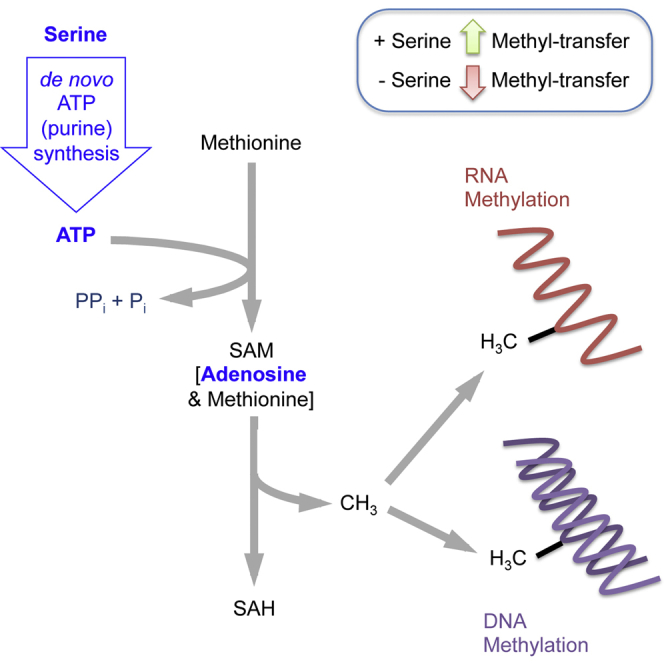
Summary
Crosstalk between cellular metabolism and the epigenome regulates epigenetic and metabolic homeostasis and normal cell behavior. Changes in cancer cell metabolism can directly impact epigenetic regulation and promote transformation. Here we analyzed the contribution of methionine and serine metabolism to methylation of DNA and RNA. Serine can contribute to this pathway by providing one-carbon units to regenerate methionine from homocysteine. While we observed this contribution under methionine-depleted conditions, unexpectedly, we found that serine supported the methionine cycle in the presence and absence of methionine through de novo ATP synthesis. Serine starvation increased the methionine/S-adenosyl methionine ratio, decreasing the transfer of methyl groups to DNA and RNA. While serine starvation dramatically decreased ATP levels, this was accompanied by lower AMP and did not activate AMPK. This work highlights the difference between ATP turnover and new ATP synthesis and defines a vital function of nucleotide synthesis beyond making nucleic acids.
Graphical Abstract
Highlights
-
•
Serine supports the methionine cycle through de novo ATP synthesis
-
•
De novo ATP-synthesis is an important source of ATP in cancer cells
-
•
Methionine fed colorectal cancer cells do not detectably re-methylate homocysteine
-
•
A method to track methyl units from amino acids onto DNA and RNA is described
The methionine cycle generates S-adenosyl methionine (SAM), the primary cellular methyl donor. Serine may provide one-carbon units to regenerate methionine from homocysteine. Maddocks et al. show that in colorectal cancer cells serine also maintains the methionine cycle through de novo ATP synthesis, which supports the conversion of methionine to SAM.
Introduction
The methionine cycle provides methyl units for a variety of reactions such as the methylation of proteins, DNA, RNA and lipids, allowing for the modulation of their biological functions (Bauerle et al., 2015, Gut and Verdin, 2013). S-adenosyl methionine (SAM) is the primary methyl donor molecule utilized in cellular methylation reactions and is synthesized directly from the essential amino acid methionine (Cantoni, 1952) (Figure S1A). The function of methylation reactions is context and substrate dependent; for example, DNA and histone methylation are epigenetic modifications that have important influences on chromatin structure and regulation of gene expression (Cedar and Bergman, 2009, Gut and Verdin, 2013). Previously, DNA methylation was perceived as a relatively stable epigenetic modification with the propensity to encode heritable epigenetic information (Dolinoy, 2007, Dolinoy et al., 2007, Suzuki and Bird, 2008). However, recent work has highlighted the importance of dynamic control of DNA methylation, for example, in embryogenesis (Guo et al., 2014a, Guo et al., 2014b, Shen et al., 2014, Smith et al., 2014), cardiomyocyte development (Gilsbach et al., 2014), and actively transcribed and regulatory regions of DNA (Kangaspeska et al., 2008, Métivier et al., 2008, Schübeler, 2015). It is therefore increasingly appreciated that existing DNA, as well as newly synthesized DNA, can be dynamically methylated and demethylated (Bhutani et al., 2011, Byun et al., 2012, Feldmann et al., 2013, Kohli and Zhang, 2013, Yu et al., 2012). Accordingly, it is important to define the cellular processes that control dynamic methylation of nucleic acids.
