Abstract
Despite the identification and characterization of four opioid receptor subtypes and the genes from which they are encoded, pharmacological data does not conform to the predications of a four opioid receptor model. Instead, current studies of opioid pharmacology suggest the existence of additional receptor subtypes; however, no additional opioid receptor subtype has been identified to date. It is now understood that this discrepancy is due to the generation of multiple isoforms of opioid receptor subtypes. While several mechanisms are utilized to generate these isoforms, the primary mechanism involves alternative splicing of the pre-mRNA transcript. Extensive alternative splicing patterns for opioid receptors have since been identified and discrepancies in opioid pharmacology are now partially attributed to variable expression of these isoforms. Recent studies have been successful in characterizing the localization of these isoforms as well as their specificity in ligand binding; however, the regulation of opioid receptor splicing specificity is poorly characterized. Furthermore, the functional significance of individual receptor isoforms and the extent to which opioid- and/or HIV-mediated changes in the opioid receptor isoform profile contributes to altered opioid pharmacology or the well-known physiological role of opioids in the exacerbation of HIV neurocognitive dysfunction is unknown. As such, the current review details constitutive splicing mechanisms as well as the specific architecture of opioid receptor genes, transcripts, and receptors in order to highlight the current understanding of opioid receptor isoforms, potential mechanisms of their regulation and signaling, and their functional significance in both opioid pharmacology and HIV-associated neuropathology.
Identification and classification of multiple opioid receptor subtypes
There is a vast amount of pharmacological evidence that suggests the existence of multiple opioid receptor subtypes, such as the unique pharmacological profiles of individual opioids as well as the more recent inconsistencies seen in genetic knockout models. Indeed, multiple binding sites, through which various opioids exert their physiological effects, had been proposed as early as the 1950s and 1960s based on rigid structure-activity relationships of opioids (Snyder and Pasternak, 2003) and were eventually identified in mammalian brain tissue in 1973 (Pasternak and Pan, 2013; Snyder and Pasternak, 2003). Since then, opioid receptors have been identified in a wide range of vertebrates. To date, four opioid receptors have been cloned: the mu (μ)-opioid receptor (named for its affinity for morphine), the kappa (κ)-opioid receptor (named for its affinity for ketocyclazocine), the delta (δ)-opioid (named for the mouse vas deferens where it was first isolated), and the nociceptin/orphanin FQ receptor (also called the opioid receptor-like receptor). The three classical opioid receptors, μ, κ, and δ, display nearly 60% homology with one another, while the newly discovered nociceptin/orphanin FQ receptor displays nearly 50% homology (Waldhoer et al., 2004). Following the identification of these four opioid receptors by selective ligand binding, it was found that each receptor is encoded by its own distinct gene. These genes, named OPRM1, OPRK1, OPRD1, and OPRL1 as they encode the μ, κ, δ, and nociceptin/orphanin FQ receptors, respectively, display a similar genomic structure, suggesting a single, common ancestral gene. This shared evolutionary history was later confirmed using a combination of positional and phylogenetic data (Waldhoer et al., 2004; Wei and Loh, 2002; Wei and Loh, 2011). Despite the high homology between opioid receptor subtypes and their encoding genes, each receptor displays unique localization, agonist selectivity, and transcriptional regulation.
The δ-opioid receptor
The δ-opioid receptor (DOR) was the first opioid receptor successfully cloned and was initially identified by its selective binding of enkephalins, a family of endogenous opioid peptides (Abbadie and Pasternak, 2002). It is encoded by the OPRD1 gene, located on chromosome 1 in humans (Zaki et al., 1996). This gene utilizes a minimal promoter sequence that lacks a TATA sequence but contains an E box sequence, which is activated by the binding of upstream stimulatory factor (USF), and a GC box, which is activated by the binding of Sp proteins. Additional regulatory elements include an Ets-binding site, which overlaps with the E box, as well as regulatory sites for AP-1 and AP-2 binding (Wei and Loh, 2002). DORs can be found abundantly throughout the brain with various densities, with the highest densities in the cerebral cortex (Abbadie and Pasternak, 2002).
The κ-opioid receptor
Although originally identified by their high affinity for benzomorphans, the κ-opioid receptor (KOR) selectively binds the dynorphin family of endogenous opioid peptides (Abbadie and Pasternak, 2002). The gene encoding κ-opioid receptors, OPRK1, is located on human chromosome 8 (Zaki et al., 1996) and is unique in that it utilizes two promoters separated by a non-coding exon. Transcription initiation by the first promoter region within OPRK1 utilizes two TATA boxes, although this is not well conserved between species. Transcription initiation by the second promoter region is localized to an intronic region of OPRK1 and utilizes a CAAT box and a NF-κB transcription factor binding site (Wei and Loh, 2002; Yakovlev et al., 1995). This intronic sequence is also involved in the suppression of both OPRK1 promoters as it contains a DNA element that selectively binds the repressive transcription factor Ik-1 (Hu et al., 2001). At least two types of transcripts are produced from the OPRK1 gene, depending on the promoter utilized, with transcription of the classical transcript being initiated downstream from one of two TATA boxes while transcription of an alternative transcript begins within intron 1, resulting in its retention in the mature mRNA (Wei and Loh, 2002; Yakovlev et al., 1995). Relative to other opioid receptors, KORs are found in low abundance in the brain. The greatest density of KORs occurs within the striatum, thalamus, hypothalamus, cerebral cortex, cerebellum, brainstem, and spinal cord; however, localization and density is highly variable between species. For example, the terminal field of the mossy fiber system in the hilus of guinea pigs contains a high density of KORs while this same region is devoid of KORs in rats (Abbadie and Pasternak, 2002).
The μ-opioid receptor
The μ-opioid receptor (MOR) was cloned shortly after the δ-opioid receptor and, clinically, represents the most relevant opioid receptor as it has a high affinity for classical opioid agonists, such as morphine and heroin, and antagonists, such as naloxone. Endogenously, the MOR is the target receptor for endorphins, although MORs may also interact with opioid peptides belonging to other families, such as dynorphins. Its biological significance can be inferred from the fact that it is highly conserved across species, with more than 95% homology between the human and rat receptors. Various densities of MORs can be found in multiple structures throughout the CNS, including the cerebral cortex, striatum, thalamus, hypothalamus, cerebellum, brain stem and spinal chord, with expression seen more so in neurons than glial cells (Abbadie and Pasternak, 2002). The MOR is encoded by the OPRM1 gene, which is located on chromosome 6 in humans (Wei and Loh, 2011). This gene utilizes two promoter regions, designated as proximal and distal, that lack both TATA and CCAAT sequences. These promoter regions can function independently from each other to promote transcription, with transcription initiation by the proximal promoter region being favored (Pan, 2005; Pasternak and Pan, 2013) and accounting for almost 95% of OPRM1 activity (Wei and Loh, 2002). Regardless, multiple transcription initiation sites in the 5’ regulatory region of the distal promoter region have also been reported (Xu and Carr, 2001b) and, as such, both the proximal and distal promoter regions are subject to regulation. Several DNA elements homologous to known transcription factor binding sites have been identified within this region (Pasternak and Pan, 2013). In addition to these proximal and distal promoter regions, a third, TATA-containing promoter region has been identified upstream of exon 11 (Pan, 2005; Pasternak and Pan, 2013). Therefore, similar to OPRK1, at least three types of transcripts are produced from the OPRM1 gene depending on whether the proximal, distal, or exon 11 promoter is utilized.
The nociceptin/orphanin FQ receptor
The nociceptin/orphanin FQ receptor, also called the opioid receptor-like receptor (ORL1), is the most recently identified opioid receptor. The gene that encodes ORL1, designated OPRL1, is located on human chromosome 20 and has nearly 70% sequence homology to other opioid receptor genes (Levran et al., 2012; Waldhoer et al., 2004). While being a weak target for classical opioids, ORL1 selectively binds the newly identified neuropeptide orphanin FQ/nociception (OFQ/N), an endogenous 17-amino acid peptide that is similar to dynorphin A (Chevlen, 2003; Waldhoer et al., 2004), and is suggested to mediate addiction responses by negatively regulating the mesolimbic dopaminergic system (Levran et al., 2012). ORL1 is localized to the cerebral cortex, thalamus, hypothalamus, periaqueductal gray, dorsal raphe and locus coeruleus nuclei, and the spinal cord dorsal horn as well as in peripheral sensory and sympathetic ganglia, (Xie et al., 1999). Despite the physiological and pharmacological significance of ORL1, little is known about the exact function and regulation of this receptor.
Post-transcriptional regulation by alternative splicing mechanisms
The human genome consists of roughly 25,000 protein encoding genes; however, there is a large discrepancy between the number of protein encoding genes and the repertoire of mRNA transcripts and encoded proteins they produce, with gene products far more numerous than estimates would predict. It is now understood that this discrepancy is due to the generation of multiple RNA isoforms from a single gene by several mechanisms, including alternative transcription initiation and polyadenylation site usage. The primary mechanism for the generation of multiple mRNA transcripts from a single gene is alternative splicing and, in addition to 5′ capping and 3′ polyadenylation, constitutes an important step of pre-mRNA processing. General splicing, often referred to as constitutive splicing, is a complex process through which specific portions of the precursor mRNA (pre-mRNA) sequence, referred to as introns, are removed and the remaining nucleotide sequences, referred to as exons, are ligated to form a mature mRNA transcript. Alternative splicing involves the differential inclusion and exclusion of exons, and sometimes introns, into the pre-mRNA sequence, resulting in multiple mRNA variants (Barrie et al., 2012; Hui, 2009; Keren et al., 2010; Kim et al., 2008; Markovic and Challiss, 2009). It was originally estimated that at least 75% of human genes are subject to alternative splicing, but more recent estimates suggest that 90-95% of human genes are regulated in this manner (Pan et al., 2008). Furthermore, nearly 90% of alternatively spliced genes include a minor mRNA isoform with an abundance greater than 15% of the total gene expression (Barrie et al., 2012). As such, alternative splicing is not a trivial phenomenon but represents a major regulatory mechanism for the generation of multiple mRNA transcripts and is the basis for the discrepancy between the nearly 25,000 protein encoding genes and the four-fold greater abundance of synthesized proteins (Keren et al., 2010).
