Abstract
Spatiotemporal regulation of enzyme-substrate interactions governs the decision-making steps in biological systems. Enzymes, being functional units of every living cell, contribute to the macromolecular stability of cell survival, proliferation and hence are vital windows to unraveling the biological complexity. Experimental measurements capturing this dynamics of enzyme-substrate interactions in real time add value to this understanding. Furthermore these measurements, upon validation in realistic biological specimens such as clinical biopsies – can further improve our capability in disease diagnostics and treatment monitoring. Towards this direction, we describe here a novel, high-sensitive measurement system for measuring diffusion-limited enzyme-substrate kinetics in real time. Using catalase (enzyme) and hydrogen peroxide (substrate) as the example pair, we demonstrate that this system is capable of direct measurement of catalase activity in vitro and the measured kinetics follows the classical Michaelis-Menten reaction kinetics. We further demonstrate the system performance by measuring catalase activity in living cells and in very small amounts of liver biopsies (down to 1μg total protein). Catalase-specific enzyme activity is demonstrated by genetic and pharamacological tools. Finally we show the clinically-relevant diagnostic capability of our system by comparing the catalase activities in liver biopsies from young and old mouse (liver and serum) samples. We discuss the potential applicability of this system in clinical diagnostics as well as in intraoperative surgical settings.
Keywords: Catalase activity, oxygen sensing, breast cancer, aging, oxidative stress, antioxidants
Introduction
Aerobic metabolism defines the fundamental characteristic of life for the eukaryotic organisms.[1] Harnessing the available oxygen for utilization in bioenergetic and biosynthetic processes involves complex layers of enzymatic reactions in every living eukaryotic cell.[2] Cellular respiration (oxygen intake) is tightly regulated by subcellular compartments and in particular, mitochondria – which make the cellular energy currency, the adenosine triphosphate (ATP), by complete oxidation of oxygen via a series of redox steps. Electron leaks during these steps lead to the generation of oxygen free radicals. Evolutionarily, living cells have developed sophisticated enzymes and other non-enzymatic reaction partners that continuously scavenge these free radicals thereby detoxifying the cells for their survival and proper function.[3, 4] These antioxidant enzymes vary in their subcellular location and modes of action. For instance, superoxide dismutase is a mitochondrially localized enzyme that converts toxic superoxide (negatively charged oxygen free radical) to less toxic hydrogen peroxide. Whereas catalase is the enzyme that converts hydrogen peroxide to water and predominantly is localized to peroxisomes and also in cytoplasm. By fine-tuning the rates and the magnitudes of these antioxidant enzymes, cell survival is regulated amidst dynamic changes in pro-oxidant balance. Deregulation of this balance has been implicated in a variety of diseases including diabetes, cancer and aging.[5] Experimental methods to monitor the pro-anti-oxidant balance in various tissues add significant value to not only disease diagnostics but also in treatment monitoring.[6–9] In this paper, we describe a novel method to directly measure the activity of a representative antioxidant enzyme, catalase in real time.
2. Materials and Methods
2.1 Cells & Reagents
The normal mammary epithelial MCF10A cells and the breast cancer cells, MDA-MB-231, MCF7 and BT-549 - were originally from ATCC and were cultured in DMEM with high glucose (4.5 g/l) and 10% FBS and antibiotics. MDA-MB-231 cells were transduced with lentiviral particles containing human Catalase expression vector and the MDA-MB-231 cells overexpressing catalase were selected using neomycin (3μg/ml) antibiotics at least for 10 consecutive passages and the catalase overexpression was confirmed by western blotting. Immunofluorescence slides were prepared by standard procedure from monolayer cultures of parental (231-P) and catalase-overexpressing (231-CAT) breast cancer cells.
2.2 Animal tissues
Male FVB/N-J mice were originally purchased from Jackson Laboratory and further breeding was carried out in-house as approved by the institutional animal care and use committee. Just before the tissue enzyme activity measurements, animals of two age groups (4 weeks & 40 weeks) were sacrificed and the liver tissues were surgically excised. Immediately after the surgery, the liver tissues were homogenized in 2ml microcentrifuge tube containing hypotonic buffer using a tissue lyser (Qiagen, 5 minutes, 30Hz or 1800 oscillations/minute). Protein lysates were then used immediately for measuring catalase activity as described. In another set of experiments, 50mg fresh liver biopsy was used to measure catalase activity in the presence and in the absence of catalase-specific pharmacological inhibitor 3AT (6mM 3-Amino, 1,2,4, triazole, Sigma Aldrich, USA).
