Summary
The RNA polymerase II (Pol II) transcribes all mRNA genes in eukaryotes and is among the most highly regulated enzymes in the cell. The classic model of mRNA gene regulation involves recruitment of the RNA polymerase to gene promoters in response to environmental signals. Higher eukaryotes have an additional ability to generate multiple cell types. This extra level of regulation enables each cell to interpret the same genome by committing to one of the many possible transcription programs and executing it in a precise and robust manner. Whereas multiple mechanisms are implicated in cell type-specific transcriptional regulation, how one genome can give rise to distinct transcriptional programs and what mechanisms activate and maintain the appropriate program in each cell remains unclear. This review focuses on the process of promoter-proximal Pol II pausing during early transcription elongation as a key step in context-dependent interpretation of the metazoan genome. We highlight aspects of promoter-proximal Pol II pausing, including its interplay with epigenetic mechanisms, that may enable cell type-specific regulation, and emphasize some of the pertinent questions that remain unanswered and open for investigation.
Keywords: Transcription, Gene Regulation, Epigenetics, Pol II, RNA polymerase pausing
Gene transcription: a moving target
Promoter-proximal RNA polymerase II (Pol II) pausing involves a temporary halt of transcription elongation within the first ~100 nucleotides downstream of the transcription start site (TSS) (Figure 1). A key hallmark of promoter-proximal pausing is accumulation of Pol II near the promoter without the corresponding enrichment within the gene body (Kim et al. 2005; Guenther et al. 2007; Muse et al. 2007). Originally discovered on heat shock genes in Drosophila (Gilmour and Lis, 1986; Rougvie and Lis, 1988; Giardina et al. 1992; Rasmussen and Lis, 1993), pausing is now known to be widespread in metazoans (Core et al. 2012) (reviewed in (Adelman and Lis, 2012)) and is implicated in many regulatory processes including organism development, cellular responses to signals, and differentiation (Muse et al. 2007; Zeitlinger et al. 2007; Min et al. 2011; Saha et al. 2011; Chen et al. 2013a; Lagha et al. 2013; Williams et al. 2015). Its original discovery on environmentally responsive, exceptionally highly inducible heat shock genes suggested that accumulation of paused Pol II prepares these, and by extension other genes, for future activation. However, recent reports from multiple groups suggest that poising genes for activation may be but one function of pausing. For example, it is now well established that the presence of paused Pol II is not repressive (reviewed in (Nechaev and Adelman, 2008; Adelman and Lis, 2012)). In fact, Pol II pausing is generally associated with active genes (Guenther et al. 2007; Core et al. 2008), and can even be retained on genes during their activation (Danko et al. 2013; Samarakkody et al. 2015). Furthermore, work in human breast cancer cells demonstrated that the presence of paused Pol II prior to activation does not correlate with how rapidly a gene would be activated by the hormone beta-estrogen (E2) (Hah et al. 2011). On the other hand, whereas pausing is associated with active genes, its correlation with gene activity across the genome is rather poor, as shown in Drosophila and mammalian cells (Nechaev et al. 2010; Min et al., 2011) (Figure 2). These observations suggest that rather than controlling the absolute levels of transcription, pausing may “license” Pol II to proceed into synthesizing the mRNA. Borrowing an analogy from the automobile, pausing is a stop at the charging station: while it may appear to an outside observer as just an impediment that merely slows down the flow of traffic, it is in fact beneficial, and one may argue essential for the enzyme to proceed to the destination.
Figure 1. Promoter-proximal Pol II pausing as a checkpoint in gene regulation.
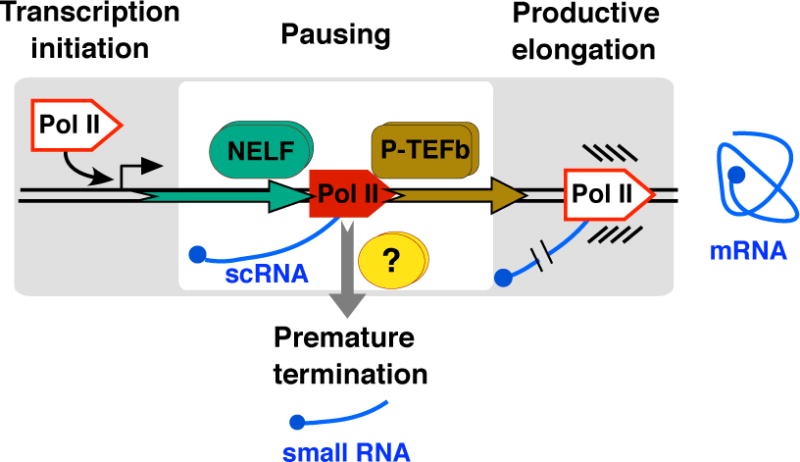
The scheme shows the initially transcribed region of a gene, with pausing occurring within the first 100 nucleotides of the gene to generate a short capped RNA (scRNA). Pol II pausing is established through interaction with NELF and can be resolved by release into productive elongation by P-TEFb or to premature transcription termination by factors that are still not fully known (shown by a question mark). Transcription factors necessary for transcription initiation as well as nucleosomes are omitted.
Figure 2. Pol II pausing does not correlate with gene expression in human cells.
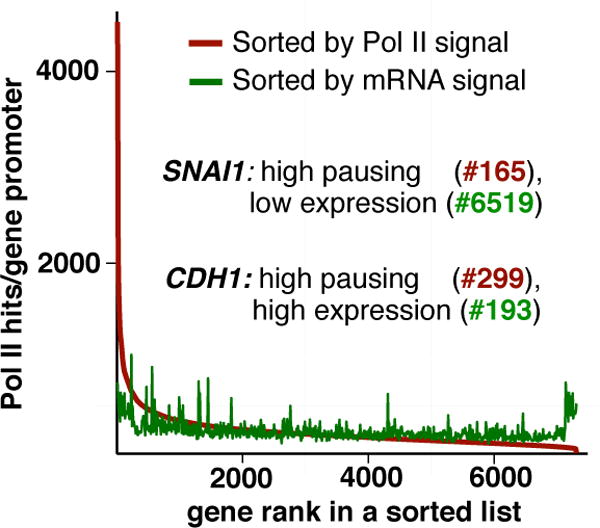
Pol II pausing was analysed in breast cancer MCF-7 cells by ChIP-sequencing after precipitation with anti-Pol II antibody (Samarakkody et al. 2015). The list of 7302 genes with Pol II enrichment (Samarakkody et al. 2015) was sorted by promoter Pol II enrichment or by gene expression signal, which was obtained from our analysis of RNA-sequencing data in MCF-7 cells combined from accession numbers SRR787327 and SRR787328 (Vanderkraats et al. 2013), SRR805877(Yamamoto et al. 2014b), SRR882016 (Jin et al. 2013), and SRR925723(Daemen et al. 2013). Numerical ranks for Pol II pausing and mRNA levels of two individual genes previously analysed by us (Samarakkody et al. 2015) are indicated.