Alternative splicing of pre-mRNA can occur through multiple, mutually exclusive processes. The most common processes, exon skipping and mutually exclusive exon selection, account for nearly 40% of alternative splicing events in higher eukaryotes. Exon skipping, as the name implies, involves the selective exclusion of a cassette exon along with its flanking introns in the mature mRNA transcript. Mutually exclusive exon selection involves the inclusion of an alternative exon into the mRNA transcript or the replacement of a constitutive exon with an alternative exon through a process similar to exon skipping. Alternative splicing may also occur due to differential selection of alternative 3′ and/or 5’ splice sites due to the presence of multiple splicing sequences within an exon. This process accounts for roughly 25% of alternative splicing events and results in the partial exclusion of a cassette exon or a partial inclusion of its flanking intronic region into the mRNA transcript. Intron retention, in which an intronic region between two constitutive exons is incorporated into the mRNA transcript, is also possible, although rare, accounting for less than 5% of alternative splicing events. Interestingly, intron retention is a favored mechanism of alternative splicing in plants, fungi and protozoa, suggesting an evolutionary shift in the splicing process and regulation. Finally, production of alternative mRNA transcripts may also occur through infrequent processes such as alternative promoter usage and alternative polyadenylation (Keren et al., 2010; Kim et al., 2008). Collectively, these processes result in, on average, the excision of more than 90% of the pre-mRNA sequence as introns and the synthesis of two to three isoforms for a given gene (Stamm et al., 2005).
Alternative splicing of opioid receptors
The existence of multiple opioid receptor subtypes and their unique patterns of expression in various tissues and species suggest that multiple regulatory mechanisms exist in order to maintain proper localization and density of receptor expression. Multiple mechanisms have been suggested to explain spatial and temporal regulation of opioid receptor expression as well as for pharmacological data that suggests the existence of additional opioid receptor subtypes. Epigenetic mechanisms represent a major factor through which opioid gene expression is regulated (Regan et al., 2011; Wei, 2008; Wei and Loh, 2002). Likewise, short nucleotide polymorphisms (SNPs) of opioid receptor genes have been found to alter opioid pharmacology (Levran et al., 2012; Pasternak and Pan, 2013; Shabalina et al., 2009; Shi et al., 2002). However, differential levels of gene activity and permanent genetic polymorphisms do not account for the agonist-selective differences in opioid receptor activity. Therefore, mechanisms of post-transcriptional regulation generating additional opioid receptor subtypes have been proposed to reconcile pharmacological inconsistencies within the single receptor subtype model. Generation of additional opioid receptor subtypes is proposed to occur via modifications of known opioid receptors as no additional opioid receptor-transcribing genes have been identified. This hypothesis was later confirmed by various studies identifying multiple mechanisms of opioid receptor post-transcriptional regulation. Of these regulatory mechanisms, alternative splicing of opioid receptor mRNA is particularly interesting given that it results in the synthesis of multiple, structurally different opioid receptor proteins (Gaveriaux-Ruff et al., 1997; Pan et al., 1999; Pasternak, 2001; Pasternak, 2014).
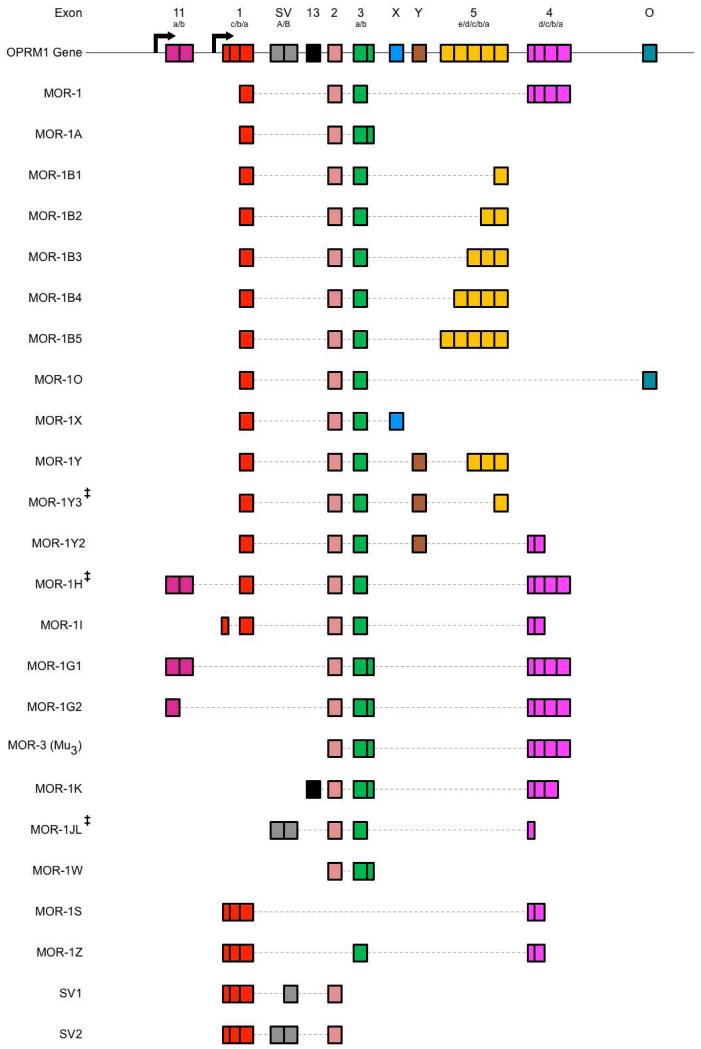
All four opioid receptors undergo alternative splicing in varying degrees. Splicing of the KOR is limited to two isoforms, generated by the existence of two promoters that differentially regulate the excision of the intronic region (Gaveriaux-Ruff et al., 1997; Wei et al., 2004; Yakovlev et al., 1995). Additionally, a third, truncated isoform has been described for the murine KOR (Wei et al., 2004). Similarly, intron inclusion between the first and second exons of the mouse OPRD1 generates two isoforms of the DOR; however, whether this splicing pattern is conserved in humans is unknown (Gaveriaux-Ruff et al., 1997). Alternative splicing of ORL1 is slightly more complex, as five exons have been identified within the OPRL1 gene. As such, five known ORL1 isoforms are alternatively spliced by mutually exclusive exon selection, alternative 3’ and/or 5’ splice site selection, and intron inclusion processes (Pan et al., 1998; Xie et al., 1999). The MOR exhibits the most extensive and complex alternative splicing patterns (Figure 1), and is further complicated by incomplete homology between species (Levran et al., 2012; Pasternak, 2014; Pasternak and Standifer, 1995). In humans, the constitutively spliced MOR-1 consists of exons 1, 2, 3, and 4. Synthesis of MOR isoforms primarily incorporates mutually exclusive exon selection, as seen in MOR-1O and MOR-1X, which replace exon 4 with exon O and exon X, respectively (Pan et al., 2003). Alternative 3’ and 5’ splice site selection also accounts for many of the MOR isoforms synthesized. This may occur in isolation, as seen in exon 1 of the MOR-1I isoform, or in conjunction with other splicing processes. The isoforms MOR-B1, MOR-1B2, MOR-1B3, MOR-1B4, and MOR-1B5 are synthesized from the replacement of exon 4 with exon 5 and are differentiated from each other by differential 3’ splice site selection within exon 5 (Pan et al., 2003). Likewise, MOR-1Y and MOR-1Y2 incorporate a new exon, exon Y, in place of exon 4 as well as incorporating different terminal exons. Exon exclusion and intron retention also account for different MOR isoforms, such as MOR-1A, in which exon 4 is excluded and the intron region following exon 3 is partially retained. Complex combinations of these four alternative splicing processes synthesize additional isoforms, including MOR-1K, MOR-1S, and MOR-1Z (Kvam et al., 2004). Utilization of a secondary promoter region, located upstream of a recently identified exon 11, facilitates exon 11 incorporation into the mRNA transcript and, in combination with additional splicing processes, results in the synthesis of additional MOR isoforms, including MOR-1G1 and MOR-1G2 (Pan et al., 2001). In total, alternative splicing of the MOR generates over 20 receptor isoforms characterized in humans, with additional isoforms predicted based on homology with the nearly 20 rat isoforms and 30 mouse isoforms known (Andersen et al., 2013; Fricchione et al., 2008; Mizoguchi et al., 2003; Pan, 2003; Pasternak, 2014; Pasternak and Pan, 2013; Xu et al., 2014b). This complexity of MOR splicing is further complicated by the genetic variability of the OPRM1 gene. Accordingly, alternatively spliced isoforms are subject to polymorphisms, with SNPs identified in multiple MOR splice variants (Diatchenko et al., 2011; Shabalina et al., 2009; Shi et al., 2002; Smith et al., 2005; Vallender et al., 2008; Xu et al., 2014a). Therefore, given their abundance and variation, the synthesis of multiple, structurally different opioid receptor proteins through alternative splicing is not merely an incidental phenomena but likely plays a key role in the enormous variability of physiological responses to various opioids and, likewise, represents a novel mechanism through which opioid pharmacology is altered. As such, regulation of opioid receptor splicing specificity is likely to have a functional impact on opioid pharmacology.