2.3 Catalase activity measurements
Viability and mitochondrial functional status were measured in all the cell lines and the tissue samples before measuring catalase activity. Mitochondrial oxygen consumption [pO2] data were obtained with a clark-type oxygen microelectrode (Strathkelvin Instruments, Scotland) in a closed-cell respirometry design. The oxygen probes consist of electrolytes separated by a polypropylene membrane. Hermetically sealed probe holder allows dissolved oxygen measurement in a constant volume (typically 2ml). The probe electrodes were calibrated with 5% sodiumthiosulfite solution (0% oxygen) and mammalian ringer solution (100% oxygen ~ 207μmol/l ) at 38°C. All measurements were performed either in phosphate buffered saline as described earlier.[10] After collecting basal respiration (in stirred medium) was monitored for about 10 minutes in the substrate-limiting conditions, 200μM H2O2 was added and catalase-specific oxygen release rates were then calculated from the initial slopes of raw pO2 data.
2.4 Statistics
Data presented are Mean ± S.E from at least three independent experiments. Statistical significance was estimated based on Student’s t-test (p<0.05).
3. Results
3.1 In vitro catalase activity profiles follow Michaelis-Menten steady state kinetics
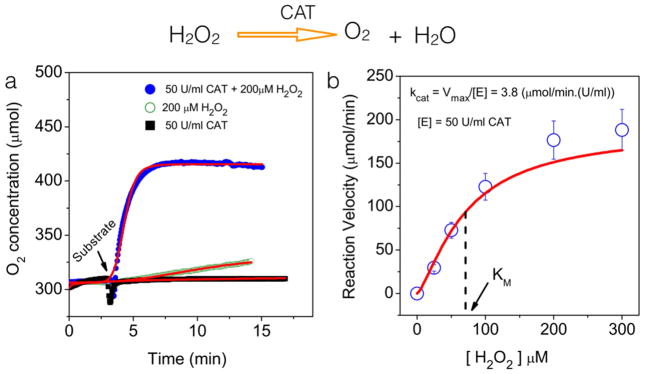
For in vitro validation study, we chose the Catalase (enzyme) –Hydrogen peroxide (substrate) system. Catalase converts the toxic hydrogen peroxide into water (H2O) and oxygen (O2). Figure 1a shows representative oxygen concentration profiles for three different situations. Detailed characterization of concentration dependence of both the enzyme and substrate were done earlier in the laboratory (data not shown) and these data suggested the optimum concentrations of hydrogen peroxide (200 μM) and Catalase (50U/ml) where one unit (U) is defined as the amount of catalase required to oxidize 1 micromole of hydrogen peroxide in one second at 24 deg C. As can be seen, the substrate-alone or the enzyme-alone did not give any appreciable oxygen release. The small change in oxygen release with hydrogen peroxide within the experimental duration is because of the known instability of the hydrogen peroxide upon dilution in micromolar range. However, the presence of catalase and hydrogen peroxide gave the largest change in oxygen release demonstrating the sensitivity of the system in measuring subtle changes in oxygen concentration in real time.
Figure 1. In vitro validation of Catalase activity assay.
(a) Representative oxygen profiles obtained from three different conditions shown. The substrate (H2O2) and enzyme (Catalase) concentrations were 200 μM and 50U/ml respectively. The kinetics of oxygen release was obtained every 6 seconds for ~ 20 minutes. All measurements were carried out at room temperature (24 deg C). (b) Oxygen release profiles during catalase activity follow the classical Michaelis-Menton equation. Catalase concentration was kept constant at 50U/ml and the substrate concentration was varied as shown. Each data point in the graph is the mean of at least 3 independent trials. The solid line is the mathematical fit to the equation (1) described in the text.
