Abstract
Although Appl1 and Appl2 have been implicated in multiple cellular activities, we and others have found that Appl1 is dispensable for mouse embryonic development, suggesting that Appl2 can substitute for Appl1 during development. To address this possibility, we generated conditionally targeted Appl2 mice. We found that ubiquitous Appl2 knockout (Appl2-/-) mice, much like Appl1-/- mice, are viable and grow normally to adulthood. Intriguingly, when Appl1-/- mice were crossed with Appl2-/- mice, we found that homozygous Appl1;Appl2 double knockout (DKO) animals are also viable and grossly normal with regard to reproductive potential and postnatal growth. Appl2-null and DKO mice were found to exhibit altered red blood cell physiology, with erythrocytes from these mice generally being larger and having a more irregular shape than erythrocytes from wild type mice. Although Appl1/2 proteins have been previously shown to have a very strong interaction with phosphatidylinositol-3 kinase (Pi3k) in thymic T cells, Pi3k-Akt signaling and cellular differentiation was unaltered in thymocytes from Appl1;Appl2 (DKO) mice. However, Appl1/2-null mouse embryonic fibroblasts exhibited defects in HGF-induced Akt activation, migration, and invasion. Taken together, these data suggest that Appl1 and Appl2 are required for robust HGF cell signaling but are dispensable for embryonic development and reproduction.
Keywords: Appl1, Appl2, Akt, HGF, T-cell, Development
INTRODUCTION
APPL1 was discovered as a binding partner to AKT2 and PI3K p110β by use of a yeast two-hybrid assay (Mitsuuchi et al., 1999). Over the past 15 years, both APPL1 and the subsequently identified APPL2 (Lin et al., 2006; Mao et al., 2006; Miaczynska et al., 2004; Ryu et al., 2014) were found to bind to and regulate many other signaling proteins such as Rab5, GIPC, adiponectin receptors, insulin receptor, and IRS1/2 to affect cell proliferation, migration and metabolism (Lin et al., 2006; Mao et al., 2006; Miaczynska et al., 2004; Ryu et al., 2014). Our group also discovered that murine Appl1 and Appl2 both bind to the p110 subunit of phosphatidylinositol-3 kinase (Pi3k), with the strongest interaction documented in thymic T cells (Tan et al., 2010a).
A role for AKT2 in glucose metabolism was first proposed in 1998, when murine Akt2 was found to be highly expressed in embryonic brown fat and its kinase product was shown to be robustly activated by insulin (Altomare et al., 1998). Subsequent work revealed that an interaction between AKT and APPL1 was required for insulin-stimulated Glut4 translocation and to promote insulin secretion in pancreatic beta cells (Cheng et al., 2012; Saito et al., 2007; Wang et al., 2013). However, the influence of Appl1/2 proteins on Akt activation appears to be highly context dependent. Specifically, some reports indicate that Appl proteins facilitate Akt activation during insulin signaling or during phagocytosis (Saito et al., 2007; Yeo et al., 2015). Other groups have reported converse findings, with Appl impairing cell migration and osteoclastogenesis by suppressing AKT (Broussard et al., 2012; Tu et al., 2011). In muscle cells, APPL2 can act as a negative regulator of adiponectin and insulin signaling by competing with APPL1 in the binding of adiponectin receptors and by sequestrating APPL1 from these two pathways (Wang et al., 2009). In contrast, Appl2 is required for Akt activation during phagocytosis of macrophages (Yeo et al., 2015).
In addition to their roles in insulin and adiponectin signaling, APPL proteins have been implicated in endocytosis. APPL proteins are enriched at the membrane of early endosomes and participate in cell trafficking via binding to RAB5 and RAB21 (Miaczynska et al., 2004; Zhu et al., 2007). Mutation of the inositol 5-phosphatase OCRL, which causes Lowe syndrome, abolishes the interaction between APPL1 and OCRL, causing trafficking failure in early endosomes (Erdmann et al., 2007). Adding even more complexity to APPL protein function, both APPL1 and APPL2 have been found to promote the survival of pancreatic cancer cells after radiation via ATM-mediated DNA repair (Hennig et al., 2014). APPL1 can also be recruited to ubiquitin-rich aggresomes in response to proteasome inhibition, which has been proposed to regulate aggresome function (Pilecka et al., 2011).
The overall complexity of the in vitro data is partially supported by in vivo knockdown studies. In lower vertebrate models, Appl1 knockdown induces neural cell death during the embryonic development of zebrafish or triggers apoptosis of pancreas and stomach progenitor cells in Xenopus (Schenck et al., 2008; Wen et al., 2010). To validate the results of mRNA knockdown studies and exclude off-target effects commonly seen in such experiments, knockout mouse models of Appl1 and Appl2 should be carefully evaluated. We previously discovered that Appl1 is not essential in mammals, as Appl1-/- knockout mice were found to be viable and phenotypically normal (Ryu et al., 2014; Tan et al., 2010a; Tan et al., 2010b), which has been confirmed by others (Cheng et al., 2012; Ryu et al., 2014). Homozygous Appl1 knockout mice showed no obvious defects during embryonic development, postnatal growth and reproduction. Moreover, in vivo Pi3k-Akt signaling was not impaired (Tan et al., 2010a). This poses an intriguing possibility that Appl1 and Appl2 may have overlapping functions, such that Appl2 alone is sufficient to compensate for Appl1 loss. To test this hypothesis, we generated Appl2 knockout mice, which were subsequently crossed with Appl1-null mice. Like Appl1-null mice, Appl2-null mice do not manifest obvious defects in embryonic development or postnatal growth. Moreover, much to our surprise, Appl1;Appl2 double knockout (DKO) mice were also found to be viable and fertile. Mice of different genotypes were born at expected Mendelian ratios, and litter size was normal in the DKO mice. Moreover, T cells from the DKO showed normal Pi3k-Akt signaling and normal development of the thymus. However, murine embryonic fibroblasts (MEFs) from Appl1;Appl2 DKO mice exhibited defects in HGF-induced Akt activation, migration and invasion, providing further support for a context-dependent role of Appl1/2 proteins in Akt activation.