Figure 1. Schematic Representation of Human OPRM1 Alternative Splicing.
The human OPRM1 gene is composed of numerous exonic and intronic regions that are selectively utilized through alternative splicing mechanisms in order to generate various MOR isoforms. Unfortunately, exon names and definition sequences are not well conserved within the literature, leading to disparities between isoform compositions. Furthermore, additional MOR isoforms have been predicted, but have not yet been verified in vitro or in vivo. As such, splicing patterns of select MOR isoforms are illustrated here according to a comparative analysis of exon and intron inclusions across multiple literature sources. Top schematic represents positions of known exonic regions within the human OPRM1 gene, with arrows indicating transcription initiation sites. Accepted names for exonic regions are annotated above each region, with sub-annotations indicating variable exon cassettes due to multiple 3’ and 5’ splice sites. Splicing patterns of known opioid receptor isoforms are schematized and aligned below OPRM1 gene, with accepted isoform names displayed to the left, incorporated exon cassettes represented by solid boxes, and excised regions represented by a thin, dotted line (Reference Sequences: NM_000914.4, NM_001008504.3, NM_001145282.2, NM_001145283.2, NM_001145284.3, NM_001145285.2, NM_001145286.2, NM_001008503.2, NM_001008505.2, NR_104351.1, NR_104349.1, AY364230.1, DQ680044.1, NM_001145279.3, NM_001145280.3, NM_001145281.2, NM_001285528.1, NM_001145287.2, GQ258059.1, NM_001285527.1, NM_001285522.1, NR_104350.1; ‡ = predicted isoform) (Andersen et al., 2013; Choi et al., 2006; Fricchione et al., 2008; Pasternak and Pan, 2013; Xu et al., 2013; Xu et al., 2009).
Regulatory mechanisms of opioid receptor splicing specificity
Overall, both transcriptional and post-transcriptional regulatory mechanisms of opioid receptor splicing specificity remain largely uncharacterized, as do the endogenous factors that mediate tissue- and cell-type-specific shifts in opioid receptor isoform expression; however, recent studies have started to identify exogenous factors that modify opioid receptor splicing patterns. Interestingly, splicing regulation of opioid receptors, specifically the MOR, is similar to other transcriptional and post-transcriptional regulatory mechanisms of opioid receptor expression in that it exhibits a level of autoregulation by opioids. Although studies are limited, methadone maintained individuals have been found to exhibit changes in the expression of alternatively spliced MOR isoforms, with the MOR-1A isoform being up-regulated while the MOR-1O isoform is down-regulated (Vousooghi et al., 2009). Additionally, chronic morphine has been shown to alter the mRNA expression of specific MOR isoforms in several CNS regions of both mice and rats (Verzillo et al., 2014; Xu et al., 2015) while acute morphine treatment has been found to increase MOR-1X mRNA expression in SH-SY5Y cells, a cell line model for human dopaminergic neurons (Regan et al., 2015; submitted). This suggests that opioids can directly regulate the profile of MOR isoforms expressed; however, specific transcriptional and post-transcriptional mechanisms through which opioids directly alter constitutive and alternative splicing have not been identified.
With regard to post-transcriptional regulation of opioid receptor isoform expression, the most promising mechanism through which opioids facilitate changes in opioid receptor splicing specificity is through altering the concentration and subcellular localization of serine-arginine rich (SR) proteins and/or heterogeneous nuclear ribonucleoproteins (hnRNPs), as these auxiliary splicing factors enhance or diminish, respectively, the utilization of the 3’ and 5’ splice sites they interact with (Stamm, 2008; Stamm et al., 2005). In support of this mechanism, chronic and acute morphine treatments were found to, respectively, increase and decrease expression of the SR protein Tra2β in the rat locus coerulus through an unknown mechanism (Li et al., 2013). Similarly, acute morphine treatment was found to increase ASF/SF2 protein expression (Regan et al. 2015; submitted). Furthermore, morphine treatment was found to increase both the cytoplasmic and nuclear expression of hnRNP K protein, but not mRNA, in rat primary cortical neurons and the HEK293 cell line, in a time-dependent and dose-dependent manner through a mechanism involving increased internal ribosome entry segment (IRES)-mediated hnRNP K translation (Lee et al., 2014). The dynamic phosphorylation of auxiliary splicing factors, important for regulating activity, substrate specificity, and subcellular localization (Hui, 2009; Long and Caceres, 2009; Stamm, 2008), may also be altered by opioids, as expression of kinase-enhanced PP1 inhibitor (KEPI), a known inhibitor of protein phosphatases, is up-regulated by both acute and chronic morphine treatment (Liu et al., 2002); however, a similar role for opioids in the regulation of SRPK and Clk family kinases has not been identified. Additional, mutually inclusive mechanisms of opioid-mediated post-transcriptional regulation may involve ~22-nucleotide non-coding RNAs, known as microRNA (miRNA), that directly down-regulate mRNA expression through the direct binding of partially complementary sequences located in the 3’ and 5’ untranslated regions (UTRs) as well as open reading frames (ORFs) of the target mRNA. To this extent, MOR signaling has been found to alter miRNA expression (Dave and Khalili, 2010) and both miR-103 and miR-107 have been found to target the 3’-UTR of the MOR-1A isoform and effectively suppress its expression (Hwang et al., 2012; Lu et al., 2014); however, a direct mechanism for selective opioid receptor isoform expression involving opioid-mediated regulation of miRNA activity has not yet been characterized.
With regard to transcriptional regulation of opioid receptor splicing specificity, potential mechanisms through which opioids facilitate changes in opioid receptor isoform expression may involve epigenetic regulation, as increased methylation of CpG sites in OPRM1 promoter regions is seen in heroin addicts as well as former heroin addicts stabilized on methadone as a therapeutic. Reduced gene expression is observed in this population, possibly as a result of reduced binding affinity for transcription factors, although increased binding of transcription factors via opioid-mediated activation of the cAMP-PKA pathway has also been observed (Wei, 2008; Xu and Carr, 2001a; Xu and Carr, 2001b). Whether this translates to differences in the utilization of the proximal, distal, and exon 11 promoters, and in the subsequent availability of different pre-mRNA transcripts for alternative splicing events, remains unclear. Therefore, while multiple transcriptional and post-transcriptional regulatory mechanisms exist, the most likely mechanisms through which opioids autoregulate the splicing specificity of opioid receptors are post-transcriptional and involve opioid-mediated modifications of auxiliary splicing protein expression, phosphorylation, and subcellular localization.
Regulation of opioid receptor splicing specificity by HIV infection
In addition to opioid-mediated mechanisms of splicing regulation, HIV infection may likewise play a significant role in regulating both opioid receptor expression and splicing specificity, as multiple pro-inflammatory cytokines are known to impact opioid gene expression (Kraus, 2009; Kraus et al., 2003; Pasternak and Pan, 2013). In support of this hypothesis, the HIV viral protein gp120 was shown to up-regulate MOR expression through autocrine/paracrine actions of TNFα, while the HIV viral protein Tat alters the expression of MOR and KOR, but not DOR, through increased inflammatory cytokine activity (Beltran et al., 2006; Fitting et al., 2010; Kraus et al., 2003; Pasternak and Pan, 2013). Furthermore, co-administration of Tat attenuates morphine-mediated down-regulation of opioid receptor expression in microglia and instead induces a significant increases in both receptor mRNA and protein, although the mechanisms through which this occurs are not yet understood (Turchan-Cholewo et al., 2008). Unfortunately, both the role of HIV infection in regulating OPRM1 activity and the mechanisms through which this may be mediated are poorly understood and, in most cases, contradictory. HIV infection has been found to both decrease and increase MOR expression depending on the cell-type investigated as well as the presence or absence of concomitant opioid abuse (Turchan-Cholewo et al., 2008). Despite this lack of consensus on HIV-mediated changes in MOR expression, it is clearly evident that HIV alters the splicing pattern of the MOR. Studies have found that HIV-1 slightly increases MOR-1, MOR-1A, and MOR-1X expression in astrocytes but down-regulates MOR-1A in neurons and microglia, although these changes were only significant for microglia (Dever et al., 2012). Astrocytes from HIV-infected individuals were also found to have elevated mRNA expression of MOR-1K (Dever et al., 2014). Therefore, HIV infection regulates the splicing specificity of the MOR, although exact transcriptional and post-transcriptional mechanisms through which this is mediated have not been identified.
Given that HIV, as a lentivirus, is known to utilize mechanisms to actively regulate host auxiliary splicing factor protein levels, both spatially and temporally, to ensure the proper balance of fully-, partially-, and unspliced mRNA transcripts, it is likely that modulation of opioid receptor splicing by HIV viral proteins utilize similar post-transcriptional mechanisms. Accordingly, HIV infection has been shown to alter the expression of several SR proteins, including SC35 (Dowling et al., 2008), 9G8 (Ryo et al., 2000), SRp75, SRp55, SRp40, SRp30c, SRp20, and ASF/SF2 (Fukuhara et al., 2006), as well as several hnRNPs, including hnRNP A1 and hnRNP H. HIV-mediated variations in auxiliary splicing factor expression exhibit both spatial regulation, with the nuclear and cytoplasmic proportions of certain factors being specifically altered, and temporal regulation, with stark differences found between early and late infection (Dowling et al., 2008). This spatial and temporal regulation is facilitated by transcripts expressed during late-stage HIV infection, such as Tat, thereby titrating SR protein expression as infection progresses from early to late stage (Dowling et al., 2008; Fukuhara et al., 2006). Therefore, while transcriptional regulatory mechanisms similar to those mediated by opioids may also exist for HIV infection, it is likely that mechanisms of HIV-mediated regulation of opioid receptor splicing are post-transcriptional and primarily involve fluctuations in the subcellular and overall abundance of auxiliary splicing factors, although this has not been explicitly shown.