Any typical biochemical reaction involving an enzyme and substrate involves an initial binding step between them to form an intermediate complex which in turn, is converted to the product and the enzyme. Under certain assumptions (e.g., enzyme concentration ≪ substrate concentration), most of the enzyme-substrate interactions follow the classical Michaelis-Menten equation which relates the reaction velocity (v) of the equation to the concentration of the substrate [S] as:
| (Eq.1) |
where Vmax is the maximum reaction velocity achievable at saturation concentration of the substrate and KM is the substrate concentration at which the reaction rate is half of Vmax. In order to test if the oxygen release assay as described in our system follows similar model, we repeated the experiments by varying the substrate (H2O2) concentration systematically and measured oxygen release rate at each of these concentrations by keeping the catalase concentration fixed. Figure 1b summarizes the results.
3.2 Direct measurement of label-free Catalase activity distinguishes normal and cancer cells
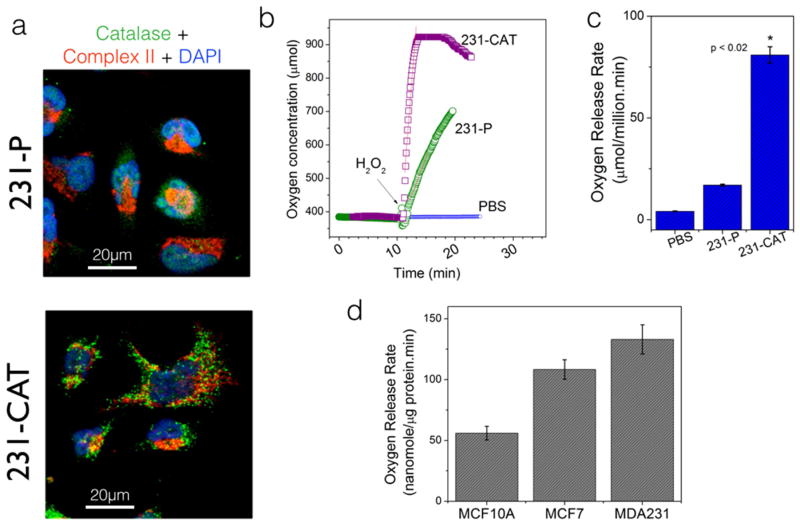
Having established a robust assay for monitoring Catalase enzyme activity in in-vitro conditions, we next tested the efficacy of the system in detecting the catalase activity in living cells. In order to determine the catalase-specific effects in the measured oxygen release, we compared the rates of oxygen release upon H2O2 stimulus – in parental and catalase-overexpressing human breast cancer (MDA-MB-231) cells. As can be seen in Figures 2a–2c, catalase-overexpressing cells indeed showed a clear increase in oxygen release rate as compared to the parental cells- thereby validating the detection specificity in our method. Next, we wanted to test if the proposed assay can give valuable information on the basal antioxidant status in living cells. Among the cancer-associated changes in epithelial cells, deregulation in redox status and the disruption of pro-oxidant/anti-oxidant balance are the common metabolic phenotypes. Figure 2d shows representative catalase activity in non-transformed human mammary epithelial cells, MCF10A and two canonical human breast cancer cells, MCF7 and MDA-MB-231 cells. Both the cancer cell lines showed a higher catalase activity as compared to the normal, MCF10A cells.
Figure 2. Demonstration of Catalase-specific activity in living cells.
(a) Immunofluorescence images of breast cancer cell line (MDA-MB-231) showing the spatial distribution of endogenous catalase in parental cells (231-P) and the lentiviral transduced overexpression (231-CAT). Co-staining with mitochondrial Complex II subunit shows that the overexpressed catalase is mainly localized in peroxisomal compartments with minimal colocalization in mitochondrial compartments. (b) Representative (n=4) oxygen concentration profiles upon H2O2 stimulus in 231-P and 231-CAT cells. In each case, 2.7 million viable living cells were added at the beginning of the experiment. (c) Summary of oxygen release rates (a measure of catalase activity) calculated from the above oxygen profiles. Note that the dramatic increase in oxygen release rate in 231-CAT cells is a clear illustration of the catalase-specific activity reported through out this paper. (d) Differential catalase activity (oxygen release rates) in non-transformed MCF10A epithelial cells and two representative human breast cancer cell lines. In contrast to the whole cells shown (b), only 50 μg of protein lysate was used for each cell line and the statistically significant increase in oxygen release rate further accentuates the high sensitivity in our measurement system.