MATERIALS AND METHODS
Generation of conditional Appl2 knockout and Appl1;Appl2 double knockout mice
Exon 5 of the Appl2 gene was flanked by two LoxP sites using conventional gene targeting strategies. In short, one LoxP sequence was inserted into Intron 4 of Appl2, and a Frt-Neo-Frt-LoxP sequence was inserted into Intron 5. The targeting construct was transfected into ES cells, and correctly targeted ES cell clones were used to generate chimeric mice. Southern blotting was carried out to identify mice with the correctly targeted allele. Appl2 floxed mice were mated with EIIA-Cre mice to generate ubiquitous Appl2 knockout mice. PCR conditions and the primer sequences for genotyping of the pre- or post-Cre Appl2-allele are listed in Table 1. Next, the Appl2-/- mice were crossed with previously reported Appl1-/- mice (Tan et al., 2010b) to generate Appl1;Appl2 DKO mice.
Table 1.
Genotyping primers for Appl2 targeted allele
| Primer name | Sequence |
|---|---|
| Single Lox P site | |
| F | TTCCTCTCAGAAGAAAGTTGC |
| R | ATCCTCATTTGACTCAAGGC |
| Neo cassette | |
| F | GCAGCTGTTCCTGAGAAGG |
| R | TTGGCTGGACGTAAACTCC |
| Appl2 knockout | |
| F | ACAGGGTACTGGTACTCATGC |
| R | ACTGTGCTCACAGGTGTACC |
| WT | |
| F | GCAGCTGTTCCTGAGAAGG |
| R | ACTGTGCTCACAGGTGTACC |
Abbreviations: F = forward; R = reverse; WT = wild type.
Reagents
Antibodies against Appl1, total and phospho-Akt, phospho-AMPK, total and phospho-Erk, p38, cleaved caspase3, and Parp were from Cell Signaling (Danvers, MA). Antibodies against β-actin, QM, and GAPDH were from Santa Cruz Biotechnology (Dallas, TX). Appl2 antibody was generated by one of us (LQD). EGF and HGF were purchased from R&D Systems (Minneapolis, MN). Tunicamycin and camptothecin were from Selleckchem (Houston, TX).
Complete blood count (CBC) and flow cytometry analysis
Mouse blood was drawn retro-orbitally and measured by VetScan HM5 (Abaxis, Union City, CA). In brief, about 50 μl of peripheral blood was collected into an EDTA-coated 0.5 ml tube (BD, San Jose CA), and CBCs were assessed within 1 h.
Flow cytometry was performed with a BD LSRII machine to analyze T cell developmental markers. Anti-CD4-APC/Cy7, anti-CD8-PE, anti-CD44-APC/Cy7, and anti-CD25-PE were obtained from BioLegend (San Diego, CA). Data were analyzed by using FlowJo software.
Cell cycle analysis
Cells were fixed with 70% ice-cold ethanol for 2 h at 4°C, washed twice with ice-cold PBS, and then stained with propidium iodine solution for 20 min. Samples were subjected to FACScan (Beckman, Brea, CA) analysis using FlowJo software.
Mouse embryonic fibroblast culture and treatment
E13.5-day embryos from wild type (WT), Appl1 KO, Appl2 KO and Appl1;Appl2 DKO mice were collected. Each carcass, minus organs and head, was subjected to 0.2% collagenase for 30 min at 37°C. Isolated cells were washed twice with DMEM supplemented with 10% FBS and 2 mM L-glutamine. Cells were seeded at 1 × 105/well in a 6-well plate overnight, starved in serum-free media for 1 h, followed by stimulation with 20 ng/ml EGF, 10% FBS, or 50 ng/ml HGF for the indicated times.
Migration and invasion assays
To evaluate cell migration, a Transwell migration assay was used in which 2 × 104 cells were resuspended in 0.1% DMEM and then seeded on a 8.0-μM PET insert (Corning, Oneonta, NY). Inserts were placed in a 24-well plate with DMEM containing 100 ng/ml HGF and 0.1% FBS in the bottom chamber. After 6 h, inserts were fixed and stained. Cells that migrated through the membrane were photographed and counted. For the invasion assay, a growth factor-reduced Matrigel invasion chamber (Corning) was used, and cells were counted 48 h after seeding. For the wound-healing assay, each well with confluent cells was scratched with a pipette tip, and the rate for the closure of the gap was monitored.
RESULTS
Targeted knockout of Appl2 in Mice
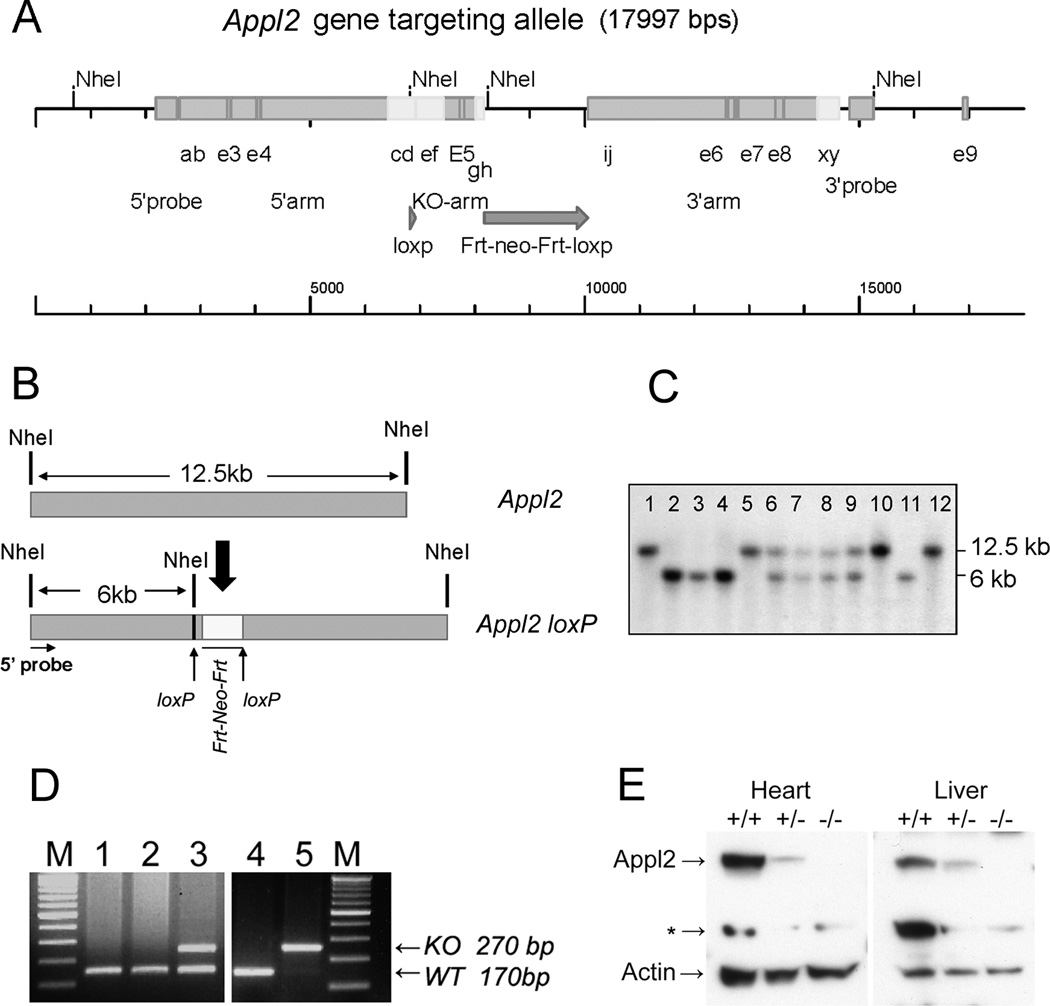
To facilitate studies of either whole-body or tissue-specific roles of Appl2, we conditionally targeted the mouse Appl2 gene. Exon 5 of the mouse Appl2 gene was flanked by two LoxP sites (Fig.1 A). Southern blot analysis was performed to identify mice with homozygously floxed Appl2 (Appl2L/L) alleles (Fig. 1B, C). To address the question about whether Appl2 is required for organogenesis and embryo development, whole-body Appl2-/- mice were generated by crossing Appl2L/L mice to EIIA-Cre mice, followed by PCR-based genotyping of tail DNA (Fig. 1D). Homozygous Appl2 knockout mice were found to be viable, and immunoblot analysis confirmed the loss of Appl2 protein expression in tissues from various organs, e.g., heart and liver, in Appl2-/- mice (Fig. 1E).