As the fields of epigenetics and cellular metabolism—particularly cancer cell metabolism—have developed in recent years, so has the appreciation of the fundamental crosstalk between these processes (Gut and Verdin, 2013, Hino et al., 2013, Nordgren and Skildum, 2015). Previous studies have shown that histone modifications are responsive to metabolite levels; for example, glucose-derived acetyl-CoA influences histone acetylation via ATP-citrate lyase (Wellen et al., 2009). In gliomas, IDH1 mutation is responsible for the generation of 2HG, which inhibits the DNA hydroxylase TET and leads to altered methylation of histones and DNA, so driving the phenotype of these tumors (Christensen et al., 2011, Figueroa et al., 2010, Turcan et al., 2012). Mouse ES cells depend on threonine to maintain SAM synthesis, with threonine starvation leading to decreased histone methylation and inhibited proliferation (Shyh-Chang et al., 2013). These findings and others illustrate the integration that exists between nutrient availability, metabolism and epigenetic control mechanisms.
The importance of folate-mediated one-carbon metabolism for cancer cell proliferation has long been appreciated (Locasale, 2013). Serine plays a key role in feeding one-carbon units to the tetrahydrofolate (THF) cycle and supports both nucleotide synthesis and NADPH production (Fan et al., 2014, Snell et al., 1987). Cancer cells have high demand for serine, which they meet by a combination of exogenous serine uptake and de novo synthesis from glucose (Locasale and Cantley, 2011, Maddocks et al., 2013, Possemato et al., 2011, Snell, 1985). Interestingly, the serine synthesis pathway enzymes can be epigenetically activated by histone H3 methyltransferase G9A to support cancer cell survival and proliferation (Ding et al., 2013). In addition to nucleotide production, one-carbon metabolism can also contribute to the methionine cycle by providing one-carbons, in the form of methyl groups, to recycle homocysteine to methionine (Herbig et al., 2002, Lu and Mato, 2012). Partitioning of one-carbon units between nucleotide synthesis and homocysteine remethylation can be controlled by cytoplasmic serine hydroxymethyltransferase (SHMT1) (Herbig et al., 2002, MacFarlane et al., 2008), with a decreased flux to thymidine synthesis associated with increased uracil incorporation into DNA and enhanced cancer risk in some models (Macfarlane et al., 2011). By contrast, modulation of MTHFD1 has been shown to lower SAM levels but decrease uracil incorporation into DNA, suggesting a switch toward nucleotide synthesis (MacFarlane et al., 2009). Nevertheless, this change in folate metabolism also increased tumorigenesis in some tumor models (Macfarlane et al., 2011). Dietary and genetic perturbation of folate mediated one-carbon metabolism have been shown to disrupt both the synthesis and methylation of DNA (Stover, 2004), and while these two pathways are intimately linked, how they are coordinated remains to be fully elucidated.
ATP is the major energy carrier used in cellular metabolism (Hardie et al., 2012). The inter-conversion of AMP, ADP, and ATP is a fundamental reaction in cellular biology allowing for the transfer of energy from energy yielding reactions (e.g., glycolysis) to energy consuming reactions (e.g., fatty acid synthesis)(Hardie et al., 2012). However, there is a clear—but potentially underappreciated—biochemical distinction between ATP turnover via energetic reactions (i.e., regeneration of ATP by re-phosphorylation of AMP and ADP) and ATP synthesis (i.e., ATP generated by de novo purine synthesis, a process that requires glycine and one-carbon units, which are both commonly derived from serine in cancer cells). Our previous studies highlighted the importance of serine in supporting purine synthesis, showing that glycine did not provide the one-carbon units necessary for the de novo synthesis of AMP, ADP, and ATP in the cancer cells we studied (Labuschagne et al., 2014). While purine synthesis provides vital nucleotides for nucleic acid synthesis, ATP generated by this biosynthetic pathway has the potential to be used in other metabolic reactions.
