Abstract
Organoid systems leverage the self-organizing properties of stem cells to create diverse multi-cellular tissue proxies. Most organoid models only represent single or partial components of a tissue, and it is often difficult to control the cell type, organization, and cell-cell/cell-matrix interactions within these systems. Herein, we discuss basic approaches to generate stem cell-based organoids, their advantages and limitations, and how bioengineering strategies can be used to steer the cell composition and their 3D organization within organoids to further enhance their utility in research and therapies.
Introduction
Model systems drive modern biological and biomedical research. These model systems aim to recapitulate body functions and processes from the molecular level to the cellular, tissue, organ, or whole organism level. The body can be viewed as a sum of a great number and wide variety of cellular and non-cellular materials formed in a highly organized manner (e.g., cell, tissue, and organ), as well as the entire interactome that includes internal (e.g., cell-cell, cell-matrix) or external (e.g., cell-environment) interactions. The hierarchical nature of all living beings suggests that multi-level recapitulation of the body could be achieved using model systems that consist of multiple cell types and their interactions (Figure 1).
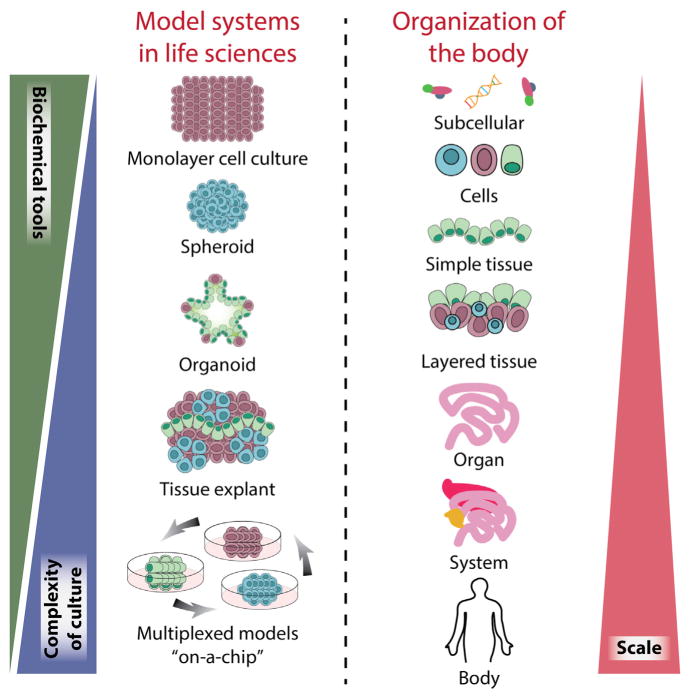
Figure 1. Model Systems in the Life Sciences.
Organisms comprise a hierarchy of systems from the subcellular level to the whole body. In the life sciences, many models have been developed across this organismal hierarchy, to address specific questions across biology and medicine. Each model system possess unique attributes; in general, with increasing scale comes increasing system complexity and challenges in cell culture and the reduced availability of biochemical and quantitative tools, which can limit study insights. Organoid models provide a unique opportunity to incorporate moderate system complexity while still affording many tools for probing structure and function. When compared to tissue explants, organoid systems can mimic similar cell-cell and cell-matrix interactions while maintaining the ability for long-term cultures thanks to maintained signaling cues important for survival.
Animal models most closely recapitulate in vivo human physiology, but they are limited by accessibility of imaging for observation, presence of confounding variables, limited throughput, limited usability, and differences between animal and human biology (Shanks et al., 2009). While simplistic models such as 2D monocultures of cell lines have their advantages, they often lack cell-cell and cell-matrix interactions that are required to maintain and define in situ phenotypes and thus fail to mimic cellular functions and signaling pathways present in tissues. Purified populations of primary cells also can lose their phenotype when cultured in 2D. 3D cell aggregate cultures of mesenchymal stem cells (MSCs) (Bartosh et al., 2010) or tumor cells (Vinci et al., 2012) exhibit improved function, though they lack relevant tissue organization present in vivo. Tissue explants or slices may transiently capture physiologically relevant cell organization and interactions, yet they tend to quickly lose their phenotype and are difficult to maintain for extended periods of time (Gähwiler et al., 1997). Other 3D culture systems include cell spheroids that often lack the presence of relevant stem or progenitor cell populations required to sustain the 3D culture and thus lack cells with the capacity for self-renewal and differentiation. While it is important to harness biological systems that can address specific scientific questions to achieve a balance between practicability and faithfulness, most current model systems exhibit a large gap between the cellular level and the tissue/organ level.
In general, stem cells exhibit an intrinsic ability to assemble into complex structures. When placed within a hydrogel (often Matrigel) and in the presence of suitable exogenous factors, the stem cells can be coaxed into forming structures that contain organized clusters of cells. The recent availability of stem cell-derived organoid systems to provide 3D self-organized tissue models provides a compelling new class of biological model to serve as both tissue and organ proxies (Lancaster and Knoblich, 2014). Organoids recapitulate a large number of biological parameters including the spatial organization of heterogeneous tissue-specific cells, cell-cell interactions, cell-matrix interactions, and certain physiological functions generated by tissue-specific cells within the organoid. Organoids bridge a gap in existing model systems by providing a stable system amenable to extended cultivation and manipulation, while being more representative of in vivo physiology.
While a wide variety of organoids have been generated, most organoid models only represent single or partial components of a tissue, and it is often difficult to control the cell type, organization, and cell-cell or cell-matrix interactions within these systems. Bioengineers have long aspired to deconstruct biological systems and manipulate or reconstruct the system in a controlled manner. Bioengineering approaches have enabled us to steer cell behavior and cell organization, which are fundamental processes in organoid formation, and improved systems are on the horizon. In this Review, we will discuss the basic principles in the process of organoid formation, their advantages and limitations, and how bioengineering approaches can be used to increase their utility in research and therapies.
Organoids: Self-Organizing Systems of Stem Cells and Their Progeny
Organoids have been generated from both pluripotent stem cells (PSCs) and adult stem cells (ASCs) by mimicking the biochemical and physical cues of tissue development and homeostasis (Lancaster and Knoblich, 2014). In a most simplified view, the development of the human body is a precisely controlled process of step-wise differentiation from the zygote and the subsequent self-organization of the cells generated in this process (Figure 2). This process can be partially reproduced when PSCs form a teratoma containing a variety of semi-organized tissues following uncontrolled differentiation and self-organization (Przyborski, 2005). Similarly, this process can be controlled in vitro with PSCs induced to differentiate down specific lineages. If provided the proper 3D scaffold and biochemical factors, differentiated cells from PSCs will self-organize to form tissue-specific organoids including the optic cup (Eiraku et al., 2011), brain (Lancaster et al., 2013), intestine (Spence et al., 2011), liver (Takebe et al., 2013), and kidney (Takasato et al., 2014). Additionally, the homeostasis of many tissues in vivo is maintained by tissue-specific ASCs through self-renewal and differentiation, followed by self-organization of the stem cells and their progeny. This process can also be reproduced in vitro under specific culture conditions to control self-renewal and differentiation, resulting in self-organized tissue organoids including intestine (Sato et al., 2009), stomach (Barker et al., 2010), liver (Huch et al., 2013), and pancreas (Huch et al., 2013).
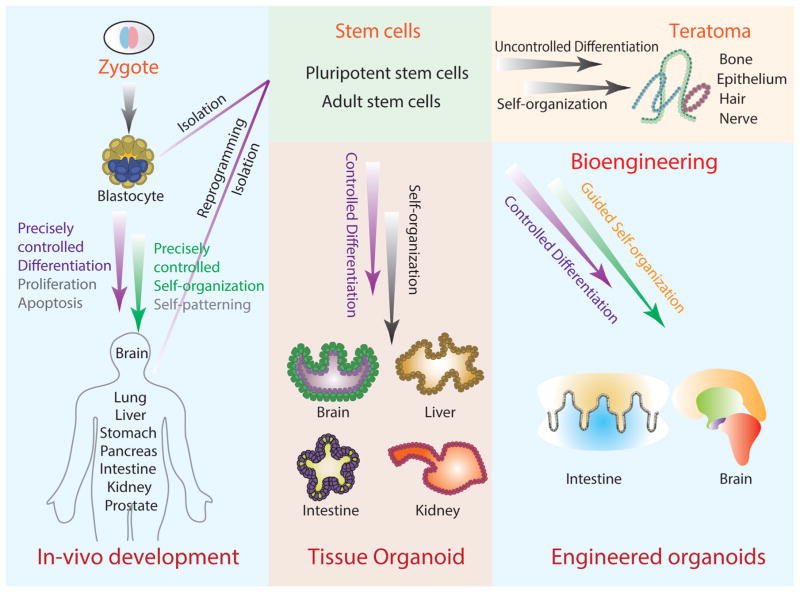
Figure 2. Organoid Development.