Consistent with pausing being a regulatory checkpoint, Pol II at promoters is increasingly implicated in multiple processes including long-distance interactions within the nucleus (Li et al. 2012), direct competition with nucleosomes at the promoter regions (Gilchrist et al. 2010) and generation of short RNAs with potentially regulatory function (Affymetrix and ENCODE Transcriptome Project, 2009; Taft et al. 2009; Kanhere et al. 2010; Zamudio et al. 2014; Carissimi et al. 2015). However, while the importance of pausing in gene transcription is no longer disputed, the fundamental roles of pausing in gene regulation remain to be understood.
Regulation of early elongation: a checkpoint on every gene?
Early transcription elongation involves multiple steps that could serve as points for regulation. The entry of Pol II into the paused state (establishment of pausing) and its exit into productive elongation to synthesize mRNA (pausing release) are directly controlled by the Negative ELongation Factor (NELF) (Yamaguchi et al. 1999) and Positive Transcription Elongation Factor b (P-TEFb), respectively (Marshall and Price, 1995; Zhu et al. 1997) (Figure 1). Because of their critical role in transcription, each of these steps is the subject of active investigation. Over the years, it has become clear that both NELF and P-TEFb activities are themselves regulated by multiple factors (Lee et al. 2008), (reviewed in (Jonkers and Lis, 2015)), the repertoire of which continues to be unravelled at a rapid pace.
Setting up pausing: Not-so-Negative Elongation Factor
Pol II pausing is established by the five-subunit NELF complex (Yamaguchi et al. 1999; Narita et al. 2003). NELF likely functions as a single complex, since RNA interference based depletion of individual NELF subunits results in the corresponding reduction of levels of other NELF subunits (Gilchrist et al. 2008; Sun and Li, 2010), with a possible exception of NELF C/D (Sun and Li, 2010), and requires an additional complex, the DRB Sensitivity Inducing Factor (DSIF) (Wada et al. 1998) to function. NELF action involves interaction of its smallest subunit, NELF-E, with RNA. This invokes earlier work in bacteria wherein folding of the nascent RNA into a stem loop was shown to directly increase the probability of RNA polymerase pausing (Artsimovitch and Landick, 1998; Toulmé et al. 2005). A similar mechanism may have been adopted by Pol II (Zamft et al. 2012), except that in the case of promoter-proximal pausing the stem loop RNA structure is functionally replaced by a dedicated factor. That NELF-E can interact with a wide variety of RNA structures (Yamaguchi et al. 2002) is consistent with a role of NELF in pausing of many if not all genes (Missra and Gilmour, 2010; Pagano et al. 2014). The regulatory role of NELF-RNA interaction remains to be fully elucidated, although recent work showed that RNAs acting in trans can displace NELF from the paused complexes and trigger the release of paused Pol II into productive elongation (Schaukowitch et al. 2014).
Despite the rapid progress in understanding Pol II pausing, many important questions on NELF function remain to be answered. First, it remains uncertain whether NELF is brought in to Pol II independently or coupled to another process during transcription initiation. In addition to its requirement for the DSIF factor (Wada et al. 1998; Yamaguchi et al. 1999), NELF complex has been shown to interact with proteins such as RNA CAP-Binding Complex (CBC) (Narita et al. 2007; Ghosh et al. 2011), BRCA-1 (Ye et al. 2001), Estrogen Receptor alpha (ERa) (Aiyar et al. 2004), or the Integrator (Stadelmayer et al. 2014; Yamamoto et al. 2014a). These observations suggest that NELF recruitment to the promoter can be coupled with transcription initiation and, further, can be regulated both in a global and gene specific manner. Second, it remains unclear whether NELF is required for transcription of all genes and, specifically, whether a gene can be sustainably transcribed without Pol II undergoing pausing or involvement of NELF. Yeast (S. cerevisiae) and worms (C. elegans) appear to lack NELF, suggesting that NELF function could in principle be dispensable. Accordingly, C. elegans shows pausing at a reduced level (Chen et al. 2013b) and yeast appear to lack pausing at the promoter-proximal regions altogether (Keaveney and Struhl, 1998; Alexander et al. 2010). However, the global distribution of NELF (the NELF-B homolog, Cofactor of BRCA-1, COBRA1) in mouse embryonic stem cells closely follows that of Pol II (Rahl et al. 2010), indicating that NELF accompanies transcription initiation events consistently and suggesting that NELF is involved in transcription of most if not all genes in higher organisms. Third, while the presence of NELF marks a paused gene, it is possible that it is involved only in a portion of individual transcription initiation events. We suggest that availability of NELF, or a functionally similar factor, may explain recently described events such as transcriptional bursts (Singh et al. 2010; Bothma et al. 2014) or transcriptional ‘memory’ (Cesbron et al. 2015). Consistent with its involvement in regulation, NELF levels are altered in cancer cells (Sun et al. 2008). Further studies of NELF dynamics will help reveal the mechanisms of Pol II pausing in different systems and environmental conditions.
If establishing pausing is the only role of NELF, then reducing NELF levels in the cells is expected to increase Pol II output, because pausing would no longer impede the passage of polymerase. However, when NELF levels are depleted using RNA interference approaches, most genes in mouse and Drosophila cells in fact exhibit a reduction in transcription (Amleh et al. 2009; Gilchrist et al. 2010), suggesting that NELF – and pausing – stimulate transcription. Accordingly, NELF knockouts are embryonic lethal and its conditional knockouts lead to spontaneous death of mice (Amleh et al. 2009). However, recent work demonstrated that mouse embryonic fibroblasts (Sun et al. 2011), cardiomyocytes (Pan et al. 2014), blastocysts (Amleh et al. 2009) or embryonic stem cells (Williams et al. 2015) can survive at least for several days in culture without a functional NELF-B gene, indicating that transcription can take place without NELF, or at least with greatly diminished amounts of residual NELF (Sun et al. 2011; Williams et al. 2015). Importantly, knockout of NELF-B resulted in failure of mouse embryonic stem cells to differentiate in vitro, suggesting that the NELF complex – and by extension Pol II pausing – is required for proper cell differentiation (Williams et al. 2015). One possibility that may explain its requirement for transcription is that NELF is a general transcription factor that must be involved in at least a minimal proportion of transcriptional events on most or all genes. The requirements for NELF in gene expression in different cell types remain to be fully understood.
