous study, phylum Bacteroidetes was 1.5 times more plentiful in the mucosal microbiota in constipated patients with single taxon analysis.12 This increase was due to a greater abundance of Flavobacterium. In addition to supporting these findings, the RF algorithm demonstrated that the genus Faecalibacterium was more abundant in constipation. Of interest, Bacteroidetes in feces is negatively associated with dietary fiber intake,42 and Faecalibacterium is among the most abundant butyrate producers in the gut.43 Some colonic effects of butyrate (e.g., decreased colonic mucin secretion,44 and increased colonic water and electrolyte absorption45), which are observed even at physiological concentrations, may predispose to constipation.
The composition of fecal microbiota measured with the weighted but not unweighted UniFrac distance was correlated with colonic transit. After adjusting for age, BMI, and constipation status, these differences were of borderline significance. These findings suggest that while people with normal and slow colonic transit had the same taxa, the abundance of taxa differed between groups. Single taxon-based testing suggested that the abundance of phylum Actinobacteria and selected genera in the phylum Firmicutes (Lactococcus and Roseburia) were correlated with faster colonic transit. Lactococcus produces serotonin,46, 47 which initiates intestinal peristalsis. The RF algorithm confirmed these associations and also identified additional taxa, for example, genera belonging to Firmicutes (Faecalibacterium and Coprococcus) and Bacteroidetes (Bacteroides) that were respectively directly and inversely correlated with colonic transit, as suggested previously in IBS.48 Of interest, obesity is also associated with a greater ratio of Firmicutes:Bacteroidetes and faster colonic transit.49–52 Perhaps this association is mediated by cholic acid, which increases the relative abundance of Firmicutes over Bacteroidetes53 and accelerates colonic transit, particularly in IBS.54 Several taxa (e.g., Faecalibacterium, Roseburia, and Coprococcus) that were correlated with faster colonic transit in this study produce butyrate,55 which has been reported to stimulate colonic motility, either directly, by stimulating 5-HT release, or by facilitating cholinergic pathways.56, 57 Further studies are necessary to resolve an apparent paradox that mucosal butyrate producers (i.e., Faecalibacterium) were associated with constipation while fecal butyrate producers (i.e., Faecalibacterium, Roseburia and Coprococcus) were associated with fast colonic transit. Indeed, butyrate has biphasic effects on colonic motility; it stimulates motility at low and inhibits motility at higher concentrations.58, 59
Even after adjusting for demographic variables, diet, constipation status, and colonic transit, the fecal, and to a lesser extent, the mucosal microbiota was associated with breath methane production. These findings suggest that the microbiota profile associated with breath methane is not explained by slow colonic transit. Indeed, up to one-third of healthy people produce methane.60 Breath methane production was associated with several genera in Bacteroidetes and Firmicutes that were different from those associated with colonic transit. Moreover, breath methane production was not correlated with colonic transit. While we did not assess for Archaea, which are methanogens, these observations do not support a link between luminal methane production and slow colonic transit. Indeed a recent study observed that both breath methane excretion and colonic methane production were not associated with clinical presentation in IBS patients, and were not correlated with symptom severity or with gastrointestinal transit.61
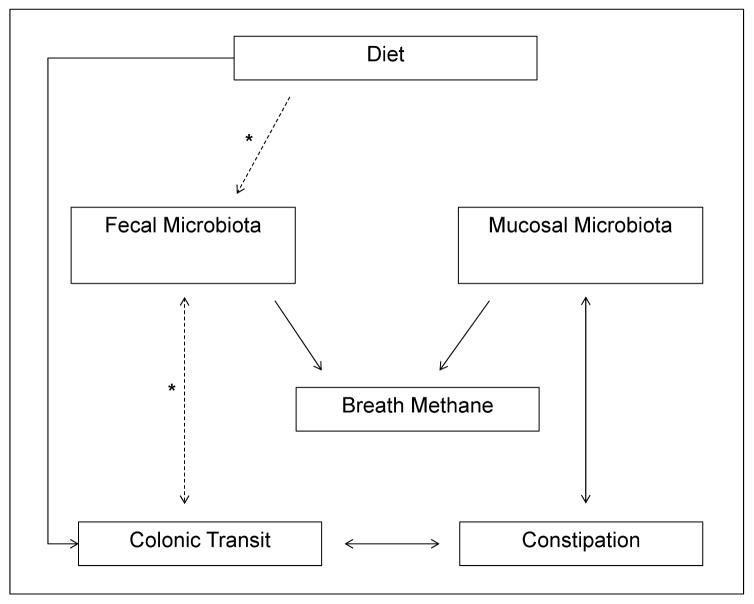
These findings substantially expand our understanding of the relationships between diet, microbiota, and GI transit, which is primarily based on studies in gnotobiotic mice and the effects of Fermentable Oligo-Di-Monosaccharides and Polyols (FODMAP) diets in IBS.62 They provide the basis for a new conceptual framework in which the mucosal microbiota are linked to constipation, independent of colonic transit, while the fecal microbiota are linked to colonic transit and breath methane production (Figure 4). One possible scenario is that a disturbance of the mucosal microbiota, e.g., more Bacteroidetes and/or Faecalibacterium,42 affects epithelial and mucosal function, e.g., by altering serotoninergic pathways or electrolyte/mucin secretion, causing symptoms of constipation (hard and/or infrequent bowel movements).11–16 In some patients, these disturbances and/or a different mechanism (e.g., dietary factors