The process of organoid formation is similar to organism development originating from a zygote and giving rise to a mature adult organism. This includes precisely controlled differentiation, proliferation, and apoptosis paired with multi-cellular self-organization and patterning, which leads to diverse mature tissues. Organoid systems are derived from ESCs (isolated from a blastocyte), iPSCs (reprogrammed from adult tissues), or ASCs (isolated from mature tissues). The driver stem cell population undergoes a similar process of culture-controlled differentiation and self-organization to give rise to tissue-specific organoids. In the case of uncontrolled differentiation (and especially following transplantation), PSCs will produce teratomas, self-organized multi-tissue tumors. Bioengineering strategies can be used to further control differentiation and organization of organoid systems to be further developed into models more representative of in vivo tissues.
In the process of organoid formation, a number of common factors are used to control the self-renewal and differentiation of stem cells or assist self-organization. Growth factors or small molecules are used to manipulate multiple signaling pathways important in cell survival, proliferation, and self-renewal, often in a tissue-specific manner. Paired with the biochemical cues, Matrigel is a common and important component of the system that provides a scaffold and additional supplementation of signaling cues via basement membrane ligands to support cell attachment and survival as well as organoid formation (Xu et al., 2001). Often, organoid systems are governed by the stem cell microenvironment (or niche) they foster, which offers a point of control. The stem cell niche contains a wide range of elements including biochemical and biophysical signals, cell-cell interactions, and the extracellular matrix (ECM) interaction (Li and Xie, 2005). Current organoid systems mostly rely on intrinsic or extrinsic biochemical signals (e.g., growth factors) and cell-autonomous or cell-cell interactions to control the stem cell fate. Although these are all essential factors to control the differentiation and organization of the cells, organoid formation is highly dependent on cell-autonomous self-organization (Lancaster and Knoblich, 2014), which is not yet easily controlled.
During the establishment of organoid model systems, several bioengineering approaches that were developed in other fields including stem cell niche engineering and tissue engineering have become available to steer the behavior and organization of organoids. Here we will discuss these strategies and highlight examples of how bioengineering approaches can be used to increase control over organoid behavior, from tools at the subcellular level to those affecting system physiology.
Bioengineering Organoid Systems
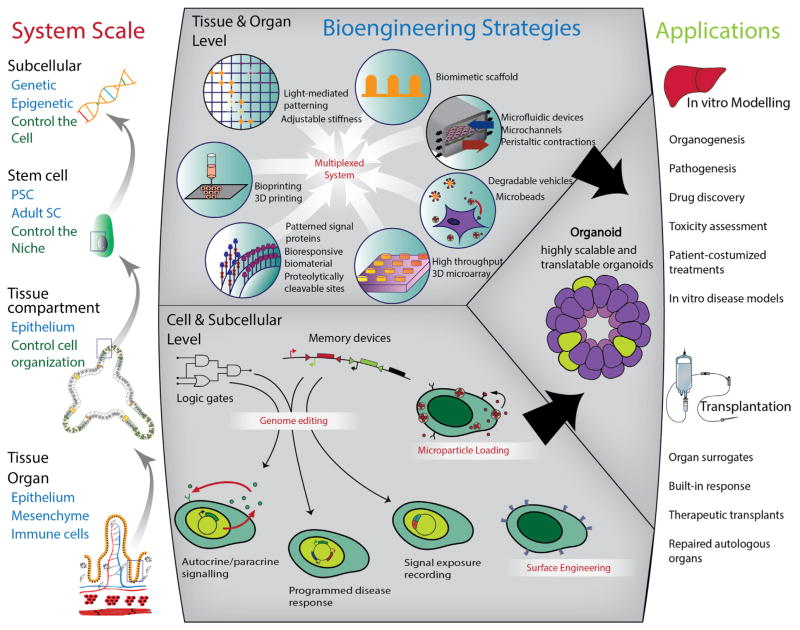
Organoid systems offer one of the most promising platforms for harnessing stem cells, specifically because they are capable of recapitulating many important properties of a stem cell niche and its resulting tissue. However, like any model system, gaps between in vitro and in vivo remain, which may in part be addressed with directed bioengineering efforts. These efforts may enhance the utility of organoids in drug screening, regenerative therapy, or studies of physiological and pathological processes. Bioengineering approaches can also be applied to develop bottom-up synthetic organoid constructs, or multiplexed organoid components, enabling improved system control and the development of additional models for basic and translational stem cell niche and organoid research.
Organoid systems leverage the self-renewal and differentiation capability of stem cells and the intrinsic self-organization ability to form organized structures. While stem cells are the “work horse” of the organoid system, the behavior of stem cells is controlled by the microenvironment. In organoids, niche components are derived by the cells (e.g., in the case of autocrine, paracrine, or juxtacrine signals) or exogenously added to the system (e.g., in the case of ECM substrates, small molecules, and growth factors). The interplay of these creates a dynamic environment in structure and function that is spatially and temporally coordinated and instructs the self-renewal/differentiation of the stem cells and self-assembly of the cells in organoids. To enhance control of organoids and further modulate the system for downstream applications, systematic engineering approaches are needed to manipulate each structural layer during the process of organoid formation. This may be achieved through modulating the cells generated by controlling stem cell self-renewal and differentiation, by directly modifying the stem cells, or by indirectly controlling them via manipulation of the microenvironment, as well as by modulating the organization of cells in the system. Recent advances in biomaterials, micro/nanotechnology, and stem cell-driven tissue assembly have enabled extensive progress toward such systems. By combining the novel approaches from these fields, it will be possible to design microenvironments that resemble in vivo structure and function, giving rise to several dynamic and self-assembled organoid tissues.
Design Process
A major aim of engineering organoid systems is to improve system utility in downstream applications. Thus, effort is required to create better proxies for in vivo tissues and organs and improve organoid system modularity to accommodate high-throughput formats or even multi-tissue organoid compatibility in larger multiplexed systems (e.g., human-on-a-chip). However, in practice, tradeoffs must be made to optimize the design of the system for its intended application while acting within technological constraints.
The intended function of an in vitro organoid system dictates the design specifications. For instance, modeling developmental processes or homeostasis in tissues dictates that closely mimicking the in vivo tissue is top priority. In that case, recapitulating complex niche components and interactions will be essential. For expanding human cell cultures or tissues for transplantation purposes, expanding stem cells in a simplified yet efficient system will be more suitable. System design will have to account for parameters such as scalability, as well as simplifying the retrieval of cells and minimizing perturbations from dynamic interactions, to maintain the homogeneity of the system. For drug screening, the system should have high prediction power by recapitulating critical (if not all) aspects of the target in vivo tissue. In addition, systems should enable simple readout, a necessity for high-throughput formats. Thus, bioengineering organoid systems involves the careful selection of essential culture components (based on previous investigations concerning the specific tissue in question), as well as a thorough consideration of the objectives of the study. This way, an ideal balance among simplicity, complexity, faithfulness, and controllability can be achieved.
Engineering the Niche
Stem cell behavior in vivo is highly regulated through the extrinsic biochemical and biophysical signals from specialized microenvironments. These microenvironments consist of a complex array of signaling mechanisms from niche support cells, the ECM, and mechanical forces, as well as systemic and physiochemical conditions such as oxygen and pH levels. The components that make up the dynamic environment integrate sustained and rapid short-term signals to either maintain a stem cell state of quiescence, or instead, induce developmental pathways or regenerative responses. In addition to this, the niche can be remodeled and directed by its stem cell constituents. Engineering a biomimetic system that incorporates each of these signaling pathways and interactions will enable better control over the growth and differentiation of stem cells in vitro and enable their manipulation in accordance with the intended application.