Functional implications of opioid receptor splicing
Pluridimensional efficacy is the phenomenon through which agonist binding of G protein-coupled receptors (GPCRs), such as opioid receptors, results in the activation of multiple downstream effector pathways. Initially, pluridimensional efficacy was thought to be the result of cell type-specific expression of effectors, such as G proteins (Wong, 2003). However, there are comparatively fewer genes encoding G protein subunits than GPCRs. In addition, it is now understood that multiple G protein subtypes bind a single GPCR, allowing for the activation of multiple signaling pathways by a single receptor. Furthermore, ligand-biased signaling has been shown for many GPCRs, particularly opioid receptors, as the binding of different agonists and antagonists selectively activate distinct signaling cascades through an individual receptor. Given the promiscuity of G proteins and second messenger proteins, in addition to a limited effector pool and the potential for ligand-biased signaling, cellular mechanisms must be utilized in order to maintain GPCR signaling specificity. The major mechanism through which cells maintain signaling specificity is now recognized as receptor heterogeneity, which arises from the existence of multiple genes that encode similar, but distinct, receptors subtypes, such as those for opioid receptors, in addition to alternative splicing of gene transcripts that produce various receptor isoforms, such as those identified for opioid receptor subtypes (Maudsley et al., 2005). The ability of receptor subtypes and isoforms to mediate signaling specificity and heterogeneity is primarily due to structural differences between receptors (Hui, 2009; Markovic and Challiss, 2009). As such, a concentrated investigation of opioid receptor structural motifs as well as the downstream signaling pathways initiated by these structural components is critical in understanding the functional significance of opioid receptor splicing in opioid receptor pharmacology and in identifying the potential signaling of individual receptor isoforms.
Opioid receptor structure
The conventional opioid receptors belong to the Class A GPCR family and, as such, contain many of the key structural features necessary for receptor activation and signaling (Pasternak and Pan, 2013; Petaja-Repo et al., 2006; Surratt and Adams, 2005). A D/ERY motif, expressed as DRY in opioid receptors, as well as a XBBXXB motif, expressed as LRRITR in opioid receptors, and a NPXXY motif, expressed as NPVLY in opioid receptors, have been identified within homologous regions of the various opioid receptor subtypes and have been shown to have functional significance in opioid-mediated signaling (Connor and Traynor, 2010; Law, 2010; Surratt and Adams, 2005; Waldhoer et al., 2004). Accordingly, amino acid substitutions in the DRY and LRRITR motifs are associated with modified receptor stabilization, G protein binding, and constitutive activity (Connor and Traynor, 2010; Pasternak and Pan, 2013) while substitutions in the NPXXY motif are associated with alterations in agonist binding, recruitment of G proteins and receptor kinases, and receptor endocytosis, trafficking, and recycling (Wang et al., 2003). Likewise, the proximal C-terminal domain of the MOR contains palmitoylated cysteine residues characteristic of Class A GPCRs. For MORs, this palmitoylation is highly dynamic, with constitutive palmitoylation occurring during the translocation of newly synthesized receptors while agonist-mediated palmitoylation/depalmitoylation cycles likely regulate signaling transduction stimulated by opioid-bound receptors (Minami and Satoh, 1995; Petaja-Repo et al., 2006; Surratt and Adams, 2005). High affinity binding sites are generated by disulfide bonds between two conserved cysteine residues found in the first and second extracellular loops (Minami and Satoh, 1995). An aromatic cluster motif, with the conserved sequence FXXXWXXXH, has also been identified within the opioid receptor family. Although it is not the terminal phenylalanine often associated with the conserved GPCR motif, the histidine residue within opioid receptors is thought to function in a similar manner by facilitating agonist-mediated disruptions of electrostatic interactions, toggling the receptor between active and inactive states (Surratt and Adams, 2005). Collectively, these conserved structural motifs interact in such a way as to generate two broad regions critical in opioid receptor G protein signaling: the third intracellular loop and the C-terminal domain (Georgoussi et al., 1997). Overall, opioid receptor subtypes are about 60% identical in their primary structure, with greater conservation of approximately 73-76% and 86-100% found in the transmembrane regions and intracellular loops, respectively, and lesser conservations of approximately 9-10%, 14-72%, and 14-20% found in the N-terminal, extracellular loops, and C-terminal, respectively (Law, 2010; Minami and Satoh, 1995; Wei et al., 2004).
The conserved structural motifs between opioid receptors and other Class A GPCRs only serve to confer a similar tertiary structure and facilitate G protein coupling and signaling cascades. Additional motifs, unique to opioid receptors, complement these general GPCR family motifs in maintaining the active and inactive conformations of opioid receptors and confer specific opioid-mediated signaling mechanisms (Li et al., 2001). For example, selective binding of opioids is determined, in part, by the third extracellular loop of opioid receptors, with varying lengths altering the affinity for certain opioid agonists and antagonists. However, studies using chimeric μ/κ opioid receptors suggest that any change in structure between the center of the third intracellular loop to the C-terminal domain, specifically within MORs, may alter binding of selective opioid agonists (Xue et al., 1995). Accordingly, it is now understood that multiple structural motifs within opioid receptors interact with the chemical structure of opioid ligands to generate a selective binding pocket. Specificity of opioid receptors is mediated by differential occupancy of one of two binding pocket domains and is ligand-dependent. These binding pockets involve interactions between sixth and seventh transmembrane domain for certain ligands, while others ligands target a binding pocket involving interactions between the second, third, and seventh transmembrane domains. As such, the efficacy of a given opioid ligand is determined by several residues within the second, third, sixth, and seventh transmembrane domains (Waldhoer et al., 2004).
Following ligand binding, opioid receptor-specific signaling pathways are activated. As with most GPCRs, this is facilitated by the recruitment of G proteins, which causes conformational changes allowing for the depalmitoylation of cysteine residues, ultimately resulting in increased phosphorylation of specific motifs within the C-terminal domain (Clayton et al., 2010; Petaja-Repo et al., 2006; Surratt and Adams, 2005). Consequently, substitution of conserved cysteine residues within the C-terminal domain, particularly C348 and C353, alters the basal rate of G protein binding (Connor and Traynor, 2010). Several potential PKA and PKC phosphorylation motifs have been identified in the second and third intracellular loops as well as the C-terminal domain (Minami and Satoh, 1995). A unique agonist-induced phosphorylation motif, defined by the consensus motif TXXXPS, has also been identified for opioid receptors and phosphorylation at this site is thought to activate downstream signal cascades (Wei et al., 2004). Additional conserved tyrosine residues are phosphorylated and are thought to be involved in receptor trafficking and signaling, particularly in a switch from adenylyl cyclase inhibition to stimulation following prolonged agonist application (Clayton et al., 2010). Endocytic trafficking of MORs, which functions to remove receptors from the plasma membrane following prolonged activation and target them for degradation, can occur through two separate processes. Structural motifs within the first intracellular loop and C-terminal domain, which are unique to the MOR, mediate an ubiquitination-dependent endocytic trafficking pathway in a late stage of receptor down-regulation. Early stage down-regulation of MORs is also mediated by structural motifs localized to the C-terminal domain but is ubiquitination-independent (Hislop et al., 2011). Additional residues, including multiple leucine residues within the C-terminal domain and third intracellular loop (Wang et al., 2003), have also been implicated in the endocytosis, trafficking, and recycling of multiple opioid receptor subtypes. Therefore, multiple functions of the opioid receptor family are mediated by a combination of conserved Class A GPCR structural motifs and unique residues, with the latter conferring specific opioid ligand binding and signaling activity.
The structure/function relationship of opioid receptor variants
Translation of alternatively spliced transcripts produces various isoforms of a given protein that differ in their amino acid sequence and, as such, are differentially subjected to post-translational modifications and protein folding. Their unique amino acid sequence, post-translational modifications, and tertiary structure cause many isoforms to display enzymatic activity, ligand binding, cellular localization, and protein stability that is distinct from other isoforms of the same protein (Hui, 2009; Markovic and Challiss, 2009). Obvious functional effects are found in GPCR splicing patterns that alter conserved motifs required for classical GPCR structure, G protein binding, and activating downstream signaling cascades, such as the D/ERY, XBBXXB, and NPXXY(X) consensus sequences. However, given the low sequence homology between GPCR families, potential functional consequences are predicted based of broad structural changes within the N- and C-terminal domains and transmembrane regions instead of defined amino acid sequences. As such, the GPCR structure can be divided into functional regions and changes within these regions, due to either SNPs, mutations, or alternative splicing events, are predicted to have distinct functional consequences. The extracellular N-terminal domain, described previously, is involved in ligand binding. As such, variations or truncations of the N-terminal domain due to alternative splicing may result in altered ligand binding. If binding is altered to such a degree that ligands cannot activate the receptor, overexpression of the GPCR isoform(s) expressing this alternative N-terminal domain may result in a dominant-negative effect. Similarly, alterations of the extracellular loop structures may alter ligand binding. Transmembrane regions of GPCRs are critical for proper tertiary structure, trafficking to the cell membrane, and maintaining the inactive and active conformations of the receptor. Alternative transmembrane sequences, particularly within the seventh transmembrane domain, may impair receptor trafficking and membrane expression, decreasing receptor concentrations and resulting in dominant negative phenotypes. Additionally, the presence of premature stop codons anywhere between the first and seventh transmembrane domains generates a truncated GPCR variant. Due to the absence of one or multiple transmembrane domains, these truncated GPCR isoforms may localize as cytoplasmic or secreted soluble proteins, resulting in vastly different mechanisms of action (Markovic and Challiss, 2009). GPCR intracellular loop structures typically help facilitate G protein selectivity and binding through the establishment of activation and selectivity domains. Accordingly, variant structures at these functional domains may alter the fidelity of G protein subtype selectivity or the kinetics of G protein coupling/uncoupling and may even result in increased constitutive activity (Markovic and Challiss, 2009; Wong, 2003). Finally, the C-terminal domain represents one of the most important functional regions of GPCRs, containing a variety of palmitoylation and phosphorylation motifs that regulate receptor activity, signaling, and trafficking. GPCRs with alternatively spliced C-terminal domains exhibit distinct GPCR signaling, receptor internalization, and protein-protein interactions with receptor kinases and β-arrestins in addition to regulating constitutive activity of the receptor (Markovic and Challiss, 2009). Consequently, alternative splicing of GPCRs has the potential to greatly alter receptor function through sequence and structural changes within the transmembrane domains, extracellular and intracellular loops, and N- and C-terminal domains.