or enteric neuropathology) may lead to slow colonic transit, which is accompanied by differences in the fecal microbiota. These findings do not explain the underlying mechanism responsible for differences in the colonic mucosal microbiota in constipated patients. Further studies that evaluate the impact of accelerating colonic transit on the mucosal microbiome are necessary to assess the cause-effect relationship between the microbiome and constipation. While dietary factors may contribute, they were not, in contrast to the fecal microbiota, even univariately associated with mucosal microbiota in this study. Because the database used to estimate the nutritional values of ingested food products does not provide the fermentable oligo-dimonosaccharides and polyols (FODMAP) content for all foods, we cannot determine whether differences in ingestion of FODMAPs, which contribute to the colonic microenvironment, explain these findings.62
Figure 4. Conceptual framework of interactions between fecal and colonic mucosal microbiota, colonic transit, breath methane excretion, and constipation.
To emphasize, the lines denote associations, not causality a solid line represents an association that remained statistically significant in the multivariate analyses; the dotted line represents an association that was only univariately statistically significant (Table 2); the asterisk denotes relationships that are supported by previous literature. While the mucosal microbiota is associated with constipation, independent of colonic transit, the fecal microbiota is associated with colonic transit and breath methane production.
To ensure a representative distribution, the constipated group included patients with functional constipation and constipation-predominant IBS because these entities are not symptomatically or physiologically distinct.63, 64 Likewise, 14 constipated patients (56%) had slow colonic transit. These findings should be validated in a larger group of patients with functional bowel disorders, in particular to identify differences, if any, between functional constipation and constipation-predominant IBS. Likewise, a larger sample size may be useful because a type II error may have limited the ability to identify significant relationships between the microbiota and some parameters (e.g., between mucosal microbiota and caloric intake) in this study. Because global bacterial patterns of rectal biopsy samples sufficiently mirror those of the more proximal colon in healthy people and in inflammatory bowel disease,65, 66 mucosal samples were obtained from the sigmoid colon. However, this may not hold true in IBS.
In conclusion, constipated patients had a unique profile of colonic mucosal microbiota that discriminated between constipation and health with an accuracy of 94% independent of diet and colonic transit. By contrast, the fecal microbiota was associated with colonic transit and breath methane production rather than constipation.
Acknowledgments
Grant Support: Dr. Bharucha was supported in part by grant R01 DK78924 from the National Institutes of Health, US Department of Health and Human Services. This project was supported by the Center for Individualized Medicine at Mayo Clinic and in part by grant number UL1 TR000135 from the National Center for Advancing Translational Sciences.
Abbreviations
- AUROC
area under the receiver operating characteristic
- FODMAP
Fermentable Oligo-Di-Monosaccharides and Polyols
- FDR
false discovery rate
- IBS
Irritable bowel syndrome
- IBS-C
constipation-predominant irritable bowel syndrome
- OTU
Operational Taxonomical Units
- PERMANOVA
permutational multivariate analysis of variance
- PMSE
predictive mean squared error
- RF
random forest
- RNA
Ribonucleic acid
APPENDIX
Post-hoc power calculation for the PERMANOVA test based on UniFrac distances, using the ‘micropower’ package for R statistical software:
Parameters used in simulation:
Level of subsampling = 0.5
Number of OTUs = 50
Matched outcome: Within group distance = 0.7 ± 0.07 (mean ± SD)
For N=20 per group:
Power = 80% to detect a medium difference (Ω2=0.07) between groups
Power = 20% to detect a medium difference (Ω2=0.02) between groups
Footnotes
Disclosures: None of the authors have any conflicts of interest that are relevant to the manuscript.
None of the original citations have joint first authors.
Author contributions:
Study concept and design: Bharucha, Parthasarathy, Gaskins.
Acquisition of data: Bharucha, Parthasarathy, Chia, O’Connor.
Analysis and Interpretation of data: Bharucha, Parthasarathy, Chen.
Drafting of the manuscript: Bharucha, Parthasarathy.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical expertise: Chen, Chen.