Figure 1.
Conditional targeting of mouse Appl2 gene. Diagram depicting insertion of LoxP sites in Intron 4 and Intron 6 (A). Restriction endonuclease map (B) and Southern blot analysis (C) performed to identify mice with floxed Appl2 allele(s). Mice represented by lanes 6, 7, 8, and 9 are heterozygous (Appl2+/L), whereas those depicted in lanes 2, 3, 4, and 11 are homozygous (Appl2L/L). Appl2 floxed mice were crossed to EIIA-Cre mice to obtain whole-body excision of the Appl2 gene. Genotyping performed by PCR on tail DNA signify wild type (lanes 1, 2, and 4), heterozygous (lane 3) and homozygous Appl2 knockout alleles (lane 5) (D). Immunoblot analysis demonstrating loss of Appl2 protein expression in heart and liver tissues of Appl2-/- mice (E). * = nonspecific band.
Appl2 is dispensable for embryonic development and reproduction
Genotyping of litters from Appl2+/- × Appl2+/- mice revealed normal Mendelian ratios of genotype distributions, suggesting that loss of Appl2 does not impact embryonic development (Sup Fig. 1A). Moreover, Appl2 loss did not impair reproductive function, given that similar litter sizes were observed among Appl2+/+ × Appl2+/+, Appl2+/- × Appl2+/- and Appl2-/- × Appl2-/- matings (Suppl. Fig. 1B).
Dual loss of Appl1 and Appl2 does not impair organogenesis and reproductive function
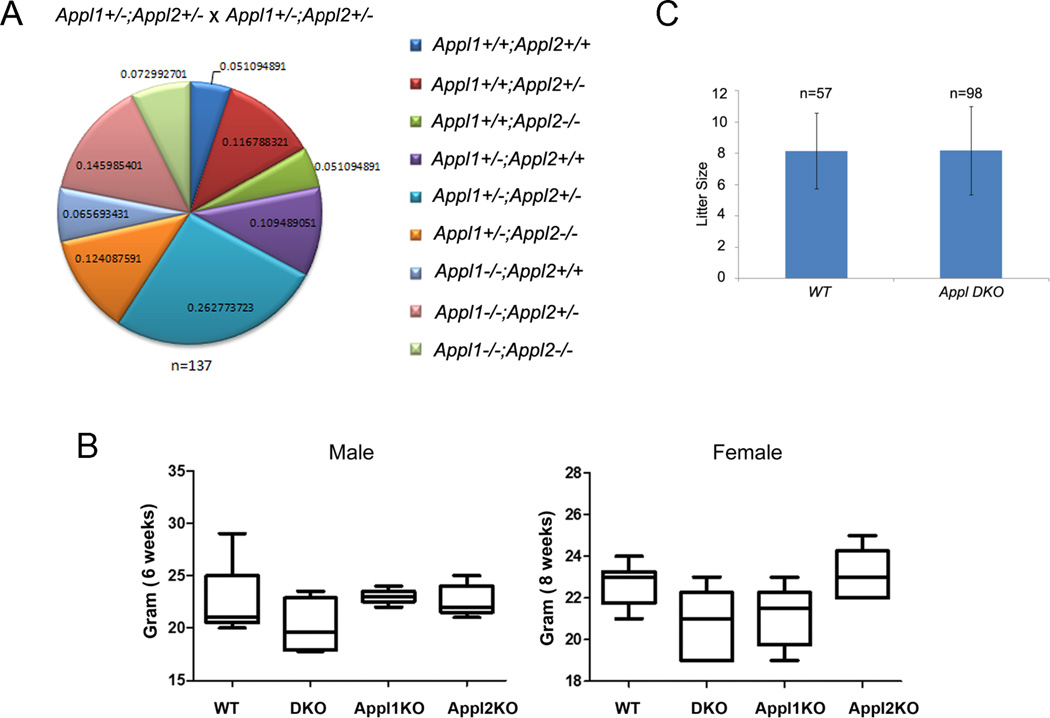
To predict the tissues that might be adversely affected upon the co-deletion of both Appl1/2 genes, we determined the abundance of Appl1 and Appl2 using real-time PCR (Suppl. Fig. 2). To further delineate the role of Appl1 and Appl2 in development, we set up a series of crosses to generate Appl1-/-;Appl2-/- DKO mice. Unexpectedly, genotyping of offspring from Appl1+/-;Appl2+/- × Appl1+/-;Appl2+/- crosses revealed normal Mendelian ratios (Fig. 2A). Appl1/2 DKO mice showed grossly normal postnatal growth with slightly lower average body weight, which did not reach statistical significance (Fig. 2B). Moreover, Appl1;Appl2 DKO mice have unimpaired reproductive function when compared to normal littermates (Fig.2C).
Figure 2.