Although all opioid receptors are alternatively spliced to some extent, only the MOR exhibits extensive splicing within the coding regions of every major GPCR structural domain, including the transmembrane domains, extracellular and intracellular loops, and N- and C-terminal domains. As such, MOR isoforms can be categorized into multiple classes based on their structural and functional similarities. The first class consists of MOR isoforms that are typically generated through alternative 3’ splice site selection and comprise full-length receptor variants. These isoforms share identical N-terminal domains, transmembrane regions, and intra- and extracellular loop structures. In following, these isoforms are highly selective for μ-selective opioids and show little variability in receptor binding affinities, as their conserved sequences encode the binding pocket domain. However, the distal C-terminal domains of these isoforms are altered due to the utilization of alternative, suboptimal 3’ splice sites within the C-terminal domain encoding exon 4, the substitution of exon 4 with an alternative exon, or the exclusion of exon 4 entirely followed by intron retention. The majority of MOR isoforms, including MOR-1A and MOR-1X, belong to this class. The variant C-terminal domains of these isoforms are differentially palmitoylated, ubiquitinated, and phosphorylated and, as a result, display distinct regional and cell-type specific localizations, G protein coupling, desensitization, internalization, and post-endocytic sorting. Additionally, C-terminal splicing greatly impacts both the potency and efficacy of μ-selective agonists. The selective incorporation or exclusion of putative phosphorylation sites in the C-terminal domain of MOR isoforms alters ligand-biased signal transduction pathways, most likely due to differential recruitment of receptor kinases and β-arrestins. For example, morphine and DAMGO are typically regarded as stimulating different levels of receptor phosphorylation and internalization; however, certain variants show equal levels of phosphorylation and internalization by these two compounds. Similar differences in ligand-mediated internalization can be seen for many C-terminal domain variants of the MOR. As a result, some MOR isoforms modulate second messenger signaling, including adenylate cyclase, Ca2+/NFAT, and MAPK activity, with distinct differences (Pan, 2005; Pasternak, 2010; Pasternak and Pan, 2013; Surratt and Adams, 2005; Waldhoer et al., 2004).
A second class of MOR isoforms is generated by transcription initiation from the exon 11 promoter region and the subsequent substitution of exon 1 with exon 11, which frequently results in the loss of the first transmembrane region and the development of a unique N-terminal domain that protrudes into the intracellular region as opposed to the extracellular environment. As expected, expression of these N-terminal variants produces a diminished response to certain opioids, with no effect seen to others, suggesting a change in the selective binding of certain ligands. Evidence from in vivo studies using exon 1 and exon 11 knockout mice suggests that these variants are particularly important for heroin and M6G activity as well as mediating the actions of novel opioid compounds, such as 3-Iodobenzoylnaltrexamide 1 (IBNtxA), that lack side-effects of respiratory depression, physical dependence, and rewarding behavior typically associated with μ-selective compounds (Abbadie et al., 2004; Klein et al., 2009; Pan, 2002; Pan et al., 2009; Pan et al., 2001; Pasternak, 2010; Pasternak and Pan, 2013; Waldhoer et al., 2004; Xu et al., 2014b; Xu et al., 2013; Xu et al., 2009; Xu et al., 2011). Additionally, the substitution of exon 1 for exon 11 may result in MOR variants with cellular and molecular functions completely opposite of classical MORs. This is demonstrated by the exon 11 variant MOR-1K, which is retained in the cytoplasm where it couples Gαs, as opposed to Gαi/o. In following, activation of MOR-1K was found to enhance cAMP, Ca2+, and NO production and resulted in cellular excitation, as opposed to the classical MOR-mediated cellular inhibition (Gris et al., 2010).
A group of severely truncated MOR receptors comprises the third class of MOR isoforms. These variants are produced by transcription initiation at the exon 11 promoter region but excise the majority of recognized exons, including exons 2 and/or 3, from their final mRNA transcript. Given that exons 2 and 3 encode the majority of the MOR transmembrane domains, these variants only contain a single transmembrane region. As such, these membranes do not bind opioid ligands nor are they expressed in a typical manner within the plasma membrane. Instead, these variants are thought to function as receptor chaperones to reduce ER-associated degradation of full-length MOR variants, enhancing receptor expression (Pan, 2002; Pan et al., 2001; Pasternak and Pan, 2013; Xu et al., 2014b; Xu et al., 2013). In support of this, MOR-1 and the SV1 and SV2 isoforms of the MOR have been found to dimerize either during or shortly after translation resulting in inhibited ligand binding (Choi et al., 2006). Interestingly, certain single transmembrane MORs appear to compliment certain six transmembrane MORs and their physical interaction has been shown; however, the functional significance of this association has yet to be characterized (Abbadie et al., 2004).
Opioids isoforms & HIV pathogenesis
In addition to their antinociceptive effects, many opioids stimulate profound immunosuppression, thereby increasing the susceptibility of individuals chronically using opioids, both clinically and illicitly, to various infections. The mechanism of opioid-mediated immunosuppression is thought to involve cross-talk between immune system and CNS regulatory pathways; however, it is now understood that opioids also modulate immune system activity by directly interacting with immune cells (Taub et al., 1991). While both endogenous and exogenous opioids modulate immune system activity and increase susceptibility to infection, the illicit use of opioids such as heroin, serves as an additional risk factor for the transmission of blood-borne pathogens, particularly HIV, through the sharing of contaminated syringes. Furthermore, injection drug abuse, specifically of opioids, alters the pathogenesis of HIV as well as enhances HIV-associated systemic and neurological complications collectively known as HIV-associated neurocognitive dysfunction (HAND) (Hauser et al., 2005). Currently, two mutually inclusive models account for neurodegeneration and the development of neurological symptoms in HAND: the indirect model and the direct model. The indirect model proposes that HIV-mediated neurodegeneration is a secondary effect of the inflammatory responses and deregulation of glial function caused by infected and non-infected glial cells within the CNS. The direct model proposes that HIV-mediated neurodegeneration is a primary effect of the interaction between neurons and HIV viral proteins secreted from infected monocyte-derived cells within the CNS (Kaul et al., 2001). Opiate drug abuse is now recognized to interact synergistically with both direct and indirect mechanisms of HIV-associated neurotoxicity. This synergy is primarily mediated through activation of the MOR, although limited evidence suggests roles for KORs and DORs as well. With respect to the indirect mechanism of HIV-associated neurotoxicity, opioids are considered to exacerbate HAND pathology by synergistically decreasing trophic and metabolic support and enhancing marginally neurotoxic, excitotoxic, and inflammatory cellular stressors secreted by dysfunctional glial cells, all of which may occur in the absence of glial cell loss (Hauser et al., 2012). Similarly, with respect to the direct mechanism of HIV-associated neurotoxicity, concomitant HIV viral protein and chronic opioid exposure is typically accompanied by altered patterns of gene activation, decreased neurotrophic signaling, increased ROS production and oxidative stress, and enhanced mitochondrial dysfunction within neurons due to synergistic disruption of second messenger cascades, including adenylyl cyclase, SDF-1, PKA, PKB, PI3K/Akt, p38 MAPK, ERK1/2, JNK, GSK-3β, IKK-α/Iκ-Bα/NF-κB, CREB, p53, PTEN, calcineurin, Bcl-2 family member proteins, endonuclease G, and caspase-1, -3, and -7 (Hauser et al., 2005; Hauser et al., 2012). Therefore, in addition to indirect, glia-dependent mechanisms, marginally toxic opioids directly exacerbate the excitotoxic, dendrotoxic, and neurotoxic events triggered by secreted HIV viral proteins through the synergistic disruption of numerous signaling cascades. This, in turn, triggers sublethal synaptodendritic injury that culminates in the activation of caspase-dependent and caspase-independent apoptotic mechanisms, ultimately resulting in the neuronal cell loss characteristic of HAND. While the exact contribution of individual MOR isoforms to the exacerbation or alleviation of HIV-associated neurotoxic mechanisms is currently unknown, a recent study found a correlation between HIV-mediated increases in an excitatory MOR isoform, MOR-1K, and the severity of neurocognitive deficits in HAND pathology (Dever et al., 2014), suggesting a dynamic relationship between opioid receptor isoforms and HIV-associated neuropathology.