Obtained funding and study supervision: Bharucha.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Malinen E, Rinttila T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 2.Kassinen A, Krogius-Kurikka L, Makivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Lyra A, Rinttila T, Nikkila J, et al. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol. 2009;15:5936–5945. doi: 10.3748/wjg.15.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tana C, Umesaki Y, Imaoka A, et al. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–519. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 6.Carroll IM, Ringel-Kulka T, Siddle JP, et al. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–530. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeffery IB, O’Toole PW, Ohman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 8.Chassard C, Dapoigny M, Scott KP, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012;35:828–838. doi: 10.1111/j.1365-2036.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 9.Ohman L, Tornblom H, Simren M. Crosstalk at the mucosal border: importance of the gut microenvironment in IBS. Nat Rev Gastroenterol Hepatol. 2015;12:36–49. doi: 10.1038/nrgastro.2014.200. [DOI] [PubMed] [Google Scholar]
- 10.Simren M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sartor RB. Gut microbiota: Optimal sampling of the intestinal microbiota for research. Nat Rev Gastroenterol Hepatol. 2015;12(5):253–254. doi: 10.1038/nrgastro.2015.46. [DOI] [PubMed] [Google Scholar]
- 12.Parkes GC, Rayment NB, Hudspith BN, et al. Distinct microbial populations exist in the mucosa-associated microbiota of sub-groups of irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:31–39. doi: 10.1111/j.1365-2982.2011.01803.x. [DOI] [PubMed] [Google Scholar]
- 13.Codling C, O'Mahony L, Shanahan F, et al. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci. 2010;55:392–397. doi: 10.1007/s10620-009-0934-x. [DOI] [PubMed] [Google Scholar]
- 14.Carroll IM, Chang YH, Park J, et al. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010;2:19. doi: 10.1186/1757-4749-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durban A, Abellan JJ, Jimenez-Hernandez N, et al. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ Microbiol Rep. 2012;4:242–247. doi: 10.1111/j.1758-2229.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- 16.Carroll IM, Ringel-Kulka T, Keku TO, et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashyap PC, Marcobal A, Ursell LK, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoppi G, Cinquetti M, Luciano A, et al. The intestinal ecosystem in chronic functional constipation. Acta Paediatr. 1998;87:836–841. doi: 10.1080/080352598750013590. [DOI] [PubMed] [Google Scholar]
- 19.Khalif IL, Quigley EM, Konovitch EA, et al. Alterations in the colonic flora and intestinal permeability and evidence of immune activation in chronic constipation. Dig Liver Dis. 2005;37:838–849. doi: 10.1016/j.dld.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Zhu L, Liu W, Alkhouri R, et al. Structural changes in the gut microbiome of constipated patients. Physiol Genomics. 2014;46:679–686. doi: 10.1152/physiolgenomics.00082.2014. [DOI] [PubMed] [Google Scholar]
- 21.Kang DW, DiBaise JK, Ilhan ZE, et al. Gut microbial and short-chain fatty acid profiles in adults with chronic constipation before and after treatment with lubiprostone. Anaerobe. 2015;33:33–41. doi: 10.1016/j.anaerobe.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Attaluri A, Jackson M, Valestin J, et al. Methanogenic flora is associated with altered colonic transit but not stool characteristics in constipation without IBS. Am J Gastroenterol. 2010;105:1407–1411. doi: 10.1038/ajg.2009.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pimentel M, Lin HC, Enayati P, et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1089–1095. doi: 10.1152/ajpgi.00574.2004. [DOI] [PubMed] [Google Scholar]
- 24.Kunkel D, Basseri RJ, Makhani MD, et al. Methane on breath testing is associated with constipation: a systematic review and meta-analysis. Dig Dis Sci. 2011;56:1612–1618. doi: 10.1007/s10620-011-1590-5. [DOI] [PubMed] [Google Scholar]
- 25.Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 26.Bharucha AE, Pemberton JH, Locke GR., 3rd American gastroenterological association technical review on constipation. Gastroenterology. 2013;144:218–238. doi: 10.1053/j.gastro.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camilleri M, Iturrino J, Bharucha AE, et al. Performance characteristics of scintigraphic measurement of gastric emptying of solids in healthy participants. Neurogastroenterol Motil. 2012;24:1076–e1562. doi: 10.1111/j.1365-2982.2012.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deiteren A, Camilleri M, Bharucha AE, et al. Performance characteristics of scintigraphic colon transit measurement in health and irritable bowel syndrome and relationship to bowel functions. Neurogastroenterol Motil. 2010;22:415–423. e495. doi: 10.1111/j.1365-2982.2009.01441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson J, Garges S, Giovanni M, et al. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeraldo P, Chia N, Goldenfeld N. On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environ Microbiol. 2011;13:3000–3009. doi: 10.1111/j.1462-2920.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- 31.Jeraldo P, Kalari K, Chen X, et al. IM-TORNADO: a tool for comparison of 16S reads from paired-end libraries. PloS One. 2014;9:e114804. doi: 10.1371/journal.pone.0114804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1. 0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 36.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 39.Kursa MB, Jankowski A, Rudnicki WR. Boruta - A System for Feature Selection. Fundamenta Informaticae. 2010;101:271–286. [Google Scholar]