3.3 Catalase activity profiles in tissue biopsies reveal age-dependent compensatory metabolism
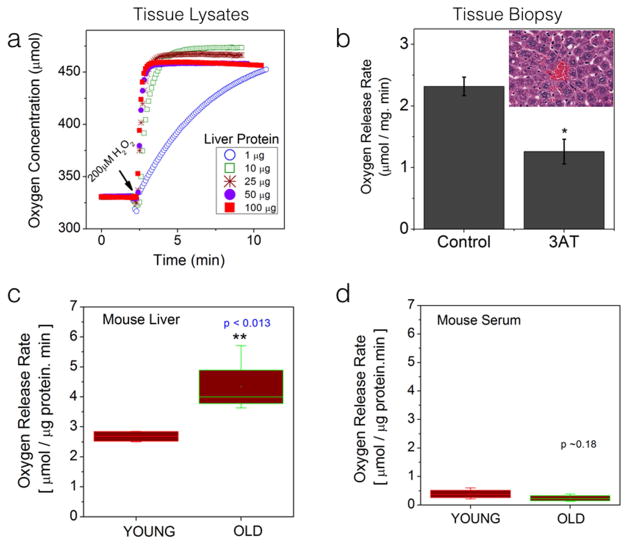
We next tested the utility of our method in measuring catalase activity in fresh liver biopsy specimens obtained from experimental mouse models. Catalase is an ubiquitous antioxidant enzyme that is present in almost all the body tissues although varying in its content and functional capacity in different tissues.[3, 11] Liver has been reported to have one of the highest catalase and other antioxidant activity owing to its role in extensive detoxification processes in the body.[12] As approved by the institutional animal care and use protocol, we collected liver tissues from 4-week old male mice and made fresh homogenates to obtain protein lysates from each liver tissue. Figure 3a shows representative oxygen concentration profiles obtained by using increasingly higher total protein concentrations. As can be seen, the current system could detect the oxygen release upon H2O2 stimulus from tissue samples with as little as 1μg total protein. In order to confirm if the measured oxygen release is specific to catalase activity, we measured the oxygen release rates (50mg of fresh liver biopsy) in the presence and absence of catalase-inhibitor (6mM 3AT). As shown in Figure 3b, acute 3AT treatment decreased catalase-activity further validating the sensitivity and specificity of the proposed method in tissue samples. In order to test if this proposed assay can yield meaningful information in differential health conditions, we compared the catalase activity (or the oxygen release rate) by substrate stimulus in liver tissue biopsies (100μg each) obtained from healthy, young male animals (4-week old mice, n=5) and from old male animals (40-week old mice, n=5). Figure 3c shows the statistical comparison between these two animal groups. Free radical theory of aging posits that cumulative acquisition of free radical-induced DNA damages and the concomitant cellular damages contribute to the decline in the organ function. A cumulative increase of free radicals in any organ will be countered by a compensatory increase in antioxidant enzyme activity in that organ.[13] Figure 3c is a clear illustration of such compensatory increase in catalase activity in old animal liver tissues as compared to those in the young animals. Interestingly the catalase activity measured in serum samples did not show any significant difference between the young and old animal groups (Figure 3d) as also reported by other groups. [14]
Figure 3.
Direct measurement of Catalase activity in freshly excised tissue specimens in real-time : (a) Oxygen release profiles upon stimulation with 200μM H2O2. Young mouse (~4 weeks old) liver tissues were homogenized to yield protein lysates. Protein concentration was measured by standard laboratory assays. Samples from these liver homogenates (as shown) were added in 2ml of phosphate buffered saline to obtain basal oxygen profile. After ~ 2.5 minutes, substrate was added as indicated to obtain the individual oxygen release profiles. (b) In an alternate experimental design, 50mg of liver tissue (within 10 minutes after surgical excision) was used to measure catalase activity as described earlier. By pre-incubating identical liver biopsy specimen in 6mM 3-amino-triazole (catalase inhibitor), a significant reduction in catalase activity was observed further confirming the catalase-specific enzyme activity in small biopsy specimens; The inset shows a representative liver tissue section showing regular arrangement of hepatocytes (liver cells) around a central blood vessel (red blood cells). (c & d) Statistical comparison of oxygen release rates in young and old mouse livers and blood serum samples. 100μg protein sample was used from each specimen. The box plots indicate the average rates calculated from 5 young and 5 old animal tissues.