Release of pausing: P-TEFb
The positive transcription elongation complex P-TEFb, which consists of the cyclin T1 and Cdk9 kinase subunits, phosphorylates several proteins including NELF, Pol II, and Spt5 component of the DSIF complex in a process that accompanies the release of paused polymerase into productive elongation (reviewed in (Peterlin and Price, 2006)), and is found on promoter regions of many genes (Schwartz et al. 2012). On Pol II, P-TEFb is suggested to phosphorylate Serine 2 of the largest polymerase subunit C-terminal Domain (CTD) repeat with YSPTSPS consensus sequence (reviewed in (Buratowski, 2009; Heidemann et al. 2013)), although in vitro work points to Serine 5 as a target of P-TEFb (Czudnochowski et al. 2012). Treatment of cells with inhibitors of Cdk9, 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole (DRB)(Chodosh et al. 1989) or flavopiridol (Chao et al. 2000) leads to the overall suppression of transcription, indicating that P-TEFb continuously functions on most if not all Pol II transcribed genes (Ni et al. 2008; Jonkers et al. 2014). Indeed, the function of P-TEFb is conserved in eukaryotes as yeast contains kinases Bur1 and Ctk1 that phosphorylate the CTD at Serine 2 (Keogh et al. 2003; Ahn et al. 2009). However, in an apparent contradiction to the global role of P-TEFb in transcription, some genes were shown to be upregulated in the presence of P-TEFb inhibitors (Gomes et al. 2006; Keskin et al. 2012; Ni et al. 2004), indicating that the requirement for P-TEFb for transcription can in principle be bypassed. On the other hand, recent work showed that this residual transcription results from redistribution of the available P-TEFb to a small number of genes and that the residual transcription in the presence of a novel specific P-TEFb inhibitor still involves CTD phosphorylation by the remaining P-TEFb (Lu et al. 2015). Whether and when the P-TEFb function can be compensated by additional kinases (Bartkowiak et al. 2010) remains to be determined.
Consistent with its importance in Pol II transcription, the activity of P-TEFb in the cell is tightly regulated. Elevated levels of P-TEFb are associated with cancer transformation (Moiola et al. 2010), and P-TEFb components, particularly CDK9, are targets for anti-cancer therapies (reviewed in (Romano, 2013)). Much of P-TEFb in the cell is found in a nucleoprotein complex that contains several dedicated protein components and a non-coding 7SK RNA (Nguyen et al. 2012), which sequesters P-TEFb away from Pol II (reviewed in (Peterlin and Price, 2006)). Activation of P-TEFb for pause release involves its dislodging from the 7SK complex and recruitment to promoters. The mechanisms for the recruitment of P-TEFb to promoters may include interaction with chromatin remodelling factors such as BRD4 (Jang et al. 2005) (reviewed in (Chen et al. 2014; Peterlin and Price, 2006)), Mediator (Donner et al. 2010; Ebmeier and Taatjes, 2010), association with transcription factors that bind DNA such as NF-KB and c-Myc, an RNA-based interaction such as the case for the Human Immunodeficiency Virus (HIV) promoter (reviewed in (Peterlin and Price, 2006)), or large protein complexes such as Super Elongation Complexes (Lin et al. 2011; Smith et al. 2011) or Integrator (Gardini et al. 2014). These findings arising from a rapidly growing field suggest that P-TEFb can act both globally and in a gene-specific manner.
Whereas the kinase activity of P-TEFb is associated with the release of paused Pol II into productive elongation, how P-TEFb phosphorylation triggers the release remains to be fully understood. The existence of several possible targets that can be phosphorylated by P-TEFb at the promoter region, including NELF, DSIF, and Pol II CTD, suggests that phosphorylation requirements of these factors could vary for different genes or at different conditions. Interestingly, genes have been shown to have different elongation velocities in human cells (Danko et al. 2013), offering an intriguing possibility that the speed of transcription elongation in the body of a gene may be pre-determined early on, including during pause release. It is also worth noting that c-Myc and NF-kB are transcription factors that have been classically considered to act on transcription initiation through Pol II recruitment to promoters (reviewed in (Dang et al. 1999; Levens, 2003)). That they also interact with P-TEFb to encourage pause release suggests that P-TEFb may be brought in to the promoter together with Pol II in a process that tightly integrates initiation and early elongation.
Pausing duration: hitting the balance?
Promoter-proximal Pol II pausing takes place within the same distance on all genes, peaking at approximately 35 nucleotides from the transcription start site. This notion is based on global sequencing of short capped RNAs generated by paused polymerase as well as on high-resolution Global Run-on sequencing (Gro-Seq) analyses in Drosophila and mammalian systems (Nechaev et al. 2010; Core et al. 2014; Samarakkody et al. 2015). These observations suggest that the basic mechanisms of promoter-proximal pausing are similar for all genes in the cell and are conserved across metazoans. Given this conservation, the molecular mechanisms that establish pausing and control its duration must also be conserved across organisms and highly robust across different cell types and environmental conditions. In particular, pausing must occur in different cell types, on genes displaying varying levels of promoter activity, and the process must be resistant to changes in the environment (Brown et al. 1996; Samarakkody et al. 2015). Measurements of the stability of paused complexes in mouse embryonic stem cells across the genome using the specific inhibitor of TFIIH, triptolide, (Vispé et al. 2009) demonstrated that most genes show a rather tight distribution of Pol II pause duration times, centered at about 7 minutes (Jonkers et al. 2014). Interestingly, recent work in human (HeLa) cells suggested that some genes, including FOS, can be outliers and undergo an order of magnitude longer pausing, with complexes remaining essentially stable even after an hour of exposure to triptolide (Chen et al. 2015). While these findings offer an exciting possibility that pausing duration can be regulated in a gene-specific manner, the extent and mechanisms of such regulation remain to be characterized.
The point of pausing: is it worth not stopping by?
Recent findings indicate that promoter-proximal pausing represents the principal form of Pol II on promoters of metazoan genes, and that other Pol II complexes at the promoter, including closed and open preinitiation complexes, are much more transient and represent at best only a small proportion of Pol II. There are at least two lines of evidence in support of this notion. First, the magnitude of Gro-seq signal, which represents Pol II actively engaged in RNA synthesis, corresponds well with the abundance of Pol II detected by ChIP at the promoter regions, suggesting that Pol II present at promoter regions is indeed engaged in elongation (Core et al. 2012). Second, genome-wide permanganate reactivity profiling shows the absence of permanganate reactivity of open complexes at transcription start sites in Drosophila S2 cells (Li et al. 2013). Notably, open promoter complexes are not detectable even during robust activation of the Drosophila (Giardina et al. 1992) and human Hsp70 gene (Samarakkody et al. 2015), suggesting that pausing remains the slowest transcriptional intermediate even at the conditions of high-level gene activation (Boehm et al. 2003). It is worth noting that because both Gro-Seq and permanganate footprinting approaches detect only functional Pol II, but not closed promoter complexes, it remains possible that some proportion of Pol II exists in a closed complex without opening the transcription bubble or synthesizing RNA. While the presence of such a complex cannot be completely ruled out, given the high degree of correspondence between different assays that measure Pol II abundance (Core et al. 2012), the putative stable closed complex must coexist with the paused Pol II on the same gene copy, which was previously deemed unlikely (Li et al. 2013). We note that this appears to be in contrast to lower organisms including yeast, where stable open complexes, but not paused complexes, are detectable in vivo (Giardina and Lis, 1993; Giardina and Lis, 1995; Guzmán and Lis, 1999; Rhee and Pugh, 2012), further indicating that Pol II pausing is a regulatory, not obligatory, step of transcription.