Appl1;Appl2 double-knockout (DKO) mice are viable and grossly normal. Normal Mendelian ratio of genotype distribution in litters from Appl1+/-;Appl2+/- × Appl1+/-;Appl2+/- matings (A). Box plots demonstrating that Appl DKO mice show normal postnatal growth base and body weight at 6 weeks (males) and 8 weeks (females) (B). Histogram showing similar average litter sizes arising in WT × WT and Appl DKO X Appl DKO matings (C).
Deficiency of Appl1 and Appl2 affects the physiology of erythrocytes, but not hematopoietic stem cell (HSC) differentiation
Complete blood counts (CBC) of 8-week-old mice revealed that deletion of Appl1 and/or Appl2 did not have an effect on the numbers and percentages of lymphocytes, monocytes, neutrophils, eosinophils, and basophils. However, hematocrit (HCT), mean corpuscular volume (MCV), and mean corpuscular hemoglobin concentration (MCHC) were slightly but significantly altered in Appl1-/-, Appl2-/- and Appl1;Appl2 DKO mice, particularly in female mice. There changes were not severe enough to cause pernicious anemia, as WBC counts as well as the sizes of spleens and livers were normal (Table 2 and Suppl. Table 1).
Table 2.
Complete blood counts from WT, Appl1 KO, Appl2 KO and DKO female mice.
| WT | DKO | Appl1 KO | Appl2 KO | |
|---|---|---|---|---|
| WBC (10^9/L) | 7.75±1.23 | 6.48±1.71 | 8.17±1.65 | 8.53±2.51 |
| LYM (10^9/L) | 6.83±0.89 | 5.47±1.08 | 6.9±1.34 | 7.18±2.00 |
| MON (10^9/L) | 0.10±0.05 | 0.21±0.17 | 0.12±0.06 | 0.09±0.05 |
| NEU (10^9/L) | 0.81±0.55 | 0.8±0.48 | 1.16±0.36 | 1.25±0.65 |
| LYM (%) | 88.7±4.88 | 85.3±5.23 | 84.57±2.75 | 84.58±4.13 |
| MON (%) | 1.47±0.69 | 3.00±1.51 | 1.47±0.65 | 1.25±0.62 |
| NEU (%) | 9.8±5.52 | 11.63±3.81 | 13.93±3.05 | 14.13±4.07 |
| RBC (10^12/L) | 11.14±0.75 | 10.96±0.23 | 10.94±0.75 | 11.24±0.46 |
| HGB (g/dl) | 16.17±0.50 | 16.2±0.69 | 16.57±1.27 | 16.58±0.40 |
| HCT (%) | 50.77±2.40 | 56.84±1.65** | 54.68±3.63* | 57.88±1.42*** |
| MCV( ft) | 46.00±1.29 | 52.00±1.00*** | 50.17±3.71 | 51.75±0.96**** |
| MCH (pg) | 14.63±0.80 | 14.8±0.60 | 15.12±0.50 | 14.75±0.41 |
| MCHC( g/dl) | 31.8±1.42 | 28.53±0.61** | 30.33±2.18 | 28.65±0.44*** |
| RDWc (%) | 17.93±0.57 | 19.13±0.5* | 20.02±0.96*** | 18.55±0.26* |
| PLT (10^9/l) | 518±88.01 | 533.67±50.5 | 547.17±82.53 | 742.25±90.7* |
| PCT (%) | 0.32±0.04 | 0.40±0.03* | 0.43±0.08 | 0.63±0.11*** |
| MPV (ft) | 6.3±0.68 | 7.57±0.21* | 7.77±0.38**** | 8.5±0.63**** |
| PDWc (%) | 30.13±2.33 | 34.27±1.81 | 37.78±2.02** | 37.05±2.12**** |
Note: Compared to WT mice.
p<0.05,
p<0.01,
p<0.005,
p<0.001
Appl proteins are not required for Pi3k-Akt signaling in thymic T-cell and other tissues
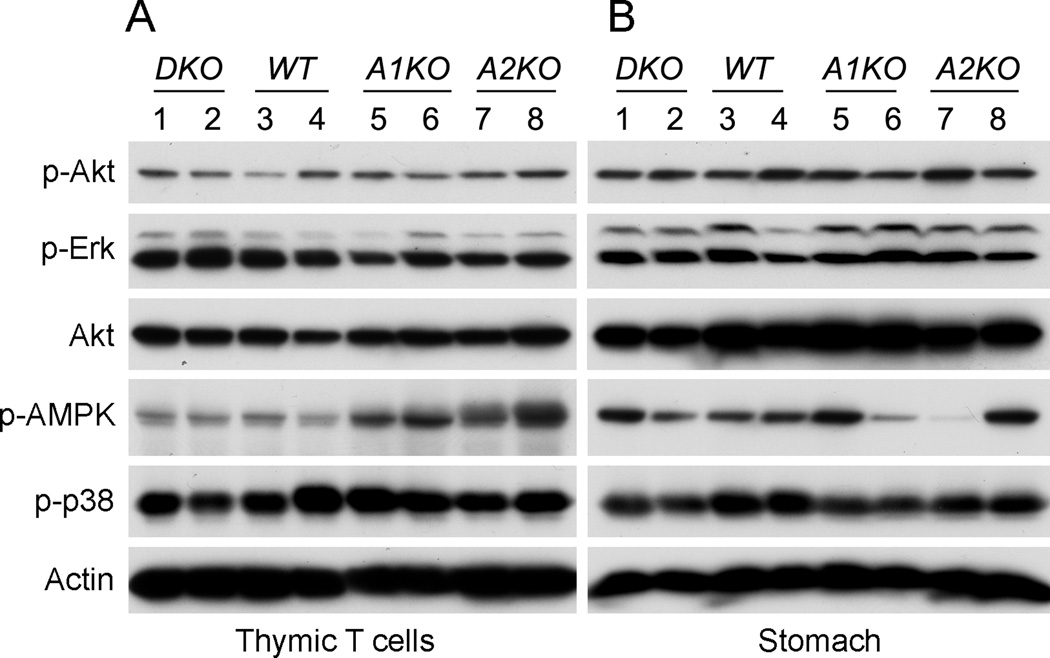
We previously found that among all tissues tested, the greatest amount of interaction between Appl1/2 and the p110β subunit of Pi3k occurs in thymic T cells. Despite this, we report here that basal level of Akt activation is not altered in thymic T-cells from DKO mice under normal husbandry conditions (Fig. 3A). Moreover, no consistent alteration of basal Akt activity was observed in tissues from stomach, liver or muscle among wild type (WT), Appl1 KO, Appl2 KO and DKO mice (stomach shown in Fig.3B, Suppl. Fig.3).