Customized biomimetic scaffolds
ECM is one of the main components of the stem cell niche, providing structural support and mediating instructive signaling for cell polarization, retention, and mobilization (Peerani and Zandstra, 2010). ECM components such as laminin, fibronectin, and collagen make up the physical framework of tissues and also influence cell behavior by engaging their integrin receptors (Vazin and Schaffer, 2010). Several approaches can be used to simulate the native ECM of a stem cell niche or mature tissue, including 3D scaffolds with microscale or nanoscale topography, producing customized biomaterials (Peerani and Zandstra, 2010). Alternatively, ECM scaffolds can also be produced from decellularized matrices. For example, upon reaching confluency in vitro, the underlying ECM matrix of bone marrow stromal cells can be decellularized and used to mimic endosteal or vascular niches along with the appropriate growth factors (Tan and Barker, 2013).
Recently introduced stem cell-derived organoids have been used to mimic aspects of human organogenesis and have achieved notable morphological similarities to their native counterparts (Eiraku et al., 2011; Lancaster et al., 2013; Stange et al., 2009). Matrigel is often a critical component of organoid culture. However, using Matrigel, which is generally not well defined, disregards the specific ECM cues required by different tissue types. Additionally, given its heterogeneous composition, it does not allow the morphogenetic processes, which are tightly governed in vivo by specific spatio-temporal cues, to be easily manipulated. As an alternative, essential signals from native ECMs can be incorporated into synthetic polymer matrices to produce designer ECMs, with specifically tailored compositions. For example, glycosaminoglycans such as hyaluronic acid (HA) are important ECM molecules that have shown to play an important part in modulating neural stem cell (NSC) and hematopoietic stem cell (HSC) behavior in their niche (Vazin and Schaffer, 2010), making them potentially suitable components of biomaterial-based systems for emulating NSC and HSC niche structure. Biomimetic scaffolds can be constructed from either synthetic polymers (such as polyacrylamine and polyethylene glycol [PEG]) or natural macromolecules (for instance, agarose or collagen) that can then be used to make them permissive to biological processes. For example, a biomimetic hydrogel scaffold constructed from pullulan (a polysaccharide polymer) complexed with collagen to mimic the epidermal niche ECM was able to enhance vascularization and healing when delivering MSCs to wound-sites (Rustad et al., 2012). Despite this progress, synthetic scaffolds remain primitive compared to substrates like Matrigel, and they lack the critical dynamic property of cell-driven remodeling.
Synthetic environments can be engineered to more closely replicate natural ECM by decorating bioinert matrices with signaling proteins via chemical/enzymatic crosslinking through adhesive or proteolytically cleavable sites. High-throughput screening of such platforms would enable analysis of multiple variables to determine which combination of signals is most suitable for modulating the stem cell activity and help design the synthetic ECM analogs to elicit a desired response. Cellular microarrays are capable of performing such high-throughput analysis. By tethering microenvironmental signals such as ECM components, soluble factors, and cell-cell interaction proteins to discrete locations, an array of artificial scaffolds can be generated (Gjorevski et al., 2014; Gobaa et al., 2011). Another method for providing relevant ECM support and signaling cues is micro-contact printing. This technique directly deposits proteins, ECM, or cells onto a partially polymerized hydrogel substrate. This is done using a stamp often made up of poly(dimethyl siloxane) (PDMS) using soft lithography techniques (Perl et al., 2009). However, morphogenesis and organogenesis are inherently 3D processes and thus their extrinsic regulation cannot be thoroughly understood under 2D analysis. Recently, this technology has been extended to a 3D microarray platform (Ranga et al., 2014), providing a high-throughput method of unveiling the influence of signaling proteins as well as matrix elasticity and degradability on stem cell regulation in a spatially relevant context. Achieving an ECM design that has physiologically relevant topography is also important. Nanolithography strategies such as electrospinning, electron-beam, nano-imprint lithography, and selective etching have been used to form nanofibrous substrates, nanopits, nanopillars, or nanochannels on various materials. Electron-beam lithography uses a computer-guided electron gun to scan the surface of substrates, producing patterns at nanoscale resolutions, while electrospinning is used to produce ultra-fine fibers that can form randomly oriented fibrous meshes suitable for a tissue engineering scaffold. Other methods include nano-imprint lithography (pressing a rigid mold into a layer of heated polymer) and selective etching (using a chemical etchant to roughen a surface). These strategies have been used to generate surface features that approach the natural shape and dimensions of the basement membrane fiber and pore sizes, also mimicking the porosity of natural ECM. These arrangements have been shown to support human embryonic stem cell self-renewal and control human MSC differentiation, based on the various configurations of these surfaces (Murphy et al., 2014).
Mechanical signaling applied by surrounding tissues also plays an important role in modulating cellular behavior in vivo. The absence of such forces in vitro can account for important morphological differences, such as the lack of villi formation in intestinal organoids (Gjorevski et al., 2014; Shyer et al., 2013). To dynamically tune the mechanical properties of the microenvironment, light-mediated patterning technology can be employed. A PEG hydrogel containing photolabile crosslinks can undergo local degradation when exposed to light, softening the gel. Alternatively, by including photo-initiators in the gel, shining light on specific regions will cause additional crosslinking and local stiffening (Guvendiren and Burdick, 2012). These light-dependent strategies can also be implemented for producing stiffness gradients at a microscale resolution. By photo-masking UV-cross-linked materials, the amount of light exposure can be used for reconstituting the natural stiffness variations within an artificial matrix (Vincent et al., 2013).
Adaptability to cell-induced modifications
An important step toward creating materials that can interface with cells is to engineer bioactive interfaces that offer both specificity and flexibility. To accomplish this, materials and surfaces must be rationally designed to impart specific biofunctionalities. For example, to design growth-promoting surfaces, generic cell adhesion motifs such as RGD (arginine-glycine-aspartic acid), or more cell-specific adhesion-promoting proteins and complementary receptors, can be used to engage cell integrins and promote adhesion and spreading. However, such materials must also be able to accommodate the changes brought on by the cellular activities that they are hosting.
Cells can actively modify a surface as they go through rapid and dynamic processes of cell adhesion and growth. As part of their natural growth process, cells begin to pull together and rearrange surface-bound biomolecules after adhering. On an engineered surface where ligands are covalently bound, this process is impeded. To resolve this, adhesion motifs like RGD can be linked to material surfaces by long, flexible tethers, allowing the cell to pull them into clusters (Kuhlman et al., 2007). Artificial systems that incorporate carefully arranged proteins are also susceptible to enzymatic cleavage as cells try to remodel their surrounding ECM for migration or growth. It is possible to graft shorter polypeptides, which are less susceptible to proteolysis. However, an alternative approach would be to design intentionally degradable materials that take advantage of the cell-secreted proteases. For example, by crosslinking polymers such as PEG with synthetic peptides recognizable by cell proteases, the point of cleavage can be controlled. Scaffold degradation, mediated by the cell itself, can be used to promote cell migration in a pre-designed orientation (Raeber et al., 2005).
Spatio-temporal control
In addition to establishing a 3D culture scaffold that can accommodate synthetic analogs of cell-ECM interactions, it is critical to integrate other niche components for the in vitro culturing of stem cells to drive organoid formation. A major challenge in constructing an artificial scaffold in vitro is to precisely replicate the spatial presentation of signals to cells. In traditional 3D cultures, cells are flooded with biochemical signals without any spatio-temporal control. This gives rise to the major differences observed between organogenesis in vivo and in vitro. Recent advances yielded unique approaches to overcome this limitation by manipulating the ECM using light-mediated patterning. For example, by incorporating biomolecule-binding sites into a hydrogel and masking their active sites with a photo-degradable moiety, it was possible to control MSC migration within a PEG hydrogel (Gjorevski et al., 2014; Kloxin et al., 2009). The light releases the active transglutaminase factor XIII (FXIIIa) substrate, allowing ECM proteins and growth factors to be tethered within the matrix based on a light-induced pattern. This mechanism offers a powerful tool for controlling the precise pattern, location, and time when a signal can be presented to a cell.