In this study, we analyze the transfer of 13C labeled carbon from methionine and serine directly onto DNA or RNA in cancer cells. Under methionine-fed conditions, serine did not contribute one-carbon units to recycle methionine, although serine did support DNA and RNA methylation under these conditions. Surprisingly, we show that this contribution of serine is through the de novo synthesis of ATP, which is required to convert methionine to SAM. Our data show that non-energetic metabolic stress can cause a dramatic decrease in total ATP levels in rapidly proliferating cells, which can lead to changes in methyl group transfer without inducing activation of AMP-activated protein kinase (AMPK).
Results
Methionine Is the Dominant Methyl Provider in Colorectal Cancer Cells, but Its Ability to Provide Methyl Groups Is Modulated by Serine Availability
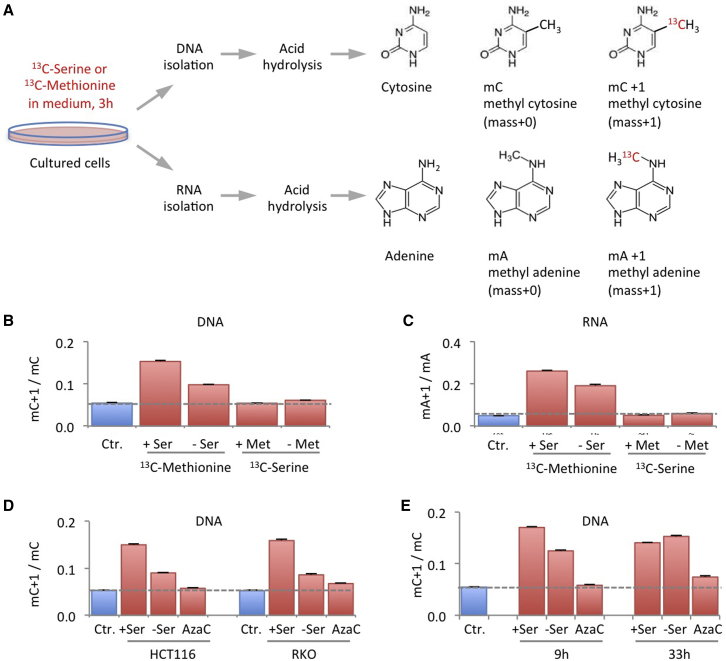
We have previously seen that serine availability has a major influence on the growth and metabolism of colorectal cancer cells (Labuschagne et al., 2014, Maddocks et al., 2013), with a clear impact on one-carbon metabolism and purine synthesis. To analyze the contribution of serine-dependent one-carbon metabolism to the methionine cycle and methylation reactions (Figure S1A) in these cells, we developed an assay to track one-carbons from extracellular nutrients, through the methionine cycle, into methylated substrates (Figure 1A). We chose to focus on the nucleic acids DNA and RNA as the methyl acceptor substrates, as the most common forms of methylated DNA and RNA (methyl-cytidine and methyl-adenosine, respectively) are well characterized (Fu et al., 2014, Jia et al., 2013, Schübeler, 2015) and amenable to analysis. A number of assays have been developed to analyze relative quantities of modified and unmodified DNA bases by mass spectrometry, including global DNA methylation (Kok et al., 2007), incorporation of deuterium labeled serine into DNA (Field et al., 2015, Herbig et al., 2002), and incorporation of deuterium-13C-labeled methionine into DNA (Bachman et al., 2014). Based on an acid hydrolysis technique (Kok et al., 2007), we developed our assay to use 13C15N-serine or 13C15N-methionine in cell culture medium for 3 hr. Using this method, we were able to track 13C-labeled serine and methionine into methyl groups on DNA and RNA. This method permitted relative quantification of the one-carbon contribution from extracellular serine and methionine onto DNA and RNA over a fixed time period (Figure 1A).
Figure 1.