Figure 3.
Appl1;Appl2 DKO mice have unaltered Pi3k signaling in vivo. Tissues from thymic T cells (A) and stomach (B) were subjected to immunoblotting using antibodies against the indicated proteins. Levels of phospho-Akt, phospho-Erk, phospho-p38 and phospho-AMPK were compared among the four genotypes. A1KO = Appl1 KO mice; A2KO = Appl2 KO mice.
Co-deletion of Appl1 and Appl2 does not affect T-cell development
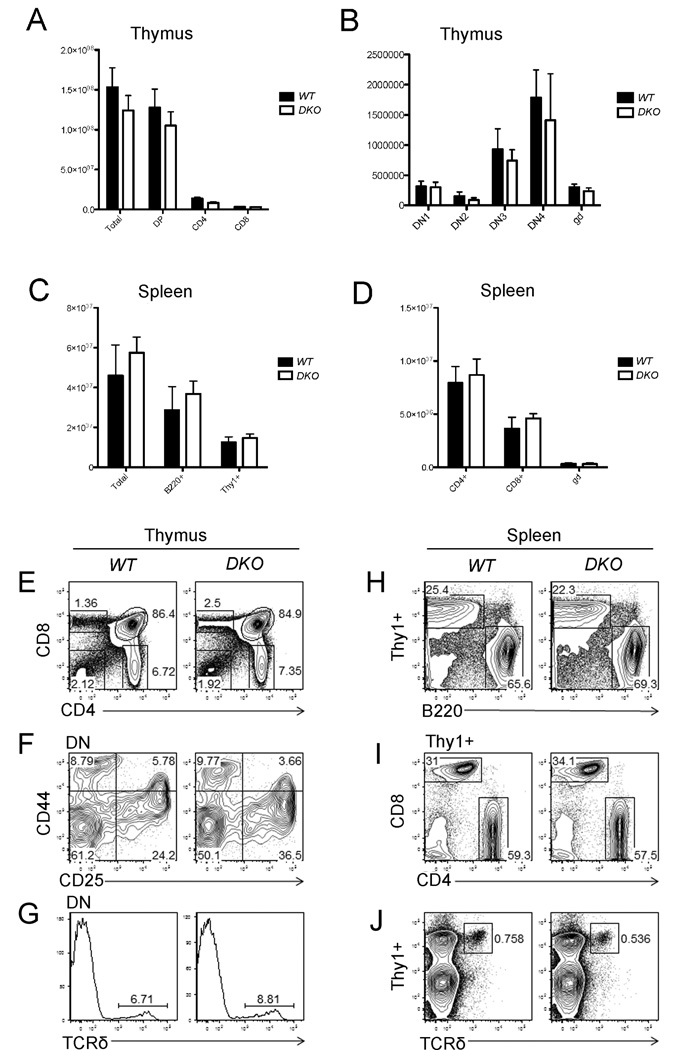
Freshly isolated thymic T cells or spleen cells from WT and Appl1;Appl2 DKO mice were subjected to flow cytometry analysis. CD4/CD8 and CD25/CD44 staining demonstrated that the total cell count of CD4/8 double positive (DP), CD4+, CD8+ and double negative (DN1, DN2, DN3 and DN4) do not differ significantly between the two genotypes (Fig. 4A, B). The total mature T cell count in the spleen also was not changed in DKO mice (Fig. 4C, D). Moreover, we observed no significant change in the percentage of CD4+, CD8+ cells, CD25+ or CD44+ cells in DKO thymus (Fig. 4E, F). The proportion of TCRδ cells also was not appreciably changed in DKO thymus (Fig. 4G). In spleen, the T/B cell ratio in DKO mice did not differ significantly from that of WT mice (Fig. 4H). Moreover, CD4+, CD8+ or TCRδ+ populations also were not changed (Fig. 4I, J). Collectively, these findings suggest that Appl1 and Appl2 do not have a crucial role in T-cell development.
Figure 4.
Co-deletion of Appl1 and Appl2 do not affect T-cell development. WT and Appl DKO mice at 6 weeks of age were sacrificed to collect thymus (A, B) and spleen cells (C, D). Cells were stained with fluorescence-labeled anti-CD4, CD8, CD44, CD25, Thy1 and B220 antibodies, and then counted using multicolor flow cytometry. (A) CD4+, CD8+ and CD4+CD8+ (DP) cells in thymus. (B) Distribution of various stages of CD4/CD8 double-negative (DN) cells in thymus of WT and Appl DKO mice. gd = γδ T cells. (C) Thy1+ and B220+ population in spleen. (D) Mature CD4+ and CD8+ T-cells in spleen. (E) Percentages of CD4+, CD8+ and CD4+CD8+ and CD4-CD8- populations in thymus of WT and DKO mice. (F) Distribution of CD44+, CD25+, CD44+CD25+, and CD44-CD25- cells. (G) TCR δ population in thymus cells from WT and DKO mice. (H, I, J) Percentages of Thy1+ and B220+ populations, mature CD4+ or CD8+ T-cells, and TCR δ T cells in spleen, respectively.