Soluble growth factors can also be delivered using microbe-ads or degradable vehicles within the culturing scaffold. Nano-particles loaded with growth factors can also be directly conjugated on the surface of the cell, slowly releasing molecules that will primarily be recaptured by the particle-carrying cells in an autocrine signaling loop (Stephan and Irvine, 2011). These methods allow some control over the release kinetics of signaling cues based on the design of the material. Bioresponsive biomaterials can also be constructed, where growth factors are released based on the release of matrix metalloproteinases from cells. PEG-based hydrogels have been fabricated with peptide sequences sensitive to proteolytic degradation, and they have been successfully used to deliver human fibroblast cells with bioactive molecules to help regenerate bone in vivo (Vazin and Schaffer, 2010). Microfluidic devices can deliver ligands as well, forming ligand gradients through the manipulation of the flow rate and the flow profile. Although this method may not readily allow macroscale architecture, it requires small sample sizes, which renders it a promising tool for high-throughput cell culture screening and analysis.
Bio-printing and bottom-up approaches to engineer organoids
To engineer organoids with controlled cell-cell and cell-matrix interactions, cells and materials can be directly deposited onto surfaces to produce 3D co-cultures of two or more cell types with customized geometries that can be pre-defined by imaging data via CT or MRI scans of the desired tissues or organ. Bio-printing evolved from 2D inkjet printing; instead of depositing drops of black ink onto a substrate, the ink comprises a biomaterial with living cells that is precisely positioned in an additive layer-by-layer approach to create 3D biological structures that mimic the structure and function of native tissues and organs (Atala and Yoo, 2015; Murphy and Atala, 2014). Current printers can be used to create complex components of the ECM with multiple materials containing multiple cell types (Atala and Yoo, 2015; Murphy and Atala, 2014). Hydrogel-based bio-inks containing nutrients to support cell survival and function, for example, are used to place cells in 3D printers, resulting in 3D tissue-like masses that are attempts to resemble native tissues (Atala and Yoo, 2015). For example, a beating heart organoid was recently created that was able to respond to electrical and chemical cues by altering its beating patterns. Other examples of bio-printing-created organoids are human liver, muscle, and blood microvessel organoids (Atala and Yoo, 2015). Despite great progress, several challenges remain, including the ability to print in high resolution, to achieve relevant and controllable cell densities, and to achieve long-term cell functionality of the bio-inks. In addition to bio-printing, bottom-up approaches have been demonstrated to provide microscale spatial control of cell-cell interaction. By first assembling microscale cell-laden constructs individually, and then inducing controlled multi-construct organization, it is possible to assemble spatially controlled cell aggregates (Du et al., 2008). Bottom-up approaches present an interesting possibility for the construction of controlled stem cell niches or the construction of multi-tissue organoid systems.
Vascularization
Permitting sufficient nutrient and oxygen supply is an additional important consideration in developing functional in vitro tissue structures and organoids. In vivo, organs consist of hierarchically branched vascular networks and almost all cells are within a few hundred microns of a capillary to ensure a sufficient supply through diffusion. Integrating a vascular structure that allows adequate delivery of oxygen and nutrients is a necessary step in fully recapitulating larger diffusion-limited organoids. One strategy for achieving this consists of a cell-based approach, where endothelial cells are seeded within the system in order to form new blood vessels, in a process known as neoangiogenesis. The other strategy is scaffold-based, where synthetic scaffolds are used to create micro-engineered 3D structures that are tunable in geometrical, mechanical, and biological properties. Many attempts to create in vitro vascularization have combined these approaches. Microfluidic devices that allow uniform distribution of flow and mass transfer can be produced using soft lithographic and micro-molding processes, with polymers such as PDMS, poly-lactic (co-glycolic acid) (PLGA), and poly-glycerol sebacate (PGS). Bio-printing methods can then be used for seeding the channels with multiple types of vascular cells (Golden and Tien, 2007; King et al., 2004; Visconti et al., 2010). An additional strategy involves a modular assembly process, wherein cells are seeded in collagen constructs small enough to avoid diffusion limitations, which are then coated with endothelial cells and combined to create larger perfusion-capable structures (McGuigan and Sefton, 2006). It should be noted that for organoid systems, these strategies will need to be modified so they would be able to integrate within the 3D macroscale tissue structures and allow perfusion and the specialized physiological functions of that tissue.
Scaffolds can also be functionalized with a combination of proangiogenic biomolecules, such as vascular endothelial growth factors (VEGFs), platelet-derived growth factors (PDGGFs), and basic fibroblast growth factors (BFGFs) for rapid formation of mature vascular networks (Richardson et al., 2001; Zisch et al., 2003). These angiogenic growth factors can not only trigger neo-vascularization, but they can also direct endothelial progenitor cell migration via gradients and promote cell assembly. Using a time-dependent release from biodegradable porous scaffolds or microparticles, these immobilized proteins can be delivered in a spatio-temporally controlled and sustained manner (Karal-Yilmaz et al., 2011; Layman et al., 2012).
Cell retrieval
Multiple strategies have emerged to release cells from surfaces with minimal impact on cell phenotype. For example, thermoresponsive polymers such as poly(N-isopropylacrylamide) and co-polymers such as di(ethylene glycol) methacrylate and a 9-mer oligo(ethylene glycol) methacrylate can change from a collapsed state to a hydrated, extended state (Lutz et al., 2006; Wischerhoff et al., 2008). In the extended state, the material becomes resistant to proteins, forming a layer that allows only a weak attraction between the cell and the surface, gently releasing the cells without the use of harsh chemical treatments or proteolytic cleavage.
Indeed, there is a significant need for new approaches that enable efficient retrieval of cells. For example, to produce sufficient quantities of human PSCs (hPSCs) for stem cell-based therapies, hPSCs must be rapidly and robustly expanded in culture and then collected. Similar to the example above, other synthetic polymer hydrogel systems are being developed to achieve this manufacturing goal. Poly(N-isopropylacrylamide)-co-poly(ethylene glycol) (PNIPAAm-PEG) is another hydrogel that possesses thermoresponsive properties. This hydrogel enables simple encapsulation and retrieval of hPSCs by going from liquid to a solid gel with temperature switches between 4°C and 37°C. This specific hydrogel matrix also demonstrates significantly increased cell expansion from 2D adherent formats, as well as a more homogenous population that retains its pluripotent phenotype (McDevitt, 2013), making it a scalable and compatible platform for manufacturing practices.
Monitoring niche components in vitro
Certain components of the stem cell niche, such as cell-cell interactions, oxygen distribution, local pH levels, and nutrient transport, are difficult to investigate in vivo, although there has been considerable recent progress. Sensors and devices that can be incorporated into the in vitro niche, with minimal perturbations to the natural processes, can enable precise monitoring of culturing conditions and help us better understand such parameters. Some of these capabilities have already been demonstrated in vivo. For example, using two-photon phosphorescence lifetime microscopy, local oxygen levels were measured in the bone marrow of live mice with micrometer spatial resolution (Spencer et al., 2014). The influence of cell-contact-mediated signaling in the stem cell niche can be further analyzed with the use of micro-scale devices. For example, microfabricated structures can be used to precisely control the spatial positioning of cells and study their interactions (Hui and Bhatia, 2007).
Additionally, 3D microfluidic devices, which can be readily monitored with several imaging modalities, are able to closely imitate key physiological and structural features of small functional units of organs and provide a platform for studying such biological systems in more detail. For example, microfluidic features like microchannels can permit fluid flow at rates that are observed in vivo. Microfluidic devices such as organ-on-a-chip technology can also mimic peristaltic contractions and the essential function of blood vessels for delivering oxygen and nutrients, while removing waste (Bhatia and Ingber, 2014; Huh et al., 2010). Miniaturized models of functional biological units have already been fabricated on a chip, including models for lung, liver, kidney, intestine, heart, fat, bone marrow, cornea, skin, and the blood-brain barrier (see Bhatia and Ingber, 2014 for review). Organ-on-a-chip systems are amendable to high-resolution, real-time imaging as well as analysis of biochemical, genetic, and metabolic processes under conditions that closely resemble in vivo conditions.