Pol II pausing presents multiple opportunities for regulatory inputs (Figure 1). At an average half-life of 5 to 10 minutes (Jonkers et al. 2014), individual pausing events are sufficiently long to enable several proteins to interact with the same paused Pol II molecule even if these interactions are separated in time. Pausing can thus enable integration of “hit and run” regulatory inputs such as distinct transcription factors (Henriques et al. 2013) that would release paused Pol II only if interactions take place in a certain sequence that forms the “molecular password”. Indeed, observation of transcription factor dynamics offers support for this provocative “hit and run” model of gene regulation (Sung et al. 2014; Stavreva et al. 2015). Another mechanism may involve interaction of paused Pol II with other loci in trans. The involvement of a structural factor cohesin (Fay et al. 2011; Lin et al. 2013; Schaaf et al. 2013) in the interaction with paused Pol II suggests that Pol II complexes can directly connect distant gene loci in a dynamic, transcription-dependent manner (Kagey et al. 2010). Furthermore, the recently described superenhancers have been proposed to provide an integrated platform that enables concerted, cell type specific regulation of genes across the genome (Hnisz et al. 2013; Lovén et al. 2013; Whyte et al. 2013). A third mechanism involves direct competition of Pol II with chromatin, as paused Pol II has been shown to compete with nucleosomes for binding to the initially transcribed regions of genes in Drosophila (Gilchrist et al. 2008; Gilchrist et al. 2010). Lastly, it is possible that the paused Pol II itself can serve as a transcription factor to encourage recruitment of additional Pol II molecules such as that during signal response. This model was proposed previously (Rasmussen and Lis, 1995). Indeed, imaging of Drosophila polytene chromosomes using fluorescence recovery after photobleaching (Yao et al. 2006) showed that Pol II on heat shock Hsp70 gene became locally recycled for subsequent rounds of transcription at the conditions of activation by heat shock. These results suggest that transcription during signal responses may be governed by mechanisms that are different from steady state transcription before activation (Yao et al. 2006). In this regard, finding that pausing can be retained on genes during activation (Brown et al. 1996; Lis, 1998; Samarakkody et al. 2015) suggests that paused Pol II can continue to function during gene activation, to coordinate interaction with distinct sets of transcriptional factors in basal versus activated conditions, or moderate the extent of gene activation (Figure 3).
Figure 3. A proposed role of Pol II pausing in regulating transcriptional responses.
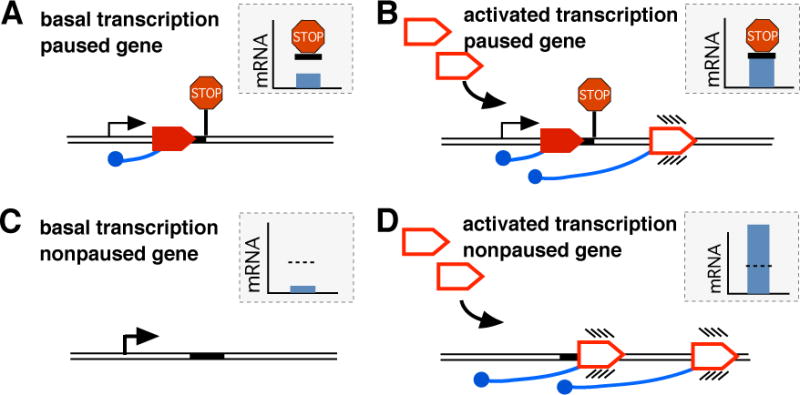
A scheme with responses of a paused (A, B) and non-paused gene (C, D) to the same activator. The colors are as in Figure 1. Insets show levels of the mRNA, in each condition. A gene with paused Pol II (A) can be activated while retaining Pol II pausing (B) (Samarakkody et al. 2015). The retention of pausing limits the extent of gene activation (horizontal bar, inset). On a nonpaused gene (C), the same signal can lead to higher-level activation in the absence of the limit imposed by pausing (D). Release of Pol II pausing during activation, changes in pausing status of a gene during differentiation or following exposure to environmental stress can alter the magnitude of its transcriptional response to the same activator.
Despite the widespread occurrence of Pol II pausing, the question of whether genes can be transcribed without undergoing pausing remains open. First, available data cannot distinguish whether “non-paused” genes (for example, (Chen et al. 2015)) in fact always bypass pausing. At least some, but not all, non-paused genes may have been designated as non-paused because of the low intrinsic activity of their promoters, and in fact become paused as the gene is activated, such as the SNAI2 gene in human MCF-7 cells (Samarakkody et al. 2015). Analysis of Pol II pausing by global sequencing of short RNAs indicated that non-paused genes show the same paused signature, albeit at lower levels (Nechaev et al. 2010), indicating that non-paused genes undergo low-level pausing in the same location in respect to the transcription start site. However, it is still unclear whether the “low-level” pausing involves lower duration of pausing for individual transcription events or, alternatively, a lower probability of pausing for each transcription event, but with the same duration. Second, and conversely, available data cannot exclude a possibility that even on highly paused genes, some individual transcription events bypass pausing. Analysis of several individual genes in Drosophila S2 cells (Henriques et al. 2013) estimated the fractional occupancy of Pol II at over 50%, suggesting that most if not all copies of these genes can be occupied by Pol II at a given time. However, the occupancy of Pol II on the highly paused Snail transcription factor (SNAI1) gene in human breast cancer MCF-7 cells was estimated at less than 40%, suggesting that most genes in MCF-7 cells, including highly paused genes, are not fully occupied by Pol II. Some of this lower occupancy may be explained by transcriptional events that bypass the pausing step.
One important question relating to the dynamics of pausing is whether the same gene can change its pausing status, such as during cell differentiation or as a result of exposure to stimuli, and what factors can trigger such a change. We propose that a biological role of Pol II pausing is to moderate the extent of a transcriptional response by limiting the turnover of molecules during activation (Figure 3). Taken genome-wide, the presence of paused Pol II enables the cell to maintain expression levels of key regulatory genes and thus maintain the transcription network (Gilchrist et al. 2012; Henriques et al. 2013). We propose further that normal cell differentiation is accompanied by concerted changes in Pol II pausing status across the genome. Consequently, a spurious change in pausing status resulting from an exposure to an environmental challenge can alter the ability of cells to respond to a signal and contribute to unwanted changes including cancer. It has been proposed that genes can change their “pausing class“ between different cell types (Min et al. 2011), but the scope of such changes during cell differentiation as well as mechanisms underlying such changes remain to be fully determined.
A word with many meanings: Roles of the DNA sequence in dynamic regulation of Pol II pausing
Analysis in Drosophila S2 cells showed that paused genes contain distinct DNA sequence signatures within their initially transcribed regions (Kutach and Kadonaga, 2000; Hendrix et al. 2008; Nechaev et al. 2010). The probability of Pol II to pause on a given gene depends on the nucleotide sequence of its initially transcribed region and is directly dependent on the stability of the DNA-RNA hybrid in the enzyme’s active center (Kireeva et al. 2000). Importantly, this mechanism transmits the sequence context information within the initially transcribed regions of genes directly to Pol II. As a caveat, however, we note that because C+G context is different between Drosophila and mammals (which have CpG islands around promoters of many genes) (Core et al. 2008; Rozenberg et al. 2008), regulation of pausing in mammalian systems might be different from that in Drosophila.