Appl1 and Appl2 potentiate HGF-induced Pi3k-Akt signaling and motility in primary MEFs
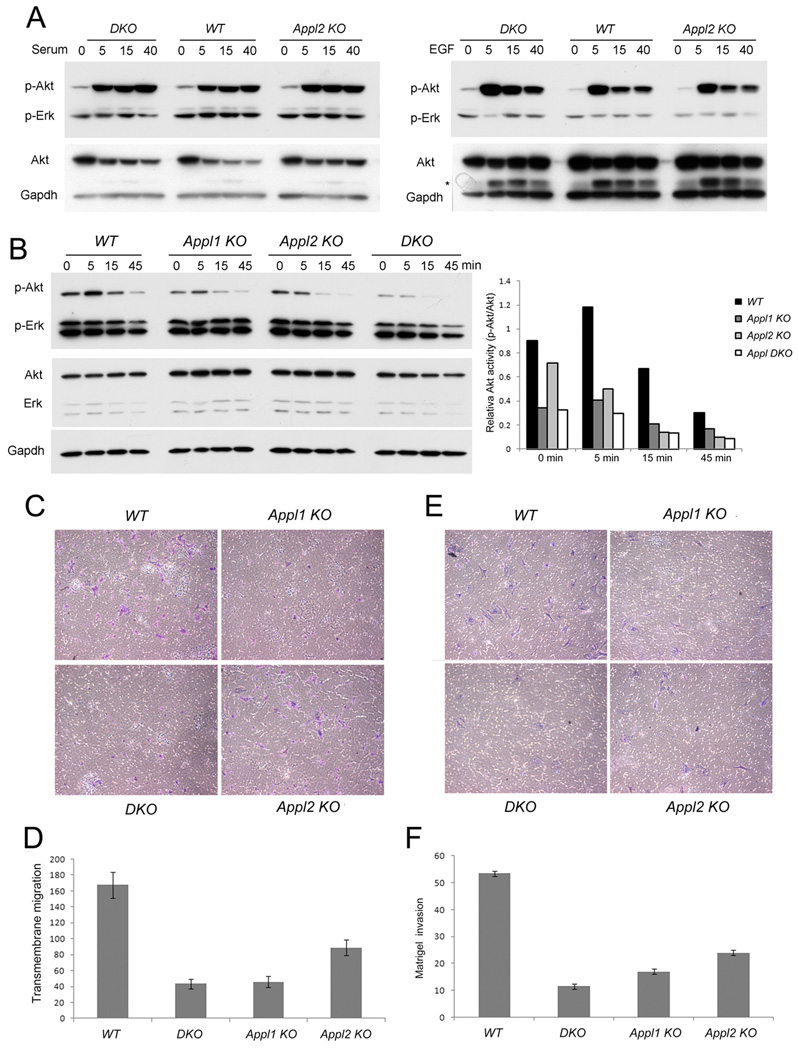
We previously showed that EGF, insulin or serum trigger robust Akt activation in serum-starved MEFs independent of Appl1 status, whereas Appl1 is required for moderate Akt activation triggered by HGF (Tan et al., 2010b). Here, we further demonstrate that Appl2 is not required for robust activation of Akt by EGF or serum. MEFs generated from E13.5-day-old embryos were harvested and used in early passages. MEFs from Appl2 KO or Appl1/2 DKO mice showed no appreciable difference in the level of Akt activation upon stimulation with 10% serum or 20 ng/ml EGF for 5–40 min (Fig. 5A). Interestingly, Appl1 KO, Appl2 KO, and especially Appl1/2 DKO MEFs showed consistently decreased Akt activation upon stimulation with HGF (Fig. 5B). Further study revealed that loss of Appl proteins results in reduced HGF-triggered transmembrane migration (Fig. 5C, D and Suppl. Fig. 4A) and decreased movement in a wound-healing assay (Suppl. Fig. 4B). Additionally, Appl-deficient MEFs showed markedly decreased HGF-triggered invasion in a matrigel assay, indicating that loss of Appl proteins impairs cell invasion following stimulation with HGF (Fig. 5E, F). However, no difference in invasive ability was observed between WT and Appl1/2 knockout cells in response to EGF (Sup Fig. 4C, D), presumably due to unaffected EGF-Akt signaling in Appl KO cells.
Figure 5.
Appl proteins are required for HGF-induced, but not FBS- or EGF-induced, Akt activation and migration in MEFs. (A) Primary MEFs from WT, Appl2-/- and Appl DKO embryos were seeded overnight, starved with serum-free media, and then stimulated with 10% FBS (upper panel) or 20 ng/ml EGF (lower panel) for the indicated times. (B) MEFs from WT, Appl1-/-, Appl2-/- and Appl DKO embryos were starved and treated with 50 ng/ml HGF. The histogram depicts the relative densitometry levels of phosphorylated Akt normalized against total Akt protein levels. (C, D) MEFs were seeded on 8.0 μM PET inserts with DMEM plus 0.1% FBS at the top chamber. The lower chamber were filled with 100 ng/ml HGF in DMEM plus 0.1% FBS. Cells migrating to the other side of the membrane were fixed, stained and counted after 6 h. (E, F) Results of invasion assay using Matrigel invasion chambers, with cells counted 48 h after seeding. Migrated or invaded cell numbers were counted and normalized against the seeding cell numbers.
DISCUSSION
Recent in vitro studies on Appl1 and Appl2 by several labs, using RNAi or expression constructs, have generated paradoxical results (Broussard et al., 2012; Miaczynska et al., 2004; Saito et al., 2007; Tu et al., 2011; Yeo et al., 2015). Unbiased genetic evidence is therefore needed to better understand the function of Appl proteins. To address this need, in vivo studies using gene knockout strategies have been applied. We previously reported that Appl1 knockout mice have normal embryonic development and reproduction, with generally normal Akt signaling in various tissues tested, and we raised the possibility that Appl proteins may substitute for each other during development and in cell signaling (Tan et al., 2010a; Tan et al., 2010b). Since Appl2 knockout mice were not available at that time to test this idea in vivo, we used an RNAi approach to knock down Appl2 in Appl1-/- MEFs. We found that Appl1 and Appl2 are functionally redundant and dispensable for cell survival under normal cell culture conditions. In the present study with Appl2-/- mice, we found that Appl2 is dispensable for development and reproduction. Moreover, we discovered that Appl1;Appl2 DKO mice are viable and grossly normal without marked defects in Akt signaling in vivo. We previously reported that Appl1 and Appl2 each interacts strongly with the p110 subunit of Pi3k in thymic T cells in vivo (Tan et al., 2010a). However, this interaction does not appear to be required for basal level of Pi3k-Akt signaling in thymic T cells; moreover, Appl1-/-, Appl2-/- and DKO mice each demonstrated normal T-cell development. Altogether, these findings strongly suggest that Appl1 and Appl2 genes are functionally redundant during organogenesis and postnatal growth under normal conditions. Furthermore, our studies on MEFs from these mice revealed that only under certain conditions, specifically upon stimulation with HGF, do Appl1 and Appl2 regulate Akt activity and cell migration/invasion.