Although microfluidic-based organ-on-a-chip systems can integrate key components together and still allow precise control and measurement, certain features of conventional 3D organoid culture systems may still be more advantageous. For example, traditional 3D organoid culture systems can generate more tissue mass, allowing scientists to perform analytical experiments that usually require large samples. 3D cultures also allow the growth of macroscale architecture and highly complex and spatially heterogeneous tissues that cannot be supported at the microscale. Additionally, using microfluidic chip technology poses some experimental nuances. Fabrication of a chip requires micro-engineering capabilities, and the process is susceptible to bacterial contamination and bubble formation, which will interfere with cell health and chip function and fabrication. Nevertheless, despite these setbacks, microfluidic chips still offer an unprecedented flexibility in independently controlling and monitoring features such as cell and tissue position, fluid flow, and mechanical cues, helping to dissect their contribution to tissue and organ function. Most importantly, this technology reconciles a major drawback of macroscale 3D culturing. As functionality and complexity of 3D culture systems increases, certain tissues become more inaccessible. Hence, it becomes harder to perform high-resolution imaging and to track cell activity. Organ-on-a-chip allows cells to be easily integrated with fluorescence confocal microscopy, microflourimetry, trans-epithelial electrical resistance measurements, multiple electrode arrays, and other analytical systems (Bhatia and Ingber, 2014; Huh et al., 2010).
Bioprocessing and scale-up
One of the main challenges in translating tissue engineering to the clinic is the existing bio-processing gap including scalability and standardization issues. Scaling up the organoid platform to a production scale faces a similar hurdle, and future designs of the niche-mimetic organoid system should accommodate some degree of scale-up. Aggregate-based stirred suspension bioreactors have previously demonstrated the ability to control stem cell expansion and differentiation (Fluri et al., 2012; King and Miller, 2007; Ungrin et al., 2008). However, to further extend our control over stem cell behavior, bioreactors that can better re-create the stem cell niche using a two-phase system can be implemented (Kirouac and Zandstra, 2008). This consists of providing both bulk signals, such as those offered as physiological conditions of pH, oxygen tension, glucose levels, and temperature, and cell-level signals, such as those from cell-cell and cell-ECM interactions. Bulk signals can be provided using the exchange of culture media and controlled via sensors and process control loops, while microenvironment signals can be controlled by scaffold design and the same techniques of protein patterning used in lab-scale cultures. For such 3D systems, which require both continuous flow and substrate interaction, scaffold porosity and permeability will be important properties to consider. Other system characteristics, such as the ease of cell retrieval, and stringent process monitoring methods for product quality control, will also need to be incorporated. Ultimately, before organoids can transition to the clinic and be manufactured on a large scale, their dynamic response to system parameters must first be understood. This includes the derivation of mathematical models that can accurately predict system behavior and the identification of adjustable input parameters and robust cell markers for confirming output quality.
Model Organoid Systems: Applying Bioengineering Approaches
Intestinal Organoids
The intestinal epithelium is an actively renewing tissue fueled by Lgr5 intestinal stem cells (ISCs) located at the bottom of the intestinal crypts (Barker et al., 2007). The self-renewal and differentiation of ISCs is cooperatively controlled by signals from the underlining mesenchyme (e.g., BMP and Wnt) as well as cells in the epithelium, specifically Paneth cells (e.g., Notch and Wnt signals) (Sato et al., 2011b). The identification of Lgr5 ISCs and the knowledge of the signals controlling ISC behavior has collectively led to the establishment of intestinal organoids, where isolated ISCs are cultured in Matrigel with conditions that permit the self-renewal and differentiation of the stem cells, followed by self-organization of the generated cells (Sato et al., 2009).
Intestinal organoids recapitulate many aspects of the intestine in vivo (Figure 3). Within these organoids are all major cell types of the intestinal epithelium including enterocytes, entero-endocrine cells, Paneth cells, and goblet cells. Intestinal organoids also encompass crypt-villus structures, essential cell-cell interactions including the Paneth cell-stem cell axis, and functions including absorptive and secretory activities. However, intestinal organoids do not fully recapitulate the in vivo epithelium, evidenced by the lack of BMP signaling gradients—the BMP inhibitor Noggin diffuses throughout the organoid from the culture media (Sato et al., 2009). Moreover, in colon and human intestinal organoid culture, Wnt proteins and other factors (e.g., Tgf-β inhibitor, p38 inhibitor, Nicotinamide, etc.) are added, which essentially prevents the differentiation of the stem cells in culture, resulting in reduced diversity of cell types (Jung et al., 2011; Sato et al., 2011a). The incorporation of spatially controlled growth factor gradients or patterned ECM represents an attractive strategy to more closely mimic physiological conditions.
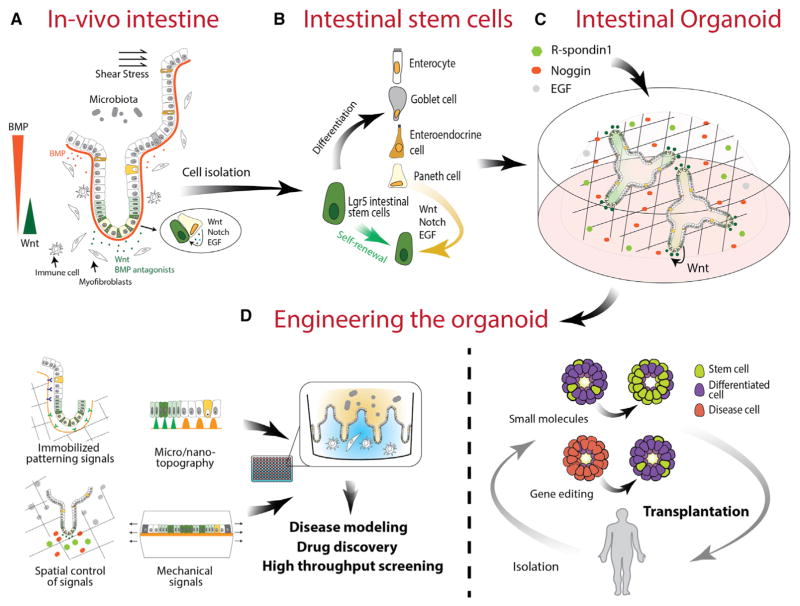
Figure 3. Intestinal Organoids.
(A) The intestinal epithelium encompasses a dynamic environment in which multiple cues drive rapid and sustained tissue turnover, which is emulated in part in organoid culture. The spatial arrangement of signaling cues and neighboring cells in vivo facilitates sustained epithelial regeneration from Lgr5+ stem cells.
(B) Lgr5+ stem cells are directly responsible for the generation of terminally differentiated epithelial cells including the Paneth cell, which directly provides cues for sustaining the stem cell niche.
(C) In organoid cultures, growth factors provided by the stem cell microenvironment and surrounding mesenchyme are supplemented with exogenous factors to sustain Lgr5+ stem cells and the organoid.
(D) By applying multiple engineering strategies, it is possible to further emulate the in vivo intestinal epithelium to build improved systems for disease modeling, drug discovery, and screening. Additionally, it is possible to use directed gene editing or small molecule treatment to drive organoids down particular paths of enrichment, thereby enabling the potential for organoid transplantation therapy.
In addition, modeling intestinal diseases often requires the presence of additional tissues of the intestine (i.e., immune cells and mesenchymal cells) and their interactions, which may have important disease implications (Lindemans et al., 2015). Direct access to the luminal compartment is also needed for studies on drug absorption, or microbe-epithelium interactions (Wilson et al., 2015). Such studies would also require a continuous and intact epithelial layer, which is absent in the organoid system. By combining spatial-temporal control of signals presented to the cells, and guided cell organization with structured scaffolds, these important targets could be incorporated. Directed organization of the cells will also have the potential to introduce additional cell types from lineages other than the epithelium, including immune and mesenchymal cells, enabling improved disease modeling.
Also of interest is the direct use of intestinal organoids as therapy. Intestinal organoids have been transplanted into damaged colon for tissue repair (Yui et al., 2012), but only with limited engraftment success. For this purpose, highly efficient expansion of a pure population of ISCs in a biochemically defined system (free of Matrigel) would be the goal, along with delivery techniques to improve targeting of ulcers, graft survival, and engraftment. Recently we have shown that small molecules can be used to significantly increase the expansion efficiency of ISCs (Yin et al., 2014).