Two models could in principle contribute to interpreting the sequence context of promoter-proximal regions by the transcriptional machinery. The first is the kinetic model, wherein the probability of pausing on a gene depends on the velocity of the elongating Pol II. As a result, NELF, which is known to slow down the transcribing Pol II (Cheng and Price, 2007; Li et al. 2013), can induce pausing in a gene-specific manner depending on the initially transcribed DNA sequence, through mechanistically the same interaction. Another model is the “molecular ruler” model, wherein the location at which Pol II pauses is determined by interaction with a protein independent of the DNA sequence, and probability of pausing at given conditions is determined by the availability of this factor. The possibility for kinetic control of pausing has been demonstrated through mutations in Pol II and depletion of NELF factor (Li et al. 2013) in Drosophila. However, Pol II pausing location at the heat shock gene did not change with activation in human cancer cells (Brown et al. 1996; Samarakkody et al. 2015), indicating that regulation of pausing can vary between species or genes. The importance of the DNA sequence in setting up pausing was directly demonstrated in Drosophila Hsp70 transgene, as the precise location of pausing in the transgenic flies followed the sequence if it was moved 5 nucleotides downstream of its original location (Kwak et al. 2013). However, extension of the pausing sequence by an additional 5 nucleotides for the total of 10 restored Pol II pausing to its original distance from the TSS. This result suggests that Pol II pausing is controlled through multiple mechanisms that sense the local sequence context by monitoring the efficiency of Pol II elongation, but additionally, limit the “degrees of freedom” through sequence-independent mechanisms. The responsiveness to signals that are hard-wired in the genomic sequence and dynamic inputs from transcription and epigenetic factors places Pol II pausing as a key step in context-dependent interpretation of the genome.
Parting the chromatin: Friend or foe?
Gene transcription takes place in the context of chromatin (reviewed most recently in (Venkatesh and Workman, 2015)), and Pol II can interact with nucleosomes through multiple mechanisms. First, gene promoters are known to serve as center points, relative to which the nucleosomes are positioned (Mavrich et al. 2008a; Afek et al. 2011). However, nucleosome-free regions (NFRs) are also well defined in yeast, which appear to lack ubiquitous promoter-proximal pausing (Rhee and Pugh, 2012), indicating that it is not pausing, but the promoter itself that may be responsible for establishing the NFR. On the other hand, nucleosomes have been proposed to enhance promoter-proximal pausing (Fuda et al. 2015; Jimeno-González et al. 2015) as also evidenced by shifting of nucleosome locations by paused Pol II (Mavrich et al. 2008b), indicating their direct physical interaction. In support of this notion, recent work demonstrated that the chromatin remodeler Chd1 is required for the positioning of promoter-proximal nucleosomes and for the escape of paused Pol II into productive elongation (Skene et al. 2014). Second, paused Pol II can compete with nucleosomes for binding in the promoter-proximal regions, as shown in Drosophila. In particular, the very same sequences that encourage Pol II pausing at the initially transcribed regions of genes were also shown to favour positioning of nucleosomes (Gilchrist et al. 2010), as depletion of NELF using RNA interference in Drosophila S2 cells resulted in the replacement of Pol II at previously paused genes with nucleosomes, leading to repression of the genes. Crucially, non-paused genes in the same cells did not show the increase in nucleosome occupancy upon NELF depletion (Gilchrist et al. 2010), indicating that the initially transcribed regions of genes specify the dynamic competition between Pol II and chromatin, but only on a subset of genes. Taken together, these observations suggest that the static genome can encode distinct regulatory states of a gene (Adelman and Lis, 2012) and, taken globally, specify the gamut of alternative states of the transcriptome through initially transcribed sequences of mRNA genes.
While nucleosomes have been shown to compete with paused Pol II, it remains unclear whether this competition takes place as dynamic replacement (on a minute time scale) on the same DNA molecule or, alternatively, reflects stable differences between paused and inactive, chromatin-occupied, gene states and could potentially account for natural heterogeneity in cell populations, including stem cells (Graf and Stadtfeld, 2008; Marks et al. 2012). In this regard, promoter-proximal nucleosomes have been shown to be enriched in H2AZ (and H3.3) variant histones (reviewed in (Jin and Felsenfeld, 2007)), which intrinsically bind less stably to DNA, perhaps facilitating their dynamic exchange with Pol II during transcription.
In the wake: covalent modifications in early transcription elongation
In addition to physical rearrangement of nucleosomes, the process of transcription involves changes in covalent histone modifications (Venkatesh and Workman, 2015). A number of histone modifications have been shown to selectively mark active or inactive genes, leading to the hypothesis of the “histone code” as a mechanism that enables interpretation of the genome in a cell type-specific manner. As H3K4Me3, or H3K9/K14 acetylation is associated with promoter-proximal regions, it is possible that Pol II pausing plays an important role in establishing these marks. Indeed, H3K4me3 and H3K9/K14ac marks have been associated with promoter enrichment of Pol II (Guenther et al. 2007; Rahl et al. 2010), not necessarily with transcriptional activity (Vastenhouw et al. 2010). This is in contrast to active elongation-specific marks in downstream regions of genes such as H3K36me3 (Rahl et al. 2010). However, just as the case with nucleosome positioning, whether the modification is a cause or consequence of transcriptional activity, or pausing, remains to be determined.
Histone H3K27 trimethylation is associated with heterochromatin and is generally considered a repressive gene mark (Mikkelsen et al. 2007). In stem cells, however, the so-called “bivalent” genes carry both H3K27 and H3K4 trimethylation marks (Azuara et al. 2006; Bernstein et al. 2006; Mikkelsen et al. 2007), (reviewed in (Voigt et al. 2013)), leading to a hypothesis that the bivalency at the histone level established in stem cells is resolved into active or repressive marks upon differentiation (Rodriguez et al. 2008). Importantly, it is the same nucleosome that can carry both marks (Bernstein et al. 2006), suggesting that distinct mechanisms are involved in establishing the bivalent chromatin state. Mass-spectrometry analysis (Voigt et al. 2012) demonstrated that nucleosomes can contain both symmetric and asymmetric modifications (different marks in each copy of H3 histone within the same nucleosome). Because H3K27 and H3K4 marks were not found on the same peptide within one nucleosome, it is likely that the histone modification ‘writers’ may be sensitive to pre-existing modifications to introduce combinatory marks, representing another level of the histone code.
Additional modifications such as H3S10 phosphorylation have been shown to occur after Pol II initiation, but before release from the paused state by P-TEFb (Ivaldi et al. 2007), suggesting that the full gamut of histone code associated with pausing is yet to be defined. Modifications such as H3K27 acetylation have been shown to be associated with super-enhancers (Achour et al. 2015), although the direct involvement of pausing with enhancers has not yet been demonstrated. It is tempting to speculate that paused Pol II interacts with super-enhancers to enable cell type specific gene rearrangement within the nucleus. As the mechanisms of generating cell-type specific enhancers remain uncertain (Pott and Lieb, 2015), exploring transcription regulation by enhancers will be the subject of future work.
Small RNAs – reading promoters between the lines?