Methyl Transfer from Methionine to DNA and RNA Is Impeded during Serine Starvation, yet Serine Does Not Contribute One-Carbon Units for Methylation under Methionine-Fed Conditions
(A) A method was developed to directly track the incorporation of labeled one-carbons from extracellular nutrients into DNA and RNA. After starvation/drug treatment, cells were fed labeled nutrients for 3 hr. Following standard DNA and RNA isolation, nucleic acids were hydrolyzed and analyzed by LCMS, revealing the extent of methyl cytosine and methyl adenine labeling.
(B and C) RKO cells were grown in the presence or absence of unlabeled methionine 0.1 mM (Met) and serine 0.8 mM (Ser) for 6 hr, after which the media were replaced with matched media containing either labeled methionine or labeled serine for 3 hr and analyzed as outlined in (A). The methionine-starved cells were supplemented with 0.8 mM homocysteine and 1 μM vitamin B12. The ratio of labeled to unlabeled methyl cytosine (mC+1/mC) in DNA and labeled to unlabeled methyl adenine (mA+1/mA) in RNA are shown. Data are averages of triplicate wells, and error bars are SD. Broken lines indicate the background labeling due to natural 13C abundance, shown as control (Ctr.) blue bars.
(D) HCT116 and RKO cells were grown with or without serine 0.8 mM (+/−Ser) for 6 hr, after which the media were replaced with matched media containing either unlabeled (Ctr.) or labeled methionine for 3 hr and analyzed as outlined in (A). Azacytidine 0.5 μM (AzaC) a DNA methyltransferase inhibitor was used as positive control. Data are averages of triplicate wells; error bars are SD.
(E) HCT116 cells were treated as described in (D), either with an initial serine starvation (with unlabeled methionine) of 6 hr or 30 hr, followed by a 3 hr period with labeled methionine. Data are averages of triplicate wells; error bars are SD.
See also Figure S1.
Using this method we found—as expected—that one-carbons from labeled methionine were readily used to methylate DNA and RNA. However, no significant contribution of one-carbons from serine was detected under the same conditions (Figures 1B and 1C). As re-methylated methionine is derived from its downstream metabolite homocysteine (Figure S1A), removal of exogenous methionine also deprives cells of the homocysteine they require for re-methylation (Shlomi et al., 2014, Tang et al., 2000). Therefore we supplemented methionine-starved cells with exogenous homocysteine and vitamin B12 (co-factor for re-methylation reaction) to ensure these nutrients were not limiting. Surprisingly, although serine did not contribute one-carbons under these conditions, serine starvation clearly decreased the contribution of one-carbon units from methionine to DNA and RNA after 9 hr (Figures 1B and 1C). Treatment with a low dose of the DNA methyltransferase inhibitor azacytidine, which directly blocks DNA methyl transfer, caused a similar decrease in the transfer of methyl groups to DNA as serine starvation (Figure 1D). After 30 hr of serine starvation, cells restored their ability to maintain the transfer of methionine-derived methyl groups to DNA (Figure 1E). Cancer cells adapt to serine starvation both by controlling serine utilization and by upregulating de novo serine synthesis, which allows recovery of serine levels (Maddocks et al., 2013), potentially explaining the recovery of methyl transfer by this later time point.
Analysis of total DNA and RNA methylation showed that serine depletion did not significantly impact total levels of methylated DNA or RNA, whereas methionine starvation caused decreased methylation (Figure S1B). This finding suggests a degree of coordination between serine availability and methyl group transfer, which is lacking for methionine. As expected, azacytidine caused a decrease in total DNA methylation, but did not alter total RNA methylation (Figure S1B). As serine starvation decreases the proliferation rate of cancer cells, we tested whether serum starvation, which also decreases proliferation rate (Figure S1C), had a similar effect. Serum starvation did not impede transfer of methyl groups to DNA to the same extent as serine starvation (Figure S1D), suggesting that the changes in methyl transfer that we detected are not simply a reflection of decreased proliferation.