Brain Organoids
The human brain embodies biological system complexity, and its development involves a high degree of coordination between the NSCs and the dynamic niche in which they exist. Through providing different levels of morphogens (i.e., BMP, Wnt, Shh, RA, and FGF), PSCs can be induced to differentiate into many different neural subtypes, such as cortical pyramidal neurons (Espuny-Camacho et al., 2013), midbrain dopaminergic neurons (Chambers et al., 2009; Lee et al., 2000; Perrier et al., 2004; Yan et al., 2005), and spinal cord motor neurons (Dimos et al., 2008; Li et al., 2005; Soundararajan et al., 2006; Wichterle et al., 2002) (see Petros et al., 2011 for review). Furthermore, the SFEBq (serum-free floating culture of embryoid body-like aggregates with quick re-aggregation) protocol has been used to generate the more complex architectures, such as sub-brain regions like the cerebral cortex (Danjo et al., 2011; Eiraku et al., 2008; Kadoshima et al., 2013; Mariani et al., 2012) and the pituitary (Suga et al., 2011). Alternatively, Lancaster et al. reported a culture system to generate heterogeneous neural organoids that contained multiple, but interdependent, brain regions within individual organoids (Lancaster et al., 2013). Here, the generated neuroectodermal tissues were maintained in 3D Matrigel for further expansion, without the addition of neural inducing or patterning factors. When transferred to a spinning bioreactor as Matrigel droplets, the cerebral organoids also showed enhanced nutrient absorption and grew as large as 4 mm in 2 months, and they generated distinct brain regions such as the dorsal cortex, ventral telencephalon, choroid plexus, hippocampus, and retina.
Although the cerebral organoid system has achieved some ability to model human brain development, several limitations still exist. Specifically, owing to the absence of surrounding tissue (and tissue cross-communication) and body axis, the current models are not organized to form the brain shape and structure as they exist in vivo. It is also worth noting that although culturing without any patterning factors has improved the developed cerebral organoids, patterning factors are necessary to ensure some level of controlled tissue organization. Bioengineering approaches such as spatial-temporal control of differentiation or cell patterning signals using customized scaffolds with immobilized signals, or signal gradient formation using control-released particles or microfluidics, will have the potential to guide the differentiation and patterning of brain regions in the organoids. Nutrient and oxygen delivery limits the size of cerebral organoids and may lead to undesired differentiation in regions of poor supply, despite the addition of agitation. This also contributes to stochastic growth patterns and limited maturation of key cell types in the brain organoids (Chambers et al., 2013). A potential solution exists in implementing co-cultures that can vascularize brain organoids or implementing microfluidic perfusion networks (Figure 4).
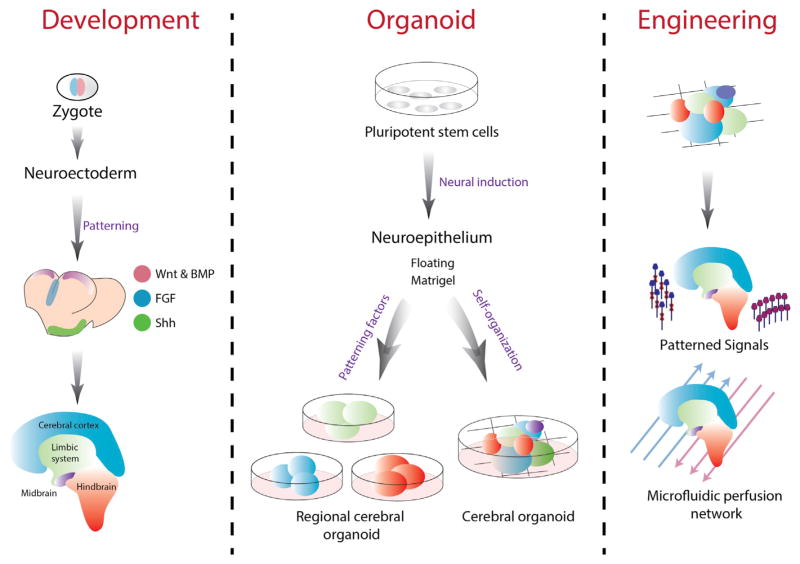
Figure 4. Brain Organoids.
Brain organoids follow a modified path of in vivo development, where ESCs develop into the brain structure following specific spatio-temporal cues, ultimately leading to the mature brain structure. In vitro this process begins with either ESCs or iPSCs, and following structural and biochemical cues, multiple organoid lineages can be produced. This includes the self-organized multi-regional organoids or the patterned induction of organoids mimicking specific regions of the brain. A promising set of tools including customized and responsive biomaterials, patterned signaling, and microfluidic networks can be applied to further refine the spatial development of brain organoids.
Future Directions: Bioengineering Strategies to Advance Organoid-Based Therapies
Targeted Genome Editing: Enabling Tighter Control of the Stem Cell Niche
A major limitation to constructing more sophisticated organoids in vitro is that current control of driver stem cell fate and behavior is primarily achieved through exogenous signals. Targeted genome editing, especially via CRISPR or TALEN technology, is an extremely powerful tool to accurately manipulate endogenous genes in clinically relevant cells and organisms. This approach can be used to enhance niche function, in which we can not only steer cell fate through exogenous niche components, but also reprogram the internal decision-making structure of driver and support cells. Reprogramming cells in organoid systems would enable user-defined “training” in how these structures self-assemble and self-regulate, driving improved studies of niche environments and their impact on organoid formation and function.
For instance, future studies may enable organoids derived from patients with single-gene hereditary diseases, such as cystic fibrosis (CF) or beta thalassemia, to undergo CRISPR genome editing to correct the mutation and then undergo transplantation as functional organ-like units back into patients to advance tissue repair and functionality. A proof-of-concept use of genome editing in the organoid field was recently demonstrated by Schwank et al. (2013). First, efficient genome editing of human stem cells in primary intestinal organoids via CRISPR/Cas9 was demonstrated. Then, using CRISPR-Cas9-mediated HDR, intestinal organoids from two CF patients were genome edited to correct the mutation (deletion of phenylalanine at position 508) of the CF transmembrane conductor receptor (CFTR), the primary cause of the disease. It was demonstrated that the genome-edited intestinal organoid systems expressed the corrected CFTR allele, which also produced fully functional proteins (involved in chloride ion channels) in these organoids. This study demonstrates genome editing of organoids as a potential gene therapy strategy, with limited risks to off-target tissue mutagenesis. Another example is microvillus inclusion disease (MVID), in which patients display microvillus inclusion and loss of brush-border microvilli, resulting in life-threatening persisting diarrhea. Mutations in myosin Vb and Syntaxin were demonstrated to cause classic and variant MVID, respectively. Establishing patient-derived intestinal organoids to be genome edited to correct those mutations and then re-transplanted into the patients may be useful in improving intestinal function. We envision that in the near future, stem cells from patients with hereditary diseases will be used to establish organoids, which will then undergo direct genome editing and be transplanted as autologous therapy (Yui et al., 2012), correcting the tissue-specific functional defects. Such a strategy may provide a cure to currently incurable hereditary diseases such as CF.
Alternatively, tools for precise gene editing in organoids can be utilized for elucidation of signaling pathways responsible for disease development. By using CRISPR gene editing, Matano et al. introduced multiple mutations in niche signaling pathways into human intestinal epithelial organoids, which were cultured in vitro and then transplanted into mice so their role in tumor progression and micrometastases could be studied (Matano et al., 2015). Overall, the rapidly evolving genome-editing approaches possess immense promise in creating next-generation organoid culture systems, advancing the study of organogenesis, pathogenesis, and drug-screening-based and organoid-based therapies. In this case, the organoid system paired with controlled editing of niche direction presented an ideal platform for the study of the stem cell niche in tumor generation and propagation.
Genetic Circuits: Programming Organoids
A major need in organoid research is to control the response of the niche cells to changing stimuli—this applies for in vitro organoid systems as well as for organoids under in vivo settings (for example, upon transplantation). Synthetic biology is emerging as a promising field with great potential for developing the next generation of therapeutics and diagnostics (Purcell and Lu, 2014). Relying on basic molecular biology components, artificial gene circuits have produced programmable and responsive systems within living cells. With continuous improvements of these biological components, as well as the construction of higher-order devices such as switches, memory elements, cascades, time-delayed circuits, oscillators, and logic gates, artificial gene circuits have achieved sophisticated cellular computational capabilities in both single-cell and multi-cellular systems (Cheng and Lu, 2012). Transcriptional regulation, cellular memory storage, and integration of logic gates are all tools that enable the complex computational abilities of genetic circuits, and they can be incorporated into organoid systems for an additional layer of control over their behavior during culture and after transplantation. For example, organoids could be programmed to reach tissue-like homeostasis, or execute diverse functions from the multiple terminal cell populations contained within.