4. Discussion
In this paper we demonstrated a novel assay for the direct measurement of label-free catalase-specific antioxidant enzyme activity in small amounts of biological specimens. The motivation for this study came from the realization of the constant need for rapid assessment of integrity and function of living cells/biological tissues in health as well as in pathological settings. Human tissues are complex and dynamic entities that are governed by multitude of enzymatic reactions distributed over varied time scales.[15] In order to delineate any specific enzymatic reaction from the rest, one needs high specificity as well as sensitivity in detection. Specificity can be achieved by eliciting a substrate-specific response whereas sensitivity is determined by the nature of assay chosen to measure the desired enzymatic reaction. As shown in Equation 1, an ideal way to monitor catalase activity is either to measure directly the removal of the substrate (H2O2) or the formation of one of the products (O2). Towards this direction, there are a number of assays available to detect catalase activity indirectly and all these assays depend on either calorimetric or fluorometric measurements of the H2O2 content in the system.[16] A reduction in H2O2 level induces a reduction in calorimetric/fluorometric signal which in turn, is interpreted as a measure of catalase activity. Although this is in principle a reliable approach, an inherent limitation with these assays is that the measured catalase activity depends on the efficiency of H2O2-mediated calorimetric reaction. The spurious background due to the basal degradation of H2O2 (as demonstrated in Figure 1a) will add uncertainty to the measured catalase activities.[17] On the other hand, methods that attempt to directly measure the spectroscopic absorbance of H2O2 (in the UV region) also do not achieve optimal sensitivity owing to the fact that many other chromophores absorb in these wavelengths.[18] When dealing with biological specimens, high energy of light at these wavelengths (250–380nm) can cause unwanted phototoxicity. As demonstrated in this study, direct measurement of catalase activity via sensitive detection of oxygen release eliminates the aforementioned limitations.
It is to be mentioned here that an increase in antioxidant enzyme levels (i.e., only the protein expression) can be due to either a functional increase in organ’s capacity to combat free radicals or due to a stress-response without any significant improvement in its activity.[13] More correlative measurements of organ-specific function may be needed to discern between these two possibilities. We suspect that the increase in catalase activity found in old animals is a measure of not just the enzyme levels but a functional improvement in enzyme activity. Since free radical processes have been implicated in the etiology of multiple human diseases such as obesity, diabetics and cancer, our method of rapid assessment of catalase activity can serve as a first line of evidence in disease diagnostics. A common initial step in the diagnosis of many diseases is the collection of biopsy specimens (~ 3 mm3 punch biopsies). Despite its well-established utilities, not all the organ types (e.g., brain) can accommodate such a collection of large tissue samples without losing their normal functional capacity. In these situations, we envision that our proposed assay can be of utmost significance since it needs only very small biological material (~ typically hundredth of the conventional punch biopsy). Our future developments will include expanding the scope of the current enzymatic measurement capability to include multiple enzyme-substrate partners. By augmenting this system with traditional fluorescence-based assays, the present configuration can be developed into a multi-spectral imaging system capable of achieving single cell resolution in minute tissue samples. Finally such a multi-modality platform can find utility in intraoperative settings where it can enable surgeons in real-time evaluation of disease processes as well as in enabling delineating disease tissues from healthy tissues in real time.
Supplementary Material
HIGHLIGHTS.