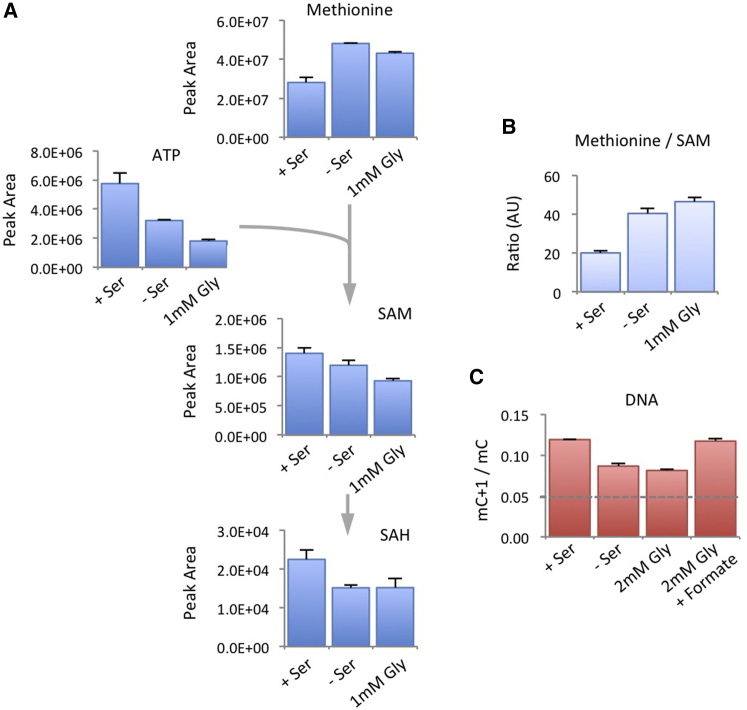
De novo ATP Synthesis Influences Methionine/SAM Ratio and Transfer of Methyl Groups to DNA
To understand why serine availability influenced the transfer of methyl groups to DNA and RNA, despite not directly contributing one-carbons for methylation, we analyzed metabolite levels in the methionine cycle by liquid chromatography mass spectrometry (LCMS). Addition of 13C15N-labeled methionine showed that serine-starved cells had increased levels of methionine but lower levels of downstream metabolites SAM and SAH (Figures 2A and 2B), suggesting a decrease in the conversion of methionine into the methyl donor molecule SAM. Given that conversion of methionine into SAM is ATP dependent, we hypothesized that methylation could be influenced by cellular ATP levels.
Figure 2.
Contribution of Methionine to the SAM Cycle Is Impeded by Serine Starvation
(A and B) HCT116 and RKO cells were grown in the presence of unlabeled methionine 0.1 mM with or without serine 0.8 mM (+/−S) for 6 hr, after which the media were replaced with matched media containing labeled methionine, then harvested after a further 5 hr. Intracellular metabolites were analyzed by LCMS. Data are averages of triplicate wells; error bars are SD.
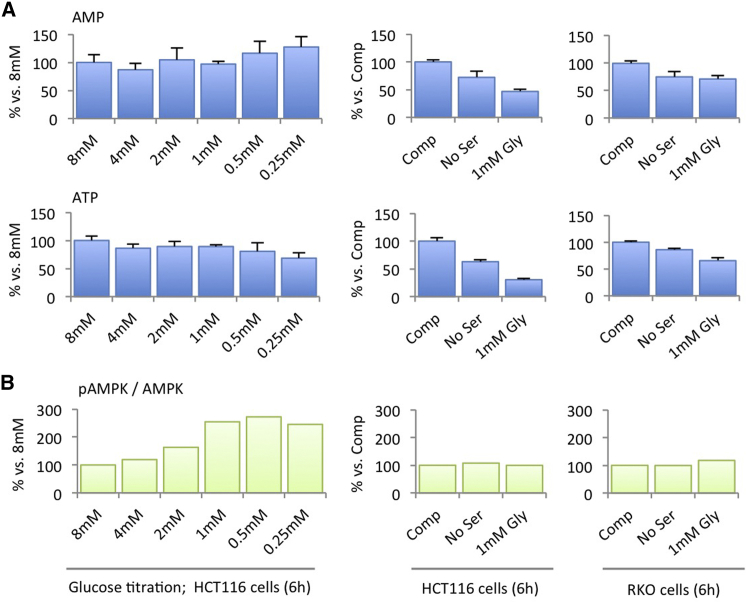
To test this, we assessed the impact of serine depletion on ATP levels and compared this to glucose starvation as a positive control. Glucose is a major cellular energy source, and as expected, glucose depletion caused increased levels of AMP and lower levels of ATP (Figure 3A). This corresponded with activation (by phosphorylation) of the cellular energy sensor AMPK (Figure 3B). By comparison, we noted that serine starvation alone decreased both ATP and AMP levels (Figure 3A). This is a reflection of the requirement of serine-derived one-carbon units for de novo purine synthesis, and as reported previously (Labuschagne et al., 2014), this effect was further exacerbated by both removing serine and increasing glycine, which further inhibits nucleotide synthesis. In previous work, we found that pyruvate partially rescued cells from serine starvation and showed that acute depletion of exogenous pyruvate and serine resulted in activation of AMPK (Maddocks et al., 2013). In the present study, cells were routinely grown without pyruvate, hence were adapted to its absence. In this context, the subsequent removal of serine led to a coordinated decrease