Many synthetic circuits have been designed using digital logic gates, relying on transcriptional control using activators, repressors, and other novel mechanisms (Lohmueller et al., 2012). Furthermore, synthetic biologists are now able to connect many single logic gates into multiple configurations to achieve more sophisticated genetic programs. One successful approach to interconnecting logic gates is to use diffusible signaling molecules. AHL (a diffusible quorum-sensing molecule) was used to connect logic gates between Escherichia coli cells, producing a complex network of light-sensitive “edge-detectors” (Tabor et al., 2009), an achievement that can potentially one day be incorporated in organoid systems to direct cellular organization. While unwanted crosstalk among the synthetic devices, signaling delays between layered circuits, and other limitations may limit the exclusive use of digital gates (Purcell and Lu, 2014), integrating both analog and digital processing may eventually achieve efficient cellular computation by reducing the size of genetic circuits and thus alleviating the cellular burden.
Although performed thus far mostly in bacteria, such as E. coli, the field of synthetic biology is continuously advancing, and soon mammalian cells, including niche-establishing PSCs, will be successfully engineered using similar synthetic biology approaches. We envision that genetic circuit-based coordination between multiple cell types has the potential to revolutionize organoid engineering. Using genetic circuits to engineer multiple types of responsive cells, an organoid structure may be generated rapidly upon administration of specific cues. In vitro niche designs can also be simplified by using reprogramed cells that produce their own signals. As organoids grow in size, it will be possible to initiate an autocrine signaling pathway for cells that are inaccessible to exogenous signals. Applying logic gates in these genetic systems will help to fine-tune such responses. Furthermore, organoids may be programmed to respond to specific signals upon their transplantation (i.e., differentiation of cells in the organoids based on a decline of a specific hormone or the presence of other cues during the course of disease). These organoids could gauge information about their local environment and have a pre-programmed built-in response based on their intended function (e.g., therapeutic), further advancing organoid-based therapies.
In a system such as an organoid, multiple niche interactions take place in a highly dynamic and transient manner. Orchestrating the delivery of such a complex array of signaling cues in vitro poses a significant engineering challenge. Applying cellular memory devices within niche cells can help to alleviate this problem. Cellular memory is an essential function that enables the storage of otherwise transient responses. A key development in the field of synthetic biology has been the creation of sophisticated memory devices, which are able to mimic cellular memory via genomically integrated circuits. Certain synthetic memory networks use positive feedback loops to achieve stable states and switch between these states based on repressive or activating inputs (Purcell and Lu, 2014). Other approaches involve the recombinase-catalyzed reconfiguration of DNA (recently demonstrated in E. coli), which can serve as both a method of implementing logic and a way to record information by embedding a memory of the received inputs into the DNA—a stable storage medium that persists even after cell death (Siuti et al., 2013). This platform can also be used for creating biosensors that record the history of cellular exposure to either individual or a sequence of environmental signals (Siuti et al., 2013). Such tools can be applied to niche engineering, as a way to record and analyze niche input or to program cellular response. For example, by storing certain signaling cues in the cellular memory, it is possible to simplify the delivery of exogenous signals and apply them in sequence rather than simultaneously.
Additionally, because organoid systems arise from clonal stem cell populations, the details of circuit construction are simplified to targeting the “driver” stem cells, and pairing circuitry with the transcriptional programs that naturally drive differentiation within organoids to obtain multiple circuits acting in synchrony. Also, for large-scale culturing processes that require a continuous supply of growth factors, memory devices can be used to activate the desired response with singular/discreet exposure, cutting manufacturing costs. Future transplantation of memory-equipped organoids can also be extremely useful to increase organoid survival and maintain an extended organoid response in the body post-transplantation. For example, organoid systems can be “trained” to bypass certain physiological conditions upon transplantation, ignoring an anticipated sequence of signals based on a pre-programmed response.
Conclusions
The use of organoid platforms has led to advancements in in vitro organogenesis and disease modeling, and subsequently, it has created exciting possibilities for the development of innovative new therapies. Important characteristics of multiple tissues and organ sub-regions, such as the formation of distinct brain regions of the dorsal cortex, ventral telencephalon, choroid plexus, or hippocampus and crypt-villus structures similar to those in the intestinal epithelial lining have been successfully recapitulated in 3D organoid models. In addition to the development of new biological models and tools for studying and manipulating tissue regeneration, organoid models can be used to model disease states, and they can also potentially be used to develop more predictive drug screening platforms and patient-specific treatments.
The utility of existing spheroid and tissue explant cultures can be attributed to their ability to mimic the complex niche interactions present in situ. Organoids provide a more advanced in vitro tool that enables more physiologically relevant experiments to be performed that cannot be conducted in animals or people. With the currently available extensive arsenal of bioengineering methods, it is possible to extend the utility of organoids with improved control over external cues and with an unprecedented opportunity to monitor and manipulate cellular behavior. Influencing how cells internally process exogenous signals offers a new layer of control, allowing the fine-tuning of organoids with genome editing and genetic circuits (Figure 5).
Figure 5. Bioengineering Approaches to Advance Organoid-Based Research and Therapy.
With the right combination and sequence of input signals that the niche relays to the cell, it is possible to obtain the desired output (i.e., the in vitro disease model or tissue-specific organoid) and there are multiple bioengineering tools that can be harnessed to modify these signals and monitor relevant responses. Based on the same principle, following elucidation of organoid biology, there is potential to harness new knowledge to create synthetic niches. The niche can be engineered by combining multiple bioengineering techniques that mimic specific niche components (e.g., biomimetic scaffolds, tunable stiffness, appropriate topography, and spatio-temporally controlled signaling cues). The stem cells used to seed the organoid culture can also be engineered. In addition to exogenous signaling mechanisms, cell activity can be controlled though genome editing and surface modifications including drug delivery nano/microparticles. Using these methodologies, we can gain better control over the organoid to maximize functionality and sustainability in culture and ideally more closely mimic in vivo biology.
However, this is not without its challenges. While it is clearly important to not only continue to expand this library of bioengineering methods and determine how best to partner relevant technologies with unmet needs in the creation and manipulation of organoids, moving from in vitro cell monolayer model systems, which can be readily standardized, to multi-phenotype models that are combined with diverse bioengineering tools will create a standardization nightmare. This will undoubtedly make it difficult for groups to compare results between systems, as has been observed for products derived from iPSCs. It is also critical to consider challenges associated with using bioengineering strategies without interfering with the system’s natural ability to individually tune each parameter, and to consider challenges to simultaneously and simply control organoids from micro-to-macroscopic levels and to maintain them in culture. Regardless, organoid systems have already found utility in many basic biological and therapeutic experiments to advance new knowledge and to advance us closer to therapies for diseases that previously appeared untouchable.
Acknowledgments
This work was supported by National Institutes of Health grant HL095722 to J.M.K. and NIH grant DE013023 to R.S.L.