- 40.Kelly BJ, Gross R, Bittinger K, et al. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics. 2015;31:2461–2468. doi: 10.1093/bioinformatics/btv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flint HJ, Scott KP, Duncan SH, et al. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barcelo A, Claustre J, Moro F, et al. Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut. 2000;46:218–224. doi: 10.1136/gut.46.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binder HJ, Mehta P. Short-chain fatty acids stimulate active sodium and chloride absorption in vitro in the rat distal colon. Gastroenterology. 1989;96:989–996. doi: 10.1016/0016-5085(89)91614-4. [DOI] [PubMed] [Google Scholar]
- 46.Kuley E, Ozogul F, Ozogul Y, et al. The function of lactic acid bacteria and brine solutions on biogenic amine formation by foodborne pathogens in trout fillets. Food Chem. 2011;129:1211–1216. doi: 10.1016/j.foodchem.2011.05.113. [DOI] [PubMed] [Google Scholar]
- 47.Clarke G, Stilling RM, Kennedy PJ, et al. Minireview: Gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajilic-Stojanovic M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 49.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 50.Finucane MM, Sharpton TJ, Laurent TJ, et al. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PloS One. 2014;9:e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delgado-Aros S, Locke GR, Camilleri M, et al. Obesity is associated with increased risk of gastrointestinal symptoms: a population-based study. Am J Gastroenterol. 2004;99:1801–1806. doi: 10.1111/j.1572-0241.2004.30887.x. [DOI] [PubMed] [Google Scholar]
- 52.Delgado-Aros S, Camilleri M, Garcia MA, et al. High body mass alters colonic sensory-motor function and transit in humans. Am J Physiol Gastrointest Liver Physiol. 2008;295:8. doi: 10.1152/ajpgi.90286.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Islam KBMS, Satoru F, Masahito H, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 54.Sadik R, Abrahamsson H, Ung K-AA, et al. Accelerated regional bowel transit and overweight shown in idiopathic bile acid malabsorption. Am J Gastroenterol. 2004;99:711–718. doi: 10.1111/j.1572-0241.2004.04139.x. [DOI] [PubMed] [Google Scholar]
- 55.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 56.Reigstad CS, Salmonson CE, Rainey JF, 3rd, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2014 doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soret R, Chevalier J, De Coppet P, et al. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology. 2010;138:1772–1782. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 58.Neunlist M, Dobreva G, Schemann M. Characteristics of mucosally projecting myenteric neurones in the guinea-pig proximal colon. J Physiol. 1999;517( Pt 2):533–546. doi: 10.1111/j.1469-7793.1999.0533t.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Squires PE, Rumsey RD, Edwards CA, et al. Effect of short-chain fatty acids on contractile activity and fluid flow in rat colon in vitro. Am J Physiol. 1992;262:G813–817. doi: 10.1152/ajpgi.1992.262.5.G813. [DOI] [PubMed] [Google Scholar]
- 60.Gibson GR, Cummings JH, Macfarlane GT, et al. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut. 1990;31:679–683. doi: 10.1136/gut.31.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Stefano M, Mengoli C, Bergonzi M, et al. Breath Methane Excretion Is not An Accurate Marker of Colonic Methane Production in Irritable Bowel Syndrome. Am J Gastroenterol. 2015;110:891–898. doi: 10.1038/ajg.2015.47. [DOI] [PubMed] [Google Scholar]
- 62.Halmos EP, Christophersen CT, Bird AR, et al. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2015;64:93–100. doi: 10.1136/gutjnl-2014-307264. [DOI] [PubMed] [Google Scholar]
- 63.Shekhar C, Monaghan PJ, Morris J, et al. Rome III functional constipation and irritable bowel syndrome with constipation are similar disorders within a spectrum of sensitization, regulated by serotonin. Gastroenterology. 2013;145:749–757. doi: 10.1053/j.gastro.2013.07.014. quiz e713–744. [DOI] [PubMed] [Google Scholar]
- 64.Wong RK, Palsson OS, Turner MJ, et al. Inability of the Rome III criteria to distinguish functional constipation from constipation-subtype irritable bowel syndrome. Am J Gastroenterol. 2010;105:2228–2234. doi: 10.1038/ajg.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lepage P, Seksik P, Sutren M, et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis. 2005;11:473–480. doi: 10.1097/01.mib.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- 66.Lavelle A, Lennon G, O'Sullivan O, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015 doi: 10.1136/gutjnl-2014-307873. [DOI] [PMC free article] [PubMed] [Google Scholar]