in both AMP and ATP levels (Figure 3A), without activation of AMPK (Figure 3B). While de novo ATP synthesis may be slower in comparison to the rate of ATP turnover (as in energy generating reactions), these results suggest that without significant de novo ATP synthesis, cancer cells do not have an adequate pool of adenine nucleotides for biosynthetic reactions (such as SAM synthesis) that require ATP to contribute adenosine.
Figure 3.
Metabolic Stress may Inhibit ATP Turnover, Increasing AMP and Decreasing ATP, or Inhibit ATP Synthesis (i.e., De Novo Nucleotide Synthesis) Decreasing Both ATP and AMP
(A) HCT116 and RKO cells were grown in varying concentrations of glucose (0.25–8 mM), or in complete medium (Comp), or without serine (No Ser), or without serine and with high glycine (1 mM Gly) for 6 hr. Intracellular pools of AMP and ATP were determined by LCMS. Data are averages of triplicate wells; error bars are SD.
(B) Western blots were performed to determine the expression of AMPK and phosphorylated AMPK; bands were quantified using a Li-Cor scanner. Ratio was calculated by dividing pAMPK band intensity by AMPK band intensity.
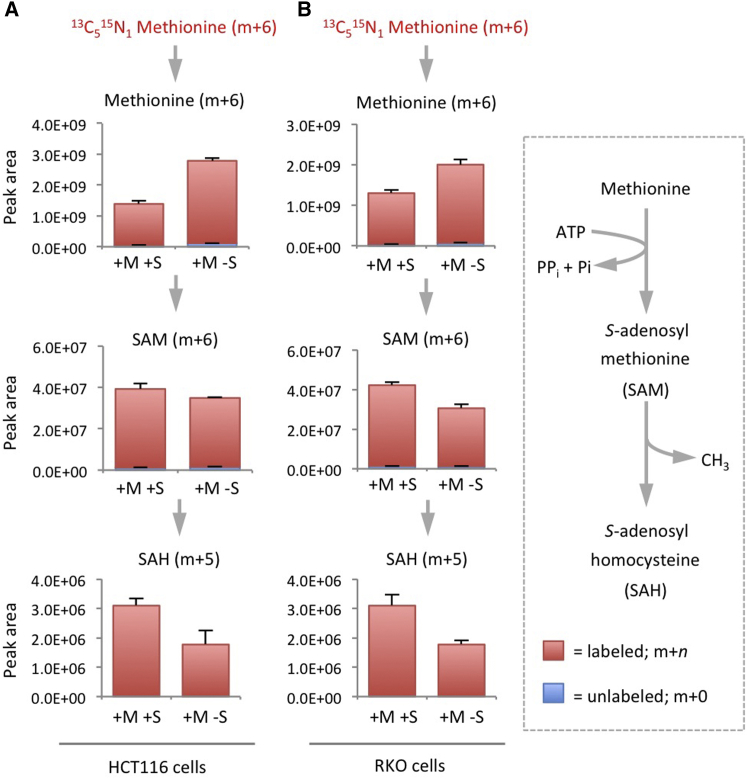
In order to test whether de novo synthesis of ATP could make a direct contribution to the methionine cycle, we analyzed SAM cycle metabolites by LCMS after feeding cells 13C15N-serine. In support of the DNA and RNA methylation data (Figures 1B and 1C), labeled serine did not contribute one-carbons to methionine re-methylation (i.e., the m+1 pool of methionine was at background levels, as seen in the control cells). However, after just 3 hr, both SAM and SAH were labeled by serine-derived carbon and nitrogen (Figures 4 and S2A). Labeling was not seen in homocysteine, reflecting the fact that the ATP-derived adenosine nucleotide is lost on conversion of SAH to homocysteine (Figure S2A). As a result, label is detected in adenosine produced at this step (Figure S2A). ATP also contributes adenosine to the synthesis other metabolites (e.g., NAD(H)). In comparison to SAM, the contribution of labeled serine to NAD(H) was smaller over 3 hr (Figure S2A), with serine starvation having a relatively modest impact on total NAD(H) levels over 6–24 hr. Excess glycine had a greater impact on NAD(H), in line with its more dramatic effects on ATP levels. These data suggest that SAM levels may be more sensitive to variations in de novo ATP synthesis than other metabolites.