Future Studies
The recent identification and characterization of several opioid receptor isoforms generated through alternative splicing of opioid receptor-encoding genes, particularly the μ-opioid receptor-encoding OPRM1, has led to a more conclusive explanation for the discrepancies observed in opioid pharmacology, as structural differences between isoforms result in unique agonist selectivity, constitutive activity, agonist-mediated signaling, and internalization. Furthermore, these isoforms, in conjunction with several SNPs, are understood to have important physiological signifigance in opioid sensitivity and addiction (Klein et al., 2009; Kreek and LaForge, 2007; Lotsch and Geisslinger, 2005), as some polymorphisms in the MOR-1K (Diatchenko et al., 2011; Shabalina et al., 2009) and MOR-1X (Pang et al., 2012) variants have been found to alter opioid drug response, while others have not (Mayer and Hollt, 2006; Smith et al., 2005). Therefore, opioid receptor isoforms must be regarded as a separate receptor subtype that collectively contributes to the overall cellular and physiological effects of opioids and differential expression of these isoforms will alter opioid pharmacology accordingly. Given this emerging importance of individual MOR isoforms in opioid pharmacology, as well as pathological conditions such as HIV-associated neurocognitive dysfunction, extensive studies are needed to further characterize the structural and functional significance of individual isoforms as well as the regulatory mechanisms through which opioid receptor splicing specificity is determined. To this extent, while recent studies have started to identify the contributions of individual opioid receptor isoforms to opioid pharmacology and have correlated the abundance of particular isoforms with the severity of pathogenic states, specifically HIV-associated neurocognitive dysfunction, additional mutagenic studies examining unique functional domains of opioid receptor isoforms predicted from their primary structure are needed to further characterize the structure/function relationship of individual opioid receptor isoforms in these processes. Furthermore, recent studies identifying opioid- and HIV-mediated alterations in opioid receptor splicing have reported potential regulatory mechanisms, particularly those involving the modification of auxiliary splicing proteins. Unfortunately, the role of individual splicing factors in specific transcripts has been complicated by the inability to identify specific binding sites within mRNA transcripts and to accurately predict splice site usage; however, recent bioinformatics approaches and experimental techniques, such as SELEX and CLIP, have been useful in identifying exon splicing enhancer (ESE) and silencer (ESS) motifs and the binding specificity of individual SR proteins, allowing for a better prediction of SR protein interaction using pre-mRNA sequence (Long and Caceres, 2009). Utilization of these assays in conjunction with new a class of indole derivative splicing inhibitors, which have selective action against the ESE-dependent activity of SR proteins (Keriel et al., 2009), and sensitive genome-wide microarrays will allow for comprehensive detection, measurement and characterization of known and predicted mRNA isoforms (Griffith et al., 2010). As such, new tools being developed to explore regulatory elements, scan previously uncharacterized exons, and predict tissue-dependent splicing patterns will be beneficial for future studies of opioid receptor splicing regulatory mechanisms and the impact of alternative splicing on opioid pharmacology (Barash et al., 2010).
ACKNOWLEDGMENTS
The authors wish to thank past and present members of the Department of Neuroscience and the Center for Neurovirology for their insightful discussion and sharing of ideas and reagents. We also thank C. Papaleo for editiorial assistance. This work was made possible by grants awarded to KK by NIH (P30 MH092177 and P01 DA023860). This investigation was supported in part by the National Institutes of Health under the Ruth L. Kirschstein National Research Service Award, (5T32MH079785) “Interdisciplinary and Translational Research Training Program in NeuroAIDS,” awarded to Temple University School of Medicine, which provided training to Patrick Regan. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.”
REFERENCES CITED
- Abbadie C, Pan Y-X, Pasternak GW. Immunohistochemical study of the expression of exon 11-containing mu opioid receptor variants in mouse brain. Neuroscience. 2004;127(2):419–430. doi: 10.1016/j.neuroscience.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Abbadie C, Pasternak GW. Peptide Receptors: Handbook of Chemical Neuroanatomy. 2002. Opioid receptors; pp. 1–29. [Google Scholar]
- Andersen S, Baar C, Fladvad T, Laugsand EA, Skorpen F. The N-terminally truncated mu3 and mu3-like opioid receptors are transcribed from a novel promoter upstream of exon 2 in the human OPRM1 gene. PLoS ONE. 2013;8(8):e71024. doi: 10.1371/journal.pone.0071024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465(7294):53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Barrie ES, Smith RM, Sanford JC, Sadee W. mRNA transcript diversity creates new opportunities for pharmacological intervention. Molecular pharmacology. 2012;81(5):620–630. doi: 10.1124/mol.111.076604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran JA, Pallur A, Chang SL. HIV-1 gp120 up-regulation of the mu opioid receptor in TPA-differentiated HL-60 cells. International immunopharmacology. 2006;6(9):1459–1467. doi: 10.1016/j.intimp.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Chevlen E. Opioids: a review. Curr Pain Headache Rep. 2003;7(1):15–23. doi: 10.1007/s11916-003-0005-5. [DOI] [PubMed] [Google Scholar]
- Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, Law P-Y, Wei L-N, Loh HH. The opioid ligand binding of human mu-opioid receptor is modulated by novel splice variants of the receptor. Biochemical and biophysical research communications. 2006;343(4):1132–1140. doi: 10.1016/j.bbrc.2006.03.084. [DOI] [PubMed] [Google Scholar]
- Clayton CC, Bruchas MR, Lee ML, Chavkin C. Phosphorylation of the mu-opioid receptor at tyrosine 166 (Tyr3.51) in the DRY motif reduces agonist efficacy. Molecular pharmacology. 2010;77(3):339–347. doi: 10.1124/mol.109.060558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor M, Traynor J. Constitutively active mu-opioid receptors. Methods Enzymol. 2010;484:445–469. doi: 10.1016/B978-0-12-381298-8.00022-8. [DOI] [PubMed] [Google Scholar]
- Dave RS, Khalili K. Morphine treatment of human monocyte-derived macrophages induces differential miRNA and protein expression: impact on inflammation and oxidative stress in the central nervous system. Journal of cellular biochemistry. 2010;110(4):834–845. doi: 10.1002/jcb.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever SM, Costin BN, Xu R, El-Hage N, Balinang J, Samoshkin A, O’Brien MA, McRae M, Diatchenko L, Knapp PE, Hauser KF. Differential expression of the alternatively spliced OPRM1 isoform mu-opioid receptor-1K in HIV-infected individuals. AIDS. 2014;28(1):19–30. doi: 10.1097/QAD.0000000000000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever SM, Xu R, Fitting S, Knapp PE, Hauser KF. Differential expression and HIV-1 regulation of mu-opioid receptor splice variants across human central nervous system cell types. Journal of neurovirology. 2012;18(3):181–190. doi: 10.1007/s13365-012-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Robinson JE, Maixner W. Elucidation of mu-opioid gene structure: How genetics can help predict responses to opioids. Eur J Pain Suppl. 2011;5(2):433–438. doi: 10.1016/j.eujps.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D, Nasr-Esfahani S, Tan CH, O’Brien K, Howard JL, Jans DA, Purcell DF, Stoltzfus CM, Sonza S. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology. 2008;5(1):18. doi: 10.1186/1742-4690-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitting S, Xu R, Bull C, Buch SK, El-Hage N, Nath A, Knapp PE, Hauser KF. Interactive comorbidity between opioid drug abuse and HIV-1 Tat: chronic exposure augments spine loss and sublethal dendritic pathology in striatal neurons. Am J Pathol. 2010;177(3):1397–1410. doi: 10.2353/ajpath.2010.090945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricchione G, Zhu W, Cadet P, Mantione KJ, Bromfield E, Madsen J, DeGirolami U, Dworetzky B, Vaccaro B, Black P, Stefano GB. Identification of endogenous morphine and a mu3-like opiate alkaloid receptor in human brain tissue taken from a patient with intractable complex partial epilepsy. Med Sci Monit. 2008;14(6):45–49. [PubMed] [Google Scholar]
- Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, Hagiwara M. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(30):11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaveriaux-Ruff C, Peluso J, Befort K, Simonin F, Zilliox C, Kieffer BL. Detection of opioid receptor mRNA by RT-PCR reveals alternative splicing for the delta- and kappa-opioid receptors. Brain Res Mol Brain Res. 1997;48(2):298–304. doi: 10.1016/s0169-328x(97)00109-5. [DOI] [PubMed] [Google Scholar]