References
- Atala A, Yoo JJ. Essentials of 3D Biofabrication and Translation. Elsevier Science; 2015. [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Bartosh TJ, Ylöstalo JH, Mohammadipoor A, Bazhanov N, Coble K, Claypool K, Lee RH, Choi H, Prockop DJ. Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their anti-inflammatory properties. Proc Natl Acad Sci USA. 2010;107:13724–13729. doi: 10.1073/pnas.1008117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Tchieu J, Studer L. Build-a-brain. Cell Stem Cell. 2013;13:377–378. doi: 10.1016/j.stem.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Cheng AA, Lu TK. Synthetic biology: an emerging engineering discipline. Annu Rev Biomed Eng. 2012;14:155–178. doi: 10.1146/annurev-bioeng-071811-150118. [DOI] [PubMed] [Google Scholar]
- Danjo T, Eiraku M, Muguruma K, Watanabe K, Kawada M, Yanagawa Y, Rubenstein JL, Sasai Y. Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. J Neurosci. 2011;31:1919–1933. doi: 10.1523/JNEUROSCI.5128-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Du Y, Lo E, Ali S, Khademhosseini A. Directed assembly of cell-laden microgels for fabrication of 3D tissue constructs. Proc Natl Acad Sci USA. 2008;105:9522–9527. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Fluri DA, Tonge PD, Song H, Baptista RP, Shakiba N, Shukla S, Clarke G, Nagy A, Zandstra PW. Derivation, expansion and differentiation of induced pluripotent stem cells in continuous suspension cultures. Nat Methods. 2012;9:509–516. doi: 10.1038/nmeth.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gähwiler BH, Capogna M, Debanne D, McKinney RA, Thompson SM. Organotypic slice cultures: a technique has come of age. Trends Neurosci. 1997;20:471–477. doi: 10.1016/s0166-2236(97)01122-3. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Ranga A, Lutolf MP. Bioengineering approaches to guide stem cell-based organogenesis. Development. 2014;141:1794–1804. doi: 10.1242/dev.101048. [DOI] [PubMed] [Google Scholar]
- Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Artificial niche microarrays for probing single stem cell fate in high throughput. Nat Methods. 2011;8:949–955. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- Golden AP, Tien J. Fabrication of microfluidic hydrogels using molded gelatin as a sacrificial element. Lab Chip. 2007;7:720–725. doi: 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- Guvendiren M, Burdick JA. Stiffening hydrogels to probe short-and long-term cellular responses to dynamic mechanics. Nature Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci USA. 2007;104:5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karal-Yilmaz O, Serhatli M, Baysal K, Baysal BM. Preparation and in vitro characterization of vascular endothelial growth factor (VEGF)-loaded poly(D,L-lactic-co-glycolic acid) microspheres using a double emulsion/solvent evaporation technique. J Microencapsul. 2011;28:46–54. doi: 10.3109/02652048.2010.523795. [DOI] [PubMed] [Google Scholar]
- King JA, Miller WM. Bioreactor development for stem cell expansion and controlled differentiation. Curr Opin Chem Biol. 2007;11:394–398. doi: 10.1016/j.cbpa.2007.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KR, Wang CCJ, Kaazempur-Mofrad MR, Vacanti JP, Borenstein JT. Biodegradable microfluidics. Adv Mater. 2004;16:2007. [Google Scholar]
- Kirouac DC, Zandstra PW. The systematic production of cells for cell therapies. Cell Stem Cell. 2008;3:369–381. doi: 10.1016/j.stem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman W, Taniguchi I, Griffith LG, Mayes AM. Interplay between PEO tether length and ligand spacing governs cell spreading on RGD-modified PMMA-g-PEO comb copolymers. Biomacromolecules. 2007;8:3206–3213. doi: 10.1021/bm070237o. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman H, Li X, Nagar E, Vial X, Pham SM, Andreopoulos FM. Enhanced angiogenic efficacy through controlled and sustained delivery of FGF-2 and G-CSF from fibrin hydrogels containing ionic-albumin micro-spheres. J Biomater Sci Polym Ed. 2012;23:185–206. doi: 10.1163/092050610X546417. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA, Zhang SC. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015 doi: 10.1038/nature16460. in press Published online December 9, 2105 http://dx.doi.org/10.1038/nature16460. [DOI] [PMC free article] [PubMed]
- Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–5187. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz JF, Akdemir O, Hoth A. Point by point comparison of two thermosensitive polymers exhibiting a similar LCST: is the age of poly(NIPAM) over? J Am Chem Soc. 2006;128:13046–13047. doi: 10.1021/ja065324n. [DOI] [PubMed] [Google Scholar]
- Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, Vaccarino FM. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- McDevitt TC. Scalable culture of human pluripotent stem cells in 3D. Proc Natl Acad Sci USA. 2013;110:20852–20853. doi: 10.1073/pnas.1320575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan AP, Sefton MV. Vascularized organoid engineered by modular assembly enables blood perfusion. Proc Natl Acad Sci USA. 2006;103:11461–11466. doi: 10.1073/pnas.0602740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peerani R, Zandstra PW. Enabling stem cell therapies through synthetic stem cell-niche engineering. J Clin Invest. 2010;120:60–70. doi: 10.1172/JCI41158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A, Reinhoudt DN, Huskens J. Microcontact Printing: Limitations and Achievements. Adv Mater. 2009;21:2257–2268. [Google Scholar]
- Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, Harrison NL, Studer L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros TJ, Tyson JA, Anderson SA. Pluripotent stem cells for the study of CNS development. Front Mol Neurosci. 2011;4:30. doi: 10.3389/fnmol.2011.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przyborski SA. Differentiation of human embryonic stem cells after transplantation in immune-deficient mice. Stem Cells. 2005;23:1242–1250. doi: 10.1634/stemcells.2005-0014. [DOI] [PubMed] [Google Scholar]
- Purcell O, Lu TK. Synthetic analog and digital circuits for cellular computation and memory. Curr Opin Biotechnol. 2014;29:146–155. doi: 10.1016/j.copbio.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. 3D niche microarrays for systems-level analyses of cell fate. Nat Commun. 2014;5:4324. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson TP, Peters MC, Ennett AB, Mooney DJ. Polymeric system for dual growth factor delivery. Nat Biotechnol. 2001;19:1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- Rustad KC, Wong VW, Sorkin M, Glotzbach JP, Major MR, Rajadas J, Longaker MT, Gurtner GC. Enhancement of mesenchymal stem cell angiogenic capacity and stemness by a biomimetic hydrogel scaffold. Biomaterials. 2012;33:80–90. doi: 10.1016/j.biomaterials.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011a;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011b;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ, Mahadevan L. Villification: how the gut gets its villi. Science. 2013;342:212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31:448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- Soundararajan P, Miles GB, Rubin LL, Brownstone RM, Rafuse VF. Motoneurons derived from embryonic stem cells express transcription factors and develop phenotypes characteristic of medial motor column neurons. J Neurosci. 2006;26:3256–3268. doi: 10.1523/JNEUROSCI.5537-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JA, Ferraro F, Roussakis E, Klein A, Wu J, Runnels JM, Zaher W, Mortensen LJ, Alt C, Turcotte R, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269–273. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan MT, Irvine DJ. Enhancing Cell therapies from the Outside In: Cell Surface Engineering Using Synthetic Nanomaterials. Nano Today. 2011;6:309–325. doi: 10.1016/j.nantod.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, Takata N, Wataya T, Muguruma K, Miyoshi H, et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature. 2011;480:57–62. doi: 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG, Little MH. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Tan S, Barker N. Engineering the niche for stem cells. Growth Factors. 2013;31:175–184. doi: 10.3109/08977194.2013.859683. [DOI] [PubMed] [Google Scholar]
- Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS ONE. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazin T, Schaffer DV. Engineering strategies to emulate the stem cell niche. Trends Biotechnol. 2010;28:117–124. doi: 10.1016/j.tibtech.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Vincent LG, Choi YS, Alonso-Latorre B, del Álamo JC, Engler AJ. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol J. 2013;8:472–484. doi: 10.1002/biot.201200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, Lomas C, Mendiola M, Hardisson D, Eccles SA. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti RP, Kasyanov V, Gentile C, Zhang J, Markwald RR, Mironov V. Towards organ printing: engineering an intra-organ branched vascular tree. Expert Opin Biol Ther. 2010;10:409–420. doi: 10.1517/14712590903563352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wilson SS, Tocchi A, Holly MK, Parks WC, Smith JG. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 2015;8:352–361. doi: 10.1038/mi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischerhoff E, Uhlig K, Lankenau A, Börner HG, Laschewsky A, Duschl C, Lutz JF. Controlled cell adhesion on PEG-based switchable surfaces. Angew Chem Int Ed Engl. 2008;47:5666–5668. doi: 10.1002/anie.200801202. [DOI] [PubMed] [Google Scholar]
- Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- Yan Y, Yang D, Zarnowska ED, Du Z, Werbel B, Valliere C, Pearce RA, Thomson JA, Zhang SC. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- Zisch AH, Lutolf MP, Hubbell JA. Biopolymeric delivery matrices for angiogenic growth factors. Cardiovasc Pathol. 2003;12:295–310. doi: 10.1016/s1054-8807(03)00089-9. [DOI] [PubMed] [Google Scholar]