Paused Pol II has been recently shown to undergo “premature” termination at the initially transcribed regions (Zamudio et al. 2014). This termination, which occurs in promoter-proximal regions to generate a free short RNA (Figure 1), is distinct from the well-known process of termination that occurs at the 3′-ends of genes to produce the mRNA, but may involve at least some of the same factors. Premature termination has been shown to take place in promoter-proximal regions of genes in yeast (Terzi et al. 2011), providing a rationale for the existence of similar mechanisms in higher organisms. Importantly, the Nrd1-Nab3-Sen1 termination pathway involved in promoter-proximal termination in yeast involves proteins that interact with nascent RNAs, indicating that transcription termination in metazoans could take place through similar mechanisms, but would also involve (or require) a stably paused complex. Indeed, the termination factor TTF2, which acts through the RNA-mediated termination pathway involving the Xrn2 exonuclease, was found at the promoter regions of many genes in mammalian cells (Brannan et al. 2012; Wagschal et al. 2012). Direct evidence for functional promoter-proximal termination in metazoans comes from the detection of short capped RNAs dissociated from paused Pol II complexes on Hsp70 gene (Buckley et al. 2014) and detection of 3′-oligoadenylated short capped RNAs through post-transcriptional processing (Preker et al. 2011; Valen et al. 2011; Henriques et al. 2013). We note that while NELF has been traditionally considered a pausing factor, studies suggest that it can also serve as a termination factor in downstream regions of genes (Egloff et al. 2009). More recently, NELF has been shown to be involved in promoter-proximal termination of the HIV RNA through recruiting the PCF11 termination factor (Natarajan et al. 2013). Taken together, these results raise an intriguing possibility that in addition to pausing, NELF may be also involved in promoter-proximal termination.
Discovery of promoter-proximal termination hints at an exciting possibility that the short noncoding RNAs generated by paused Pol II can function in the cell. Sequencing of small RNAs physically associated with miRNA processing machinery (specifically, the Argonaute 2 protein) in the mouse identified miRNAs generated from 5′-regions of many genes (Carissimi et al. 2015), lending experimental support to the idea that these RNAs that can function in trans. In addition to its processing to miRNAs, which could function translationally or at the transcription level, the unprocessed short RNAs generated by paused Pol II can in principle function as well. Indeed, short RNAs have been demonstrated to affect transcription of genes in a sequence-specific manner (Janowski et al. 2005). These exciting findings may prompt the field to revisit the role of a metazoan gene promoter as a mechanism for delivery of Pol II to the pausing site. The actual product of this transcription event, short RNA or mature, “conventional” transcript, would then be determined at the level of pause release.
Conclusions and perspectives
The perception of promoter-proximal pausing has transitioned from a peculiar phenomenon to a commonly recognized widespread step in Pol II gene transcription. Whereas great progress in understanding the role of pausing in gene regulation has been made, much more remains to be learned. We suggest that two main experimental strategies will be fruitful at least in a short perspective. The first strategy is reconstitution of Pol II pausing with purified components in vitro. Whereas a number of studies have provided key insights into the mechanisms of RNA polymerase II transcription and enzymatic mechanisms (Brown et al. 1996; Brown et al. 1998; Yamaguchi et al. 2002; Zhu et al. 2007; Chen et al. 2009; Cheng et al. 2012; Li et al. 2013), to name only a few, modelling multiple-round steady-state transcription that involves Pol II pausing in vitro remains a challenge and is expected to further improve. Indeed, recent work led to the discovery and characterization of several such components, examples of which include TRIM28 (Bunch et al. 2014) and a factor involved in premature termination, Gdown1 (Hu et al. 2006; Cheng et al. 2012; Jishage et al. 2012; Guo et al. 2014). We expect that the use of improved in vitro systems and involvement of single-molecule approaches (reviewed in (Herbert et al. 2008)) will offer further insight into the mechanisms of early transcription elongation.
Secondly, improved sequencing technologies will continue to provide more data per experiment and its use will remain widespread. However, a major limitation of sequencing approaches has been not the depth of sequencing, but rather the necessity to rely on modification enzymes whose substrate requirements limit the minimum amount of starting material required for successful preparation of a library to be sequenced. Multiple strategies are being developed to deal with this limitation. Advances in the design of libraries using current enzymes to enable reliable preparation of samples from lower amounts of starting material become available (Chu et al. 2015) and are expected to improve the sensitivity of sequencing further. The improved “spike-in” control strategies (Grzybowski et al. 2015; Hu et al. 2015) could further enhance global ChIP-sequencing based approaches (Rhee and Pugh, 2012; He et al. 2015; Skene and Henikoff, 2015). On the other hand, a limitation that cannot be overcome by increasing the depth of sequencing is the imperfect specificity of antibodies whether due to relaxed specificity of the antibody itself (Nishikori et al. 2012) or heterogeneity of the target epitope (such as the Pol II CTD). The use of tools with completely novel or vastly improved substrate specificity, including enzymes based on nucleic acids, will enable global approaches to rival and exceed the resolution and quantitative insights of individual gene experiments, eventually rendering the latter obsolete. Combining novel core technologies with their inventive interpretation beyond instruction manuals will revolutionize our ability to see into the cell’s transcriptome and will enable reprogramming of the cell fate through precise targeting of the epigenetic environment, including Pol II pausing, both globally and on individual genes.
Acknowledgments
We thank Archana Dhasarathy for critical reading of the manuscript. This work was supported by the National Institutes of Health [5P20GM104360].