AKT signal transduction pathway regulates many cellular survival mechanisms including cell cycle progression, metabolism, anti-apoptosis, invasion and metastasis (Vogiatzi and Giordano, 2007). Many growth factors can activate AKT signaling including EGF, Insulin, IGF, FGF and HGF etc (Burgering and Coffer, 1995; Forough et al., 2006; Magun et al., 1996; Sanchez-Margalet, 2000). HGF-Met signaling has been implicated in tumor growth, invasion, and metastasis, partially via Akt (Shinomiya et al., 2004). Previously, we reported that only a very small percentage of all Akt protein binds to Appl1 or Appl2 protein in MEFs. This may explain why only a weak mitogen such as HGF, not a stronger mitogen such as EGF, requires Appl1 protein to activate Akt in MEFs (Tan et al., 2010b). In the present study, we found that this is also the case with Appl2-/- MEFs and Appl DKO MEFs. When compared MEFs from WT mice, Appl-deficient MEFs exhibit impaired ability to migrate or invade Matrigel in response to HGF stimulation in vitro. Interesting, basal level of Akt activation in the liver is not altered in Appl knockout mice under normal conditions. This may be because c-Met is not required for normal liver function but is essential for regeneration after liver injury (Huh et al., 2004). Therefore, we speculate that Appl knockout mice might exhibit defects in liver regeneration after injury. Moreover, mutation of HGF results in deafness in mice (Schultz et al., 2009). Future neurological studies should be performed in Appl knockout mice to assess hearing loss. With regard to cancer, loss of HGF/c-Met signaling accelerates N-nitrosodiethylamine-induced hepatocarcinogenesis (Takami et al., 2007). Given our group’s longstanding interest in cancer research, we intend to carry out similar studies with Appl1/2-null mice to test if Appl proteins have a tumor suppressor function, and if such a potential role were associated with HGF signaling during hepatocarcinogenesis.
Appl1 and Appl2 proteins possess BAR, PH, and PTP domains. BAR domains can dimerize and interact with BAR-PH domains. The BAR, PH, and PTP motifs are each capable of binding phospholipids and possess membrane binding properties (Li et al., 2007). BAR domain proteins bind to small GTPases to induce membrane curvature (Habermann, 2004). Moreover, Appl1 and Appl2 can form homo- or hetero-oligomers via their BAR domains. Appl protein can recruit RAB5 to moving membrane structures (Chial et al., 2008). In this study, we discovered that Appl knockout mice, particularly Appl2-null and DKO mice, exhibit altered red blood cell physiology. These cells generally were larger and had a more irregular shape than erythrocytes from WT mice. Thus, we speculate that Appl proteins normally may bind to the cellular membrane of erythrocytes and facilitate the maintenance of the bioconcave disk.
Finally, the finding that Appl-deficient mice are grossly normal with regard to reproductive potential is intriguing, given that prior work demonstrated a potential link between the follicle-stimulating hormone receptor (FSHR) and APPL1 (Nechamen et al., 2004). In this earlier in vitro work, FSHR was shown to interact with APPL1, and FOXO1a, a downstream effector in the PI3K pathway tightly connected with expression of proapoptotic genes, was found to be rapidly inactivated when FSHR-expressing human cells were treated with FSH. Thus, a possible link between FSH and PI3K/Akt signaling was proposed as a survival mechanism whereby FSH selects the dominant follicle to survive. However, our current work with Appl knockout models revealed no obvious changes with regard to fertility.
Supplementary Material
Acknowledgements
We thank the Flow Cytometry and Laboratory Animal Facilities at Fox Chase Cancer Center for assistance. This work was supported by the NIH grants (NCI-CA77429, CA083638 and CA06927 to J.R.T., and NIDDK-DK102965 to L.Q.D) and by American Diabetes Association Award (Grant # 7-13-BS-043 to L.Q.D), an appropriation from the Commonwealth of Pennsylvania, and a CCSG Pilot Project from Fox Chase Cancer Center.
Footnotes
Conflict of Interest: The authors have no potential conflicts of interest to disclose.
References
- Altomare DA, Lyons GE, Mitsuuchi Y, Cheng JQ, Testa JR. Akt2 mRNA is highly expressed in embryonic brown fat and the AKT2 kinase is activated by insulin. Oncogene. 1998;16(18):2407–2411. doi: 10.1038/sj.onc.1201750. [DOI] [PubMed] [Google Scholar]
- Broussard JA, Lin WH, Majumdar D, Anderson B, Eason B, Brown CM, Webb DJ. The endosomal adaptor protein APPL1 impairs the turnover of leading edge adhesions to regulate cell migration. Molecular biology of the cell. 2012;23(8):1486–1499. doi: 10.1091/mbc.E11-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376(6541):599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- Cheng KK, Lam KS, Wu D, Wang Y, Sweeney G, Hoo RL, Zhang J, Xu A. APPL1 potentiates insulin secretion in pancreatic beta cells by enhancing protein kinase Akt-dependent expression of SNARE proteins in mice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(23):8919–8924. doi: 10.1073/pnas.1202435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial HJ, Wu R, Ustach CV, McPhail LC, Mobley WC, Chen YQ. Membrane targeting by APPL1 and APPL2: dynamic scaffolds that oligomerize and bind phosphoinositides. Traffic. 2008;9(2):215–229. doi: 10.1111/j.1600-0854.2007.00680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Developmental cell. 2007;13(3):377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forough R, Weylie B, Collins C, Parker JL, Zhu J, Barhoumi R, Watson DK. Transcription factor Ets-1 regulates fibroblast growth factor-1-mediated angiogenesis in vivo: role of Ets-1 in the regulation of the PI3K/AKT/MMP-1 pathway. Journal of vascular research. 2006;43(4):327–337. doi: 10.1159/000093198. [DOI] [PubMed] [Google Scholar]