A novel, direct measurement of Catalase enzyme activity via, oxygen sensing method;
Steady-stateprofiles of Catalase activity follow the Michaelis-Menten Kinetics
Catalase-specific activity demonstrated using genetic and pharmacological tools
Overcomes limitations of spectroscopic methods and indirect calorimetric approaches
Clear demonstration of the applicability in cancer cells and aging animal tissues
Acknowledgments
We gratefully acknowledge financial support from American Cancer Society (RSG-12-144-01-CCE), National Cancer Institute/National Institutes of Health (R21-CA124843), Komen for the Cure foundation (KG090239) and Donna & Jesse Garber Foundation all to (V.K.R). We thank Dr Bruce Gewertz and Dr Leon Fine for their intramural support and encouragement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koch LG, Britton SL. Aerobic metabolism underlies complexity and capacity. J Physiol. 2008;586:83–95. doi: 10.1113/jphysiol.2007.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storey KB. Adventures in oxygen metabolism. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:359–369. doi: 10.1016/j.cbpc.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 3.Kodydkova J, Vavrova L, Kocik M, Zak A. Human catalase, its polymorphisms, regulation and changes of its activity in different diseases. Folia Biol (Praha) 2014;60:153–167. doi: 10.14712/fb2014060040153. [DOI] [PubMed] [Google Scholar]
- 4.Jena BS, Patnaik BK. Changes in catalase activity and its thermolability in liver and kidneys of ageing male garden lizard. Gerontology. 1992;38:252–257. doi: 10.1159/000213337. [DOI] [PubMed] [Google Scholar]
- 5.Glorieux C, Zamocky M, Sandoval JM, Verrax J, Calderon PB. Regulation of catalase expression in healthy and cancerous cells. Free Radic Biol Med. 2015;87:84–97. doi: 10.1016/j.freeradbiomed.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Bekdeser B, Ozyurek M, Guclu K, Alkan FU, Apak R. Development of a new catalase activity assay for biological samples using optical CUPRAC sensor. Spectrochim Acta A Mol Biomol Spectrosc. 2014;132:485–490. doi: 10.1016/j.saa.2014.04.178. [DOI] [PubMed] [Google Scholar]
- 7.El Nashar RM. Flow injection catalase activity measurement based on gold nanoparticles/carbon nanotubes modified glassy carbon electrode. Talanta. 2012;96:161–167. doi: 10.1016/j.talanta.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Teke M. Development of a new biosensor for determination of catalase activity. Prep Biochem Biotechnol. 2014;44:608–616. doi: 10.1080/10826068.2013.854253. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Luo S, Chen W. Activity of catalase adsorbed to carbon nanotubes: effects of carbon nanotube surface properties. Talanta. 2013;113:142–147. doi: 10.1016/j.talanta.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Xu Q, Biener-Ramanujan E, Yang W, Ramanujan VK. Targeting metabolic plasticity in breast cancer cells via mitochondrial complex I modulation. Breast Cancer Res Treat. 2015;150:43–56. doi: 10.1007/s10549-015-3304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkman HN, Gaetani GF. Mammalian catalase: a venerable enzyme with new mysteries. Trends Biochem Sci. 2007;32:44–50. doi: 10.1016/j.tibs.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Muppala V, Lin CS, Lee YH. The role of HNF-1alpha in controlling hepatic catalase activity. Mol Pharmacol. 2000;57:93–100. [PubMed] [Google Scholar]
- 13.Sundaram A, Siew Keah L, Sirajudeen KN, Singh HJ. Upregulation of catalase and downregulation of glutathione peroxidase activity in the kidney precede the development of hypertension in pre-hypertensive SHR. Hypertens Res. 2013;36:213–218. doi: 10.1038/hr.2012.163. [DOI] [PubMed] [Google Scholar]
- 14.Golenia A, Leskiewicz M, Regulska M, Budziszewska B, Szczesny E, Jagiella J, Wnuk M, Ostrowska M, Lason W, Basta-Kaim A, Slowik A. Catalase activity in blood fractions of patients with sporadic ALS. Pharmacol Rep. 2014;66:704–707. doi: 10.1016/j.pharep.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Ramanujan VK, Jo JA, Cantu G, Herman BA. Spatially resolved fluorescence lifetime mapping of enzyme kinetics in living cells. J Microsc. 2008;230:329–338. doi: 10.1111/j.1365-2818.2008.01991.x. [DOI] [PubMed] [Google Scholar]
- 16.Shangari N, O’Brien PJ. Catalase activity assays. Curr Protoc Toxicol. 2006;Chapter 7(Unit 7 7):1–15. doi: 10.1002/0471140856.tx0707s27. [DOI] [PubMed] [Google Scholar]
- 17.Iwase T, Tajima A, Sugimoto S, Okuda K, Hironaka I, Kamata Y, Takada K, Mizunoe Y. A simple assay for measuring catalase activity: a visual approach. Sci Rep. 2013;3:3081. doi: 10.1038/srep03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goth L. A simple method for determination of serum catalase activity and revision of reference range. Clin Chim Acta. 1991;196:143–151. doi: 10.1016/0009-8981(91)90067-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.