References
- Achour M, Le Gras S, Keime C, Parmentier F, Lejeune FX, Boutillier AL, Néri C, Davidson I, Merienne K. Neuronal identity genes regulated by super-enhancers are preferentially down-regulated in the striatum of Huntington’s disease mice. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv099. [DOI] [PubMed] [Google Scholar]
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13(10):720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afek A, Sela I, Musa-Lempel N, Lukatsky DB. Nonspecific transcription-factor-DNA binding influences nucleosome occupancy in yeast. Biophys J. 2011;101(10):2465–2475. doi: 10.1016/j.bpj.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix, ENCODE Transcriptome Project and Cold Spring Harbor Laboratory, ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457(7232):1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SH, Keogh MC, Buratowski S. Ctk1 promotes dissociation of basal transcription factors from elongating RNA polymerase II. EMBO J. 2009;28(3):205–212. doi: 10.1038/emboj.2008.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar SE, Sun JL, Blair AL, Moskaluk CA, Lu YZ, Ye QN, Yamaguchi Y, Mukherjee A, Ren DM, Handa H, Li R. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18(17):2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40(4):582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amleh A, Nair SJ, Sun J, Sutherland A, Hasty P, Li R. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PLoS One. 2009;4(4):e5034. doi: 10.1371/journal.pone.0005034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release. Genes Dev. 1998;12(19):3110–3122. doi: 10.1101/gad.12.19.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24(20):2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- B1oehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23(21):7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc Natl Acad Sci U S A. 2014;111(29):10598–10603. doi: 10.1073/pnas.1410022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, Bentley DL. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46(3):311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10(12):1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Brown SA, Weirich CS, Newton EM, Kingston RE. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17(11):3146–3154. doi: 10.1093/emboj/17.11.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley MS, Kwak H, Zipfel WR, Lis JT. Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev. 2014;28(1):14–19. doi: 10.1101/gad.231886.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G, Taatjes DJ, Calderwood SK. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol. 2014;21(10):876–883. doi: 10.1038/nsmb.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carissimi C, Laudadio I, Cipolletta E, Gioiosa S, Mihailovich M, Bonaldi T, Macino G, Fulci V. ARGONAUTE2 cooperates with SWI/SNF complex to determine nucleosome occupancy at human Transcription Start Sites. Nucleic Acids Res. 2015;43(3):1498–1512. doi: 10.1093/nar/gku1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesbron F, Oehler M, Ha N, Sancar G, Brunner M. Transcriptional refractoriness is dependent on core promoter architecture. Nat Commun. 2015;6:6753. doi: 10.1038/ncomms7753. [DOI] [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275(37):28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 2015;29(1):39–47. doi: 10.1101/gad.246173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife. 2013a;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Yik JH, Lew QJ, Chao SH. Brd4 and HEXIM1: multiple roles in P-TEFb regulation and cancer. Biomed Res Int. 2014;2014:232870. doi: 10.1155/2014/232870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RA, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res. 2013b;23(8):1339–1347. doi: 10.1101/gr.153668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23(23):2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282(30):21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, Young RA, Price DH. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell. 2012;45(1):38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Fire A, Samuels M, Sharp PA. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem. 1989;264(4):2250–2257. [PubMed] [Google Scholar]
- Chu Y, Wang T, Dodd D, Xie Y, Janowski BA, Corey DR. Intramolecular circularization increases efficiency of RNA sequencing and enables CLIP-Seq of nuclear RNA from human cells. Nucleic Acids Res. 2015;43(11):e75. doi: 10.1093/nar/gkv213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46(12):1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the Status of RNA Polymerase at Promoters. Cell Rep. 2012;2(4):1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudnochowski N, Bösken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun. 2012;3(842) doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- Daemen A, Griffith OL, Heiser LM, Wang NJ, Enache OM, Sanborn Z, Pepin F, Durinck S, Korkola JE, Griffith M, Hur JS, Huh N, Chung J, Cope L, Fackler MJ, Umbricht C, Sukumar S, Seth P, Sukhatme VP, Jakkula LR, Lu Y, Mills GB, Cho RJ, Collisson EA, van’t Veer LJ, Spellman PT, Gray JW. Modeling precision treatment of breast cancer. Genome Biol. 2013;14(10):R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253(1):63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell. 2013;50(2):212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17(2):194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci U S A. 2010;107(25):11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Al-Rawaf H, O’Reilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol Cell Biol. 2009;29(14):4002–4013. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol. 2011;21(19):1624–1634. doi: 10.1016/j.cub.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Guertin MJ, Sharma S, Danko CG, Martins AL, Siepel A, Lis JT. GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters. PLoS Genet. 2015;11(3):e1005108. doi: 10.1371/journal.pgen.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardini A, Baillat D, Cesaroni M, Hu D, Marinis JM, Wagner EJ, Lazar MA, Shilatifard A, Shiekhattar R. Integrator regulates transcriptional initiation and pause release following activation. Mol Cell. 2014;56(1):128–139. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK, Missra A, Gilmour DS. Negative elongation factor accelerates the rate at which heat shock genes are shut off by facilitating dissociation of heat shock factor. Mol Cell Biol. 2011;31(20):4232–4243. doi: 10.1128/MCB.05930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Lis JT. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261(5122):759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- Giardina C, Lis JT. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15(5):2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Pérez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6(11):2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143(4):540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26(9):933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22(14):1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6(11):3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20(5):601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3(5):480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Grzybowski AT, Chen Z, Ruthenburg AJ. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell. 2015;58(5):886–899. doi: 10.1016/j.molcel.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Turek ME, Price DH. Regulation of RNA polymerase II termination by phosphorylation of Gdown1. J Biol Chem. 2014;289(18):12657–12665. doi: 10.1074/jbc.M113.537662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán E, Lis JT. Transcription factor TFIIH is required for promoter melting in vivo. Mol Cell Biol. 1999;19(8):5652–5658. doi: 10.1128/mcb.19.8.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol. 2015;33(4):395–401. doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829(1):55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A. 2008;105(22):7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52(4):517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Greenleaf WJ, Block SM. Single-molecule studies of RNA polymerase: motoring along. Annu Rev Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Petela N, Kurze A, Chan KL, Chapard C, Nasmyth K. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt A. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103(25):9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaldi MS, Karam CS, Corces VG. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 2007;21(21):2818–2831. doi: 10.1101/gad.1604007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol. 2005;1(4):216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- Jimeno-González S, Ceballos-Chávez M, Reyes JC. A positioned +1 nucleosome enhances promoter-proximal pausing. Nucleic Acids Res. 2015;43(6):3068–3078. doi: 10.1093/nar/gkv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21(12):1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, Chait B, Gnatt A, Ren B, Roeder R. Transcriptional Regulation by Pol II(G) Involving Mediator and Competitive Interactions of Gdown1 and TFIIF with Pol II. Molecular cell Molecular cell Mol Cell. 2012;45(1):51–63. doi: 10.1016/j.molcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16(3):167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, Pombo A, Fisher AG, Young RA, Jenner RG. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38(5):675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1(6):917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol. 2003;23(19):7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H, Garriga J, Georlette D, Graña X. Complex effects of flavopiridol on the expression of primary response genes. Cell Div. 2012;7:11. doi: 10.1186/1747-1028-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436(7052):876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275(9):6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20(13):4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339(6122):950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, Levine MS. Paused Pol II Coordinates Tissue Morphogenesis in the Drosophila Embryo. Cell. 2013;153(5):976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28(10):3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levens DL. Reconstructing MYC. Genes Dev. 2003;17(9):1071–1077. doi: 10.1101/gad.1095203. [DOI] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148(1–2):84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic Competition between Elongation Rate and Binding of NELF Controls Promoter-Proximal Pausing. Mol Cell. 2013;50(5):711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25(14):1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell. 2013;152(1–2):144–156. doi: 10.1016/j.cell.