- Habermann B. The BAR-domain family of proteins: a case of bending and binding? EMBO Rep. 2004;5(3):250–255. doi: 10.1038/sj.embor.7400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig J, McShane MP, Cordes N, Eke I. APPL proteins modulate DNA repair and radiation survival of pancreatic carcinoma cells by regulating ATM. Cell death & disease. 2014;5:e1199. doi: 10.1038/cddis.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh CG, Factor VM, Sanchez A, Uchida K, Conner EA, Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mao X, Dong LQ, Liu F, Tong L. Crystal structures of the BAR-PH and PTB domains of human APPL1. Structure. 2007;15(5):525–533. doi: 10.1016/j.str.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Lin DC, Quevedo C, Brewer NE, Bell A, Testa JR, Grimes ML, Miller FD, Kaplan DR. APPL1 associates with TrkA and GIPC1 and is required for nerve growth factor-mediated signal transduction. Mol Cell Biol. 2006;26(23):8928–8941. doi: 10.1128/MCB.00228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magun R, Burgering BM, Coffer PJ, Pardasani D, Lin Y, Chabot J, Sorisky A. Expression of a constitutively activated form of protein kinase B (c-Akt) in 3T3-L1 preadipose cells causes spontaneous differentiation. Endocrinology. 1996;137(8):3590–3593. doi: 10.1210/endo.137.8.8754791. [DOI] [PubMed] [Google Scholar]
- Mao X, Kikani CK, Riojas RA, Langlais P, Wang L, Ramos FJ, Fang Q, Christ-Roberts CY, Hong JY, Kim RY, Liu F, Dong LQ. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol. 2006;8(5):516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116(3):445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- Mitsuuchi Y, Johnson SW, Sonoda G, Tanno S, Golemis EA, Testa JR. Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene. 1999;18(35):4891–4898. doi: 10.1038/sj.onc.1203080. [DOI] [PubMed] [Google Scholar]
- Nechamen CA, Thomas RM, Cohen BD, Acevedo G, Poulikakos PI, Testa JR, Dias JA. Human follicle-stimulating hormone (FSH) receptor interacts with the adaptor protein APPL1 in HEK 293 cells: potential involvement of the PI3K pathway in FSH signaling. Biol Reprod. 2004;71(2):629–636. doi: 10.1095/biolreprod.103.025833. [DOI] [PubMed] [Google Scholar]
- Pilecka I, Sadowski L, Kalaidzidis Y, Miaczynska M. Recruitment of APPL1 to ubiquitin-rich aggresomes in response to proteasomal impairment. Experimental cell research. 2011;317(8):1093–1107. doi: 10.1016/j.yexcr.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Galan AK, Xin X, Dong F, Abdul-Ghani MA, Zhou L, Wang C, Li C, Holmes BM, Sloane LB, Austad SN, Guo S, Musi N, DeFronzo RA, Deng C, White MF, Liu F, Dong LQ. APPL1 potentiates insulin sensitivity by facilitating the binding of IRS1/2 to the insulin receptor. Cell reports. 2014;7(4):1227–1238. doi: 10.1016/j.celrep.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Jones CC, Huang S, Czech MP, Pilch PF. The interaction of Akt with APPL1 is required for insulin-stimulated Glut4 translocation. The Journal of biological chemistry. 2007;282(44):32280–32287. doi: 10.1074/jbc.M704150200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Margalet V. Stimulation of glycogen synthesis by insulin requires S6 kinase and phosphatidylinositol-3-kinase in HTC-IR cells. Journal of cellular physiology. 2000;182(2):182–188. doi: 10.1002/(SICI)1097-4652(200002)182:2<182::AID-JCP6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, Brand M, Zerial M. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133(3):486–497. doi: 10.1016/j.cell.2008.02.044. [DOI] [PubMed] [Google Scholar]
- Schultz JM, Khan SN, Ahmed ZM, Riazuddin S, Waryah AM, Chhatre D, Starost MF, Ploplis B, Buckley S, Velasquez D, Kabra M, Lee K, Hassan MJ, Ali G, Ansar M, Ghosh M, Wilcox ER, Ahmad W, Merlino G, Leal SM, Riazuddin S, Friedman TB, Morell RJ. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. American journal of human genetics. 2009;85(1):25–39. doi: 10.1016/j.ajhg.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya N, Gao CF, Xie Q, Gustafson M, Waters DJ, Zhang YW, Vande Woude GF. RNA interference reveals that ligand-independent met activity is required for tumor cell signaling and survival. Cancer research. 2004;64(21):7962–7970. doi: 10.1158/0008-5472.CAN-04-1043. [DOI] [PubMed] [Google Scholar]
- Takami T, Kaposi-Novak P, Uchida K, Gomez-Quiroz LE, Conner EA, Factor VM, Thorgeirsson SS. Loss of hepatocyte growth factor/c-Met signaling pathway accelerates early stages of N-nitrosodiethylamine induced hepatocarcinogenesis. Cancer research. 2007;67(20):9844–9851. doi: 10.1158/0008-5472.CAN-07-1905. [DOI] [PubMed] [Google Scholar]
- Tan Y, You H, Coffey FJ, Wiest DL, Testa JR. Appl1 is dispensable for Akt signaling in vivo and mouse T-cell development. Genesis. 2010a;48(9):531–539. doi: 10.1002/dvg.20657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, You H, Wu C, Altomare DA, Testa JR. Appl1 is dispensable for mouse development, and loss of Appl1 has growth factor-selective effects on Akt signaling in murine embryonic fibroblasts. The Journal of biological chemistry. 2010b;285(9):6377–6389. doi: 10.1074/jbc.M109.068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Zhang J, Dong LQ, Saunders E, Luo E, Tang J, Chen J. Adiponectin inhibits osteoclastogenesis and bone resorption via APPL1-mediated suppression of Akt1. The Journal of biological chemistry. 2011;286(14):12542–12553. doi: 10.1074/jbc.M110.152405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogiatzi P, Giordano A. Following the tracks of AKT1 gene. Cancer biology & therapy. 2007;6(10):1521–1524. doi: 10.4161/cbt.6.10.4834. [DOI] [PubMed] [Google Scholar]
- Wang C, Li X, Mu K, Li L, Wang S, Zhu Y, Zhang M, Ryu J, Xie Z, Shi D, Zhang WJ, Dong LQ, Jia W. Deficiency of APPL1 in mice impairs glucose-stimulated insulin secretion through inhibition of pancreatic beta cell mitochondrial function. Diabetologia. 2013;56(9):1999–2009. doi: 10.1007/s00125-013-2971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Xin X, Xiang R, Ramos FJ, Liu M, Lee HJ, Chen H, Mao X, Kikani CK, Liu F, Dong LQ. Yin-Yang regulation of adiponectin signaling by APPL isoforms in muscle cells. The Journal of biological chemistry. 2009;284(46):31608–31615. doi: 10.1074/jbc.M109.010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Yang Y, Wang Y, Xu A, Wu D, Chen Y. Appl1 is essential for the survival of Xenopus pancreas, duodenum, and stomach progenitor cells. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239(8):2198–2207. doi: 10.1002/dvdy.22356. [DOI] [PubMed] [Google Scholar]
- Yeo JC, Wall AA, Luo L, Stow JL. Rab31 and APPL2 enhance FcgammaR-mediated phagocytosis through PI3K/Akt signaling in macrophages. Molecular biology of the cell. 2015;26(5):952–965. doi: 10.1091/mbc.E14-10-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Chen J, Liu J, Brunzelle JS, Huang B, Wakeham N, Terzyan S, Li X, Rao Z, Li G, Zhang XC. Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J. 2007;26(14):3484–3493. doi: 10.1038/sj.emboj.7601771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.