- Georgoussi Z, Merkouris M, Mullaney I, Megaritis G, Carr C, Zioudrou C, Milligan G. Selective interactions of mu-opioid receptors with pertussis toxin-sensitive G proteins: involvement of the third intracellular loop and the c-terminal tail in coupling. Biochimica et biophysica acta. 1997;1359(3):263–274. doi: 10.1016/s0167-4889(97)00097-9. [DOI] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, Corbett R, Tang MJ, Hou YC, Pugh TJ, Robertson G, Chittaranjan S, Ally A, Asano JK, Chan SY, Li HI, McDonald H, Teague K, Zhao Y, Zeng T, Delaney A, Hirst M, Morin GB, Jones SJ, Tai IT, Marra MA. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7(10):843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- Gris P, Gauthier J, Cheng P, Gibson DG, Gris D, Laur O, Pierson J, Wentworth S, Nackley AG, Maixner W, Diatchenko L. A novel alternatively spliced isoform of the mu-opioid receptor: functional antagonism. Mol Pain. 2010;6:33. doi: 10.1186/1744-8069-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser KF, El-Hage N, Buch S, Berger JR, Tyor WR, Nath A, Bruce-Keller AJ, Knapp PE. Molecular targets of opiate drug abuse in neuro AIDS. Neurotox Res. 2005;8(1-2):63–80. doi: 10.1007/BF03033820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser KF, Fitting S, Dever SM, Podhaizer EM, Knapp PE. Opiate drug use and the pathophysiology of neuroAIDS. Curr HIV Res. 2012;10(5):435–452. doi: 10.2174/157016212802138779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hislop JN, Henry AG, von Zastrow M. Ubiquitination in the first cytoplasmic loop of mu-opioid receptors reveals a hierarchical mechanism of lysosomal down-regulation. The Journal of biological chemistry. 2011;286(46):40193–40204. doi: 10.1074/jbc.M111.288555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Bi J, Loh HH, Wei L-N. An intronic Ikaros-binding element mediates retinoic acid suppression of the kappa opioid receptor gene, accompanied by histone deacetylation on the promoters. The Journal of biological chemistry. 2001:4597–4603. doi: 10.1074/jbc.M005477200. [DOI] [PubMed] [Google Scholar]
- Hui J. Regulation of mammalian pre-mRNA splicing. Sci China C Life Sci. 2009;52(3):253–260. doi: 10.1007/s11427-009-0037-0. [DOI] [PubMed] [Google Scholar]
- Hwang CK, Wagley Y, Law PY, Wei LN, Loh HH. MicroRNAs in opioid pharmacology. Journal of neuroimmune pharmacology: the official journal of the Society on NeuroImmune Pharmacology. 2012;7(4):808–819. doi: 10.1007/s11481-011-9323-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410(6831):988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nature reviews Genetics. 2010;11(5):345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- Keriel A, Mahuteau-Betzer F, Jacquet C, Plays M, Grierson D, Sitbon M, Tazi J. Protection against retrovirus pathogenesis by SR protein inhibitors. PLoS One. 2009;4(2):e4533. doi: 10.1371/journal.pone.0004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G. Alternative splicing: current perspectives. Bioessays. 2008;30(1):38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- Klein G, Rossi GC, Waxman AR, Arout C, Juni A, Inturrisi CE, Kest B. The contribution of MOR-1 exons 1-4 to morphine and heroin analgesia and dependence. Neurosci Lett. 2009;457(3):115–119. doi: 10.1016/j.neulet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Kraus J. Regulation of mu-opioid receptors by cytokines. Front Biosci (Schol Ed) 2009;1:164–170. doi: 10.2741/s16. [DOI] [PubMed] [Google Scholar]
- Kraus J, Borner C, Giannini E, Hollt V. The role of nuclear factor kappaB in tumor necrosis factor-regulated transcription of the human mu-opioid receptor gene. Molecular pharmacology. 2003;64(4):876–884. doi: 10.1124/mol.64.4.876. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, LaForge KS. Stress responsivity, addiction, and a functional variant of the human mu-opioid receptor gene. Mol Interv. 2007;7(2):74–78. doi: 10.1124/mi.7.2.7. [DOI] [PubMed] [Google Scholar]
- Kvam TM, Baar C, Rakvag TT, Kaasa S, Krokan HE, Skorpen F. Genetic analysis of the murine mu opioid receptor: increased complexity of Oprm gene splicing. Journal of molecular medicine. 2004;82(4):250–255. doi: 10.1007/s00109-003-0514-z. [DOI] [PubMed] [Google Scholar]
- Law P-Y. Opioid receptor signal transduction mechanisms. In: Pasternak GW, editor. The Opiate Receptors. Humana Press; 2010. pp. 195–238. [Google Scholar]
- Lee PT, Chao PK, Ou LC, Chuang JY, Lin YC, Chen SC, Chang HF, Law PY, Loh HH, Chao YS, Su TP, Yeh SH. Morphine drives internal ribosome entry site-mediated hnRNP K translation in neurons through opioid receptor-dependent signaling. Nucleic Acids Res. 2014;42(21):13012–13025. doi: 10.1093/nar/gku1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Yuferov V, Kreek MJ. The genetics of the opioid system and specific drug addictions. Hum Genet. 2012;131(6):823–842. doi: 10.1007/s00439-012-1172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Huang P, Chen C, de Riel JK, Weinstein H, Liu-Chen LY. Constitutive activation of the mu opioid receptor by mutation of D3.49(164), but not D3.32(147): D3.49(164) is critical for stabilization of the inactive form of the receptor and for its expression. Biochemistry. 2001;40(40):12039–12050. doi: 10.1021/bi0100945. [DOI] [PubMed] [Google Scholar]
- Li S-J, Li Y, Cui S-C, Qi Y, Zhao J-J, Liu X-Y, Xu P, Chen X-H. Splicing factor transformer-2β (Tra2β) regulates the expression of regulator of G protein signaling 4 (RGS4) gene and is induced by morphine. PLoS ONE. 2013;8(8):e72220. doi: 10.1371/journal.pone.0072220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Zhang PW, Zhen Q, Walther D, Wang XB, Uhl GR. KEPI, a PKC-dependent protein phosphatase 1 inhibitor regulated by morphine. The Journal of biological chemistry. 2002;277(15):13312–13320. doi: 10.1074/jbc.M107558200. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417(1):15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lotsch J, Geisslinger G. Are mu-opioid receptor polymorphisms important for clinical opioid therapy? Trends in molecular medicine. 2005;11(2):82–89. doi: 10.1016/j.molmed.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Lu Z, Xu J, Xu M, Pasternak GW, Pan YX. Morphine regulates expression of mu-opioid receptor MOR-1A, an intron-retention carboxyl terminal splice variant of the mu-opioid receptor (OPRM1) gene via miR-103/miR-107. Molecular pharmacology. 2014;85(2):368–380. doi: 10.1124/mol.113.089292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic D, Challiss RA. Alternative splicing of G protein-coupled receptors: physiology and pathophysiology. Cellular and molecular life sciences: CMLS. 2009;66(20):3337–3352. doi: 10.1007/s00018-009-0093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley S, Martin B, Luttrell LM. The origins of diversity and specificity in G protein-coupled receptor signaling. The Journal of pharmacology and experimental therapeutics. 2005;314(2):485–494. doi: 10.1124/jpet.105.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer P, Hollt V. Pharmacogenetics of opioid receptors and addiction. Pharmacogenet Genomics. 2006;16(1):1–7. doi: 10.1097/01.fpc.0000182781.87932.0d. [DOI] [PubMed] [Google Scholar]
- Minami M, Satoh M. Molecular biology of the opioid receptors: structures, functions and distributions. Neurosci Res. 1995;23(2):121–145. doi: 10.1016/0168-0102(95)00933-k. [DOI] [PubMed] [Google Scholar]
- Mizoguchi H, Wu HE, Narita M, Sora I, Hall SF, Uhl GR, Loh HH, Nagase H, Tseng LF. Lack of mu-opioid receptor-mediated G-protein activation in the spinal cord of mice lacking Exon 1 or Exons 2 and 3 of the MOR-1 gene. J Pharmacol Sci. 2003;93(4):423–429. doi: 10.1254/jphs.93.423. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pan Y-X. Identification and characterization of a novel promoter of the mouse mu opioid receptor gene (Oprm) that generates eight splice variants. Gene. 2002;295(1):97–108. doi: 10.1016/s0378-1119(02)00825-9. [DOI] [PubMed] [Google Scholar]
- Pan Y-X, Xu J, Xu M, Rossi GC, Matulonis JE, Pasternak GW. Involvement of exon 11-associated variants of the mu opioid receptor MOR-1 in heroin, but not morphine, actions. Proc Natl Acad Sci USA. 2009;106(12):4917–4922. doi: 10.1073/pnas.0811586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YX. Identification of alternatively spliced variants from opioid receptor genes. Methods Mol Med. 2003;84:65–75. doi: 10.1385/1-59259-379-8:65. [DOI] [PubMed] [Google Scholar]
- Pan YX. Diversity and complexity of the mu opioid receptor gene: alternative pre-mRNA splicing and promoters. DNA Cell Biol. 2005;24(11):736–750. doi: 10.1089/dna.2005.24.736. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Abbadie C, Chang A, Zuckerman A, Rossi G, Pasternak GW. Identification and characterization of three new alternatively spliced mu-opioid receptor isoforms. Molecular pharmacology. 1999;56(2):396–403. doi: 10.1124/mol.56.2.396. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Bolan E, Xu M, Pasternak GW. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Xu M, Gilbert AK, Pasternak GW. Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochemical and biophysical research communications. 2003;301(4):1057–1061. doi: 10.1016/s0006-291x(03)00089-5. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Wan BL, Zuckerman A, Pasternak GW. Identification and differential regional expression of KOR-3/ORL-1 gene splice variants in mouse brain. FEBS letters. 1998;435(1):65–68. doi: 10.1016/s0014-5793(98)01039-4. [DOI] [PubMed] [Google Scholar]