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Xue Y, Yu GK, Arias C, Lin J, Fong S, Faure M, Weisburd B, Ji X, Mercier A, Sutton J, Luo K, Gao Z, Zhou Q. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. Elife. 2015;4 doi: 10.7554/eLife.06535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149(3):590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270(21):12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008b;453(7193):358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008a;18(7):1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25(7):742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107(25):11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiola C, De Luca P, Gardner K, Vazquez E, De Siervi A. Cyclin T1 overexpression induces malignant transformation and tumor growth. Cell Cycle. 2010;9(15):3119–3126. doi: 10.4161/cc.9.15.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39(12):1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26(3):349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, Endoh M, Yamada T, Handa H. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23(6):1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan M, Schiralli Lester GM, Lee C, Missra A, Wasserman GA, Steffen M, Gilmour DS, Henderson AJ. Negative elongation factor (NELF) coordinates RNA polymerase II pausing, premature termination, and chromatin remodeling to regulate HIV transcription. J Biol Chem. 2013;288(36):25995–26003. doi: 10.1074/jbc.M113.496489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7(11):1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327(5963):335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Krueger BJ, Sedore SC, Brogie JE, Rogers JT, Rajendra TK, Saunders A, Matera AG, Lis JT, Uguen P, Price DH. The Drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res. 2012;40(12):5283–5297. doi: 10.1093/nar/gks191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28(3):1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13(1):55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- Nishikori S, Hattori T, Fuchs SM, Yasui N, Wojcik J, Koide A, Strahl BD, Koide S. Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J Mol Biol. 2012;424(5):391–399. doi: 10.1016/j.jmb.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Kwak H, Waters CT, Sprouse RO, White BS, Ozer A, Szeto K, Shalloway D, Craighead HG, Lis JT. Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet. 2014;10(1):e1004090. doi: 10.1371/journal.pgen.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Qin K, Guo Z, Ma Y, April C, Gao X, Andrews TG, Bokov A, Zhang J, Chen Y, Weintraub ST, Fan JB, Wang D, Hu Y, Aune GJ, Lindsey ML, Li R. Negative Elongation Factor Controls Energy Homeostasis in Cardiomyocytes. Cell Rep. 2014;7(1):79–85. doi: 10.1016/j.celrep.2014.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47(1):8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A, Jensen TH. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39(16):7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90(17):7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J Mol Biol. 1995;252(5):522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483(7389):295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Muñoz M, Vives L, Frangou CG, Groudine M, Peinado MA. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105(50):19809–19814. doi: 10.1073/pnas.0810133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G. Deregulations in the cyclin-dependent kinase-9-related pathway in cancer: implications for drug discovery and development. ISRN Oncol. 2013;2013:305371. doi: 10.1155/2013/305371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Rozenberg JM, Shlyakhtenko A, Glass K, Rishi V, Myakishev MV, FitzGerald PC, Vinson C. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics. 2008;9:67. doi: 10.1186/1471-2164-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci. 2011;14(7):848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakkody A, Abbas A, Scheidegger A, Warns J, Nnoli O, Jokinen B, Zarns K, Kubat B, Dhasarathy A, Nechaev S. RNA polymerase II pausing can be retained or acquired during activation of genes involved in the epithelial to mesenchymal transition. Nucleic Acids Res. 2015;43(8):3938–3949. doi: 10.1093/nar/gkv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013;9(3):e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56(1):29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26(24):2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Razooky B, Cox CD, Simpson ML, Weinberger LS. Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophys J. 2010;98(8):L32–L34. doi: 10.1016/j.bpj.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. A simple method for generating high-resolution maps of genome-wide protein binding. Elife. 2015;4:e09225. doi: 10.7554/eLife.09225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Hernandez AE, Groudine M, Henikoff S. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. Elife. 2014;3:e02042. doi: 10.7554/eLife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25(7):661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, Sobhian B, Severac D, Rialle S, Parrinello H, Cuvier O, Benkirane M. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun. 2014;5:5531. doi: 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, Coulon A, Baek S, Sung MH, John S, Stixova L, Tesikova M, Hakim O, Miranda T, Hawkins M, Stamatoyannopoulos JA, Chow CC, Hager GL. Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res. 2015;25(6):845–857. doi: 10.1101/gr.184168.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J Biol Chem. 2010;285(9):6443–6452. doi: 10.1074/jbc.M109.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Watkins G, Blair AL, Moskaluk C, Ghosh S, Jiang WG, Li R. Deregulation of cofactor of BRCA1 expression in breast cancer cells. J Cell Biochem. 2008;103(6):1798–1807. doi: 10.1002/jcb.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Pan H, Lei C, Yuan B, Nair SJ, April C, Parameswaran B, Klotzle B, Fan JB, Ruan J, Li R. Genetic and genomic analyses of RNA polymerase II-pausing factor in regulation of mammalian transcription and cell growth. J Biol Chem. 2011;286(42):36248–36257. doi: 10.1074/jbc.M111.269167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MH, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56(2):275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, Irvine K, Arakawa T, Nakamura M, Kubosaki A, Hayashida K, Kawazu C, Murata M, Nishiyori H, Fukuda S, Kawai J, Daub CO, Hume DA, Suzuki H, Orlando V, Carninci P, Hayashizaki Y, Mattick JS. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Biol. 2011;31(17):3569–3583. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmé F, Mosrin-Huaman C, Artsimovitch I, Rahmouni AR. Transcriptional pausing in vivo: a nascent RNA hairpin restricts lateral movements of RNA polymerase in both forward and reverse directions. J Mol Biol. 2005;351(1):39–51. doi: 10.1016/j.jmb.2005.05.052. [DOI] [PubMed] [Google Scholar]
- Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol. 2011;18(9):1075–1082. doi: 10.1038/nsmb.2091. [DOI] [PubMed] [Google Scholar]
- Vanderkraats ND, Hiken JF, Decker KF, Edwards JR. Discovering high-resolution patterns of differential DNA methylation that correlate with gene expression changes. Nucleic Acids Res. 2013;41(14):6816–6827. doi: 10.1093/nar/gkt482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464(7290):922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- Vispé S, DeVries L, Créancier L, Besse J, Bréand S, Hobson DJ, Svejstrup JQ, Annereau JP, Cussac D, Dumontet C, Guilbaud N, Barret JM, Bailly C. Triptolide is an inhibitor of RNA polymerase I and II-dependent transcription leading predominantly to down-regulation of short-lived mRNA. Mol Cancer Ther. 2009;8(10):2780–2790. doi: 10.1158/1535-7163.MCT-09-0549. [DOI] [PubMed] [Google Scholar]
- Voigt P, LeRoy G, Drury WJ, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. Asymmetrically modified nucleosomes. Cell. 2012;151(1):181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27(12):1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12(3):343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, Boyer F, Parrinello H, Berkhout B, Terzian C, Benkirane M, Kiernan R. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150(6):1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell. 2015;58(2):311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22(9):2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97(1):41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Hagiwara Y, Chiba K, Isobe T, Narita T, Handa H, Yamaguchi Y. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun. 2014a;5:4263. doi: 10.1038/ncomms5263. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Wu Z, Russnes HG, Takagi S, Peluffo G, Vaske C, Zhao X, Moen Vollan HK, Maruyama R, Ekram MB, Sun H, Kim JH, Carver K, Zucca M, Feng J, Almendro V, Bessarabova M, Rueda OM, Nikolsky Y, Caldas C, Liu XS, Polyak K. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 2014b;25(6):762–777. doi: 10.1016/j.ccr.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442(7106):1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Ye Q, Hu YF, Zhong H, Nye AC, Belmont AS, Li R. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol. 2001;155(6):911–921. doi: 10.1083/jcb.200108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamft B, Bintu L, Ishibashi T, Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci U S A. 2012;109(23):8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156(5):920–934. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39(12):1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Wada T, Okabe S, Taneda T, Yamaguchi Y, Handa H. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 2007;35(12):4064–4075. doi: 10.1093/nar/gkm430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11(20):2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]