- Pang GSY, Ithnin F, Wong YY, Wang JB, Lim Y, Sia ATH, Lee CGL. A non-synonymous single nucleotide polymorphism in an OPRM1 splice variant is associated with fentanyl-induced emesis in women undergoing minor gynaecological surgery. PLoS ONE. 2012;7(11):e48416. doi: 10.1371/journal.pone.0048416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW. Insights into mu opioid pharmacology the role of mu opioid receptor subtypes. Life Sci. 2001;68(19-20):2213–2219. doi: 10.1016/s0024-3205(01)01008-6. [DOI] [PubMed] [Google Scholar]
- Pasternak GW. Molecular insights into mu opioid pharmacology: From the clinic to the bench. Clin J Pain. 2010;26(Suppl 10):S3–9. doi: 10.1097/AJP.0b013e3181c49d2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW. Opioids and their receptors: Are we there yet? Neuropharmacology. 2014;76(Pt B):198–203. doi: 10.1016/j.neuropharm.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW, Pan Y-X. Mu opioids and their receptors: evolution of a concept. Pharmacological reviews. 2013;65(4):1257–1317. doi: 10.1124/pr.112.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak GW, Standifer KM. Mapping of opioid receptors using antisense oligodeoxynucleotides: correlating their molecular biology and pharmacology. Trends Pharmacol Sci. 1995;16(10):344–350. doi: 10.1016/s0165-6147(00)89068-9. [DOI] [PubMed] [Google Scholar]
- Petaja-Repo UE, Hogue M, Leskela TT, Markkanen PM, Tuusa JT, Bouvier M. Distinct subcellular localization for constitutive and agonist-modulated palmitoylation of the human delta opioid receptor. The Journal of biological chemistry. 2006;281(23):15780–15789. doi: 10.1074/jbc.M602267200. [DOI] [PubMed] [Google Scholar]
- Regan PM, Dave RS, Datta PK, Khalili K. Epigenetics of μ-opioid receptors: 1 infection of the central nervous system. J Cell Physiol. 2011 doi: 10.1002/jcp.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Suzuki Y, Arai M, Kondoh N, Wakatsuki T, Hada A, Shuda M, Tanaka K, Sato C, Yamamoto M, Yamamoto N. Identification and characterization of differentially expressed mRNAs in HIV type 1-infected human T cells. AIDS Res Hum Retroviruses. 2000;16(10):995–1005. doi: 10.1089/08892220050058416. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K, Tchivileva IE, Belfer I, Mishra B, Kiselycznyk C, Wallace MR, Staud R, Spiridonov NA, Max MB, Goldman D, Fillingim RB, Maixner W, Diatchenko L. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Human molecular genetics. 2009;18(6):1037–1051. doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Hui L, Xu Y, Wang F, Huang W, Hu G. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum Mutat. 2002;19(4):459–460. doi: 10.1002/humu.9026. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Doyle GA, Han AM, Crowley JJ, Oslin DW, Patkar AA, Mannelli P, Demaria PA, O’brien CP, Berrettini WH. Novel exonic mu-opioid receptor gene (OPRM1) polymorphisms not associated with opioid dependence. Am J Med Genet B Neuropsychiatr Genet. 2005;133B(1):105–109. doi: 10.1002/ajmg.b.30105. [DOI] [PubMed] [Google Scholar]
- Snyder SH, Pasternak GW. Historical review: Opioid receptors. Trends Pharmacol Sci. 2003;24(4):198–205. doi: 10.1016/S0165-6147(03)00066-X. [DOI] [PubMed] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. The Journal of biological chemistry. 2008;283(3):1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Surratt CK, Adams WR. G protein-coupled receptor structural motifs: relevance to the opioid receptors. Curr Top Med Chem. 2005;5(3):315–324. doi: 10.2174/1568026053544533. [DOI] [PubMed] [Google Scholar]
- Taub DD, Eisenstein TK, Geller EB, Adler MW, Rogers TJ. Immunomodulatory activity of mu- and kappa-selective opioid agonists. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(2):360–364. doi: 10.1073/pnas.88.2.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchan-Cholewo J, Dimayuga FO, Ding Q, Keller JN, Hauser KF, Knapp PE, Bruce-Keller AJ. Cell-specific actions of HIV-Tat and morphine on opioid receptor expression in glia. Journal of neuroscience research. 2008;86(9):2100–2110. doi: 10.1002/jnr.21653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ, Priddy CM, Chen G-L, Miller GM. Human expression variation in the mu-opioid receptor is paralleled in rhesus macaque. Behav Genet. 2008;38(4):390–395. doi: 10.1007/s10519-008-9207-2. [DOI] [PubMed] [Google Scholar]
- Verzillo V, Madia PA, Liu NJ, Chakrabarti S, Gintzler AR. Mu-opioid receptor splice variants: sex-dependent regulation by chronic morphine. Journal of neurochemistry. 2014;130(6):790–796. doi: 10.1111/jnc.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousooghi N, Goodarzi A, Roushanzamir F, Sedaghati T, Zarrindast MR, Noori-Daloii MR. Expression of mu opioid receptor splice variants mRNA in human blood lymphocytes: a peripheral marker for opioid addiction studies. International immunopharmacology. 2009;9(7-8):1016–1020. doi: 10.1016/j.intimp.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Waldhoer M, Bartlett SE, Whistler JL. Opioid receptors. Annual review of biochemistry. 2004;73:953–990. doi: 10.1146/annurev.biochem.73.011303.073940. [DOI] [PubMed] [Google Scholar]
- Wang W, Loh HH, Law PY. The intracellular trafficking of opioid receptors directed by carboxyl tail and a di-leucine motif in Neuro2A cells. The Journal of biological chemistry. 2003;278(38):36848–36858. doi: 10.1074/jbc.M301540200. [DOI] [PubMed] [Google Scholar]
- Wei LN. Epigenetic control of the expression of opioid receptor genes. Epigenetics. 2008;3(3):119–121. doi: 10.4161/epi.3.3.6296. [DOI] [PubMed] [Google Scholar]
- Wei LN, Law PY, Loh HH. Post-transcriptional regulation of opioid receptors in the nervous system. Front Biosci. 2004;9:1665–1679. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LN, Loh HH. Regulation of opioid receptor expression. Curr Opin Pharmacol. 2002;2(1):69–75. doi: 10.1016/s1471-4892(01)00123-0. [DOI] [PubMed] [Google Scholar]
- Wei LN, Loh HH. Transcriptional and epigenetic regulation of opioid receptor genes: present and future. Annu Rev Pharmacol Toxicol. 2011;51:75–97. doi: 10.1146/annurev-pharmtox-010510-100605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SK-F. G protein selectivity is regulated by multiple intracellular regions of GPCRs. 2003;12(1):1–12. doi: 10.1159/000068914. [DOI] [PubMed] [Google Scholar]
- Xie GX, Meuser T, Pietruck C, Sharma M, Palmer PP. Presence of opioid receptor-like (ORL1) receptor mRNA splice variants in peripheral sensory and sympathetic neuronal ganglia. Life Sci. 1999;64(22):2029–2037. doi: 10.1016/s0024-3205(99)00150-2. [DOI] [PubMed] [Google Scholar]
- Xu J, Faskowitz AJ, Rossi GC, Xu M, Lu Z, Pan YX, Pasternak GW. Stabilization of morphine tolerance with long-term dosing: association with selective upregulation of mu-opioid receptor splice variant mRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(1):279–284. doi: 10.1073/pnas.1419183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lu Z, Xu M, Pan L, Deng Y, Xie X, Liu H, Ding S, Hurd YL, Pasternak GW, Klein RJ, Cartegni L, Zhou W, Pan YX. A heroin addiction severity-associated intronic single nucleotide polymorphism modulates alternative pre-mRNA splicing of the mu opioid receptor gene OPRM1 via hnRNPH interactions. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014a;34(33):11048–11066. doi: 10.1523/JNEUROSCI.3986-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Bolan E, Gilbert AK, Pasternak GW, Pan YX. Isolating and characterizing three alternatively spliced mu opioid receptor variants: mMOR-1A, mMOR-1O, and mMOR-1P. Synapse. 2014b;68(4):144–152. doi: 10.1002/syn.21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Brown T, Rossi GC, Hurd YL, Inturrisi CE, Pasternak GW, Pan Y-X. Stabilization of the mu opioid receptor by truncated single transmembrane splice variants through a chaperone-like action. The Journal of biological chemistry. 2013;288(29):21211–21227. doi: 10.1074/jbc.M113.458687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Hurd YL, Pasternak GW, Pan YX. Isolation and characterization of new exon 11-associated N-terminal splice variants of the human mu opioid receptor gene. Journal of neurochemistry. 2009;108(4):962–972. doi: 10.1111/j.1471-4159.2008.05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Rossi GC, Pasternak GW, Pan YX. Identification and characterization of seven new exon 11-associated splice variants of the rat mu opioid receptor gene, OPRM1. Mol Pain. 2011;7:9. doi: 10.1186/1744-8069-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Carr LG. Binding of Sp1/Sp3 to the proximal promoter of the hMOR gene is enhanced by DAMGO. Gene. 2001a;274(1-2):119–128. doi: 10.1016/s0378-1119(01)00624-2. [DOI] [PubMed] [Google Scholar]
- Xu Y, Carr LG. Transcriptional regulation of the human mu opioid receptor (hMOR) gene: evidence of positive and negative cis-acting elements in the proximal promoter and presence of a distal promoter. DNA Cell Biol. 2001b;20(7):391–402. doi: 10.1089/104454901750361451. [DOI] [PubMed] [Google Scholar]
- Xue J-C, Chen C, Zhu J, Kunapuli SP, de Riel JK, Yu L, Liu-Chen L-Y. The third extracellular loop of the μ opioid receptor is important for agonist selectivity. Journal of Biological …. 1995 [PubMed] [Google Scholar]
- Yakovlev AG, Krueger KE, Faden AI. Structure and expression of a rat kappa opioid receptor gene. The Journal of biological chemistry. 1995;270(12):6421–6424. doi: 10.1074/jbc.270.12.6421. [DOI] [PubMed] [Google Scholar]
- Zaki PA, Bilsky EJ, Vanderah TW, Lai J, Evans CJ, Porreca F. Opioid receptor types and subtypes: the delta receptor as a model. Annu Rev Pharmacol Toxicol. 1996;36:379–401. doi: 10.1146/annurev.pa.36.040196.002115. [DOI] [PubMed] [Google Scholar]