Figure 4.
Under Methionine-Fed Conditions, Serine Contributes to the SAM Cycle via De novo ATP Synthesis, but Not Homocysteine Re-Methylation
HCT116 cells were either fed unlabeled or 13C315N1-labeled serine 0.4 mM for 3 hr (in the presence of methionine 0.1 mM). Metabolites were extracted and analyzed by LCMS. The major isotopomers resulting from serine labeling are shown. Data are averages of triplicate wells, error bars are SD. See also Figure S2.
To test whether de novo nucleotide synthesis is required for incorporation of methyl units into DNA, we cultured cells under conditions which inhibited de novo nucleotide synthesis (i.e., no serine/no serine + high glycine). Analysis of total ATP, methionine, and SAM pools (without using labeled metabolites) showed that these conditions caused a dramatic decrease in cellular ATP levels concurrent with increased methionine, and lower SAM levels (Figures 5A and S3A). These metabolic changes translated to an increased methionine/SAM ratio (Figures 5B and S3B) and a decrease in the contribution of methyl units from the methionine cycle to DNA (Figure 5C). Exogenous formate directly provides one-carbons to the THF cycle and restores de novo nucleotide synthesis in the presence of excess glycine (Labuschagne et al., 2014). Consistent with the requirement for de novo ATP to support methionine to SAM conversion, the addition of formate restored transfer of methyl groups from methionine to DNA (Figure 5C).
Figure 5.
Metabolic Stresses that Suppress De novo Nucleotide Synthesis Increase the Methionine/SAM Ratio
(A) HCT116 cells were grown with or without serine 0.8 mM (+/−Ser) or without serine, plus high glycine (1 mM Gly) for 24 hr and analyzed by LCMS. Data are averages of triplicate wells; error bars are SD.
(B) The methionine/SAM ratio was calculated by dividing the peak areas of these metabolites. Data are averages of triplicate wells, error bars are SD.
(C) HCT116 cells were grown with or without serine 0.8 mM (+/−Ser) or without serine, plus high glycine (2 mM Gly) with or without formate (0.5 mM) for 6 hr. DNA was extracted and analyzed by LCMS. Data are averages of triplicate wells; error bars are SD. Broken line indicates the background labeling due to natural 13C carbon abundance.
See also Figures S3 and S4.
Wider analysis of nucleotide synthesis intermediates showed that serine starvation generally impeded the synthesis of purines (due to lack of one-carbon units, as previously reported; Labuschagne et al., 2014). By contrast, pyrimidine levels upstream of dTMP either showed little change or increased in response to serine starvation (Figure S4). As the pyrimidine nucleotides upstream of dTMP do not require one-carbon units for de novo synthesis, they can be made in the absence of serine. Furthermore, the lack of purines will inhibit nucleic acid synthesis, allowing unused pyrimidines to accumulate. The synthesis of dTMP from dUMP requires serine-derived one-carbon, and accordingly, the levels of dTMP were also lower in serine-starved cells.
One-carbon Metabolism Contributes to Methylation under Methionine-Starved, Homocysteine-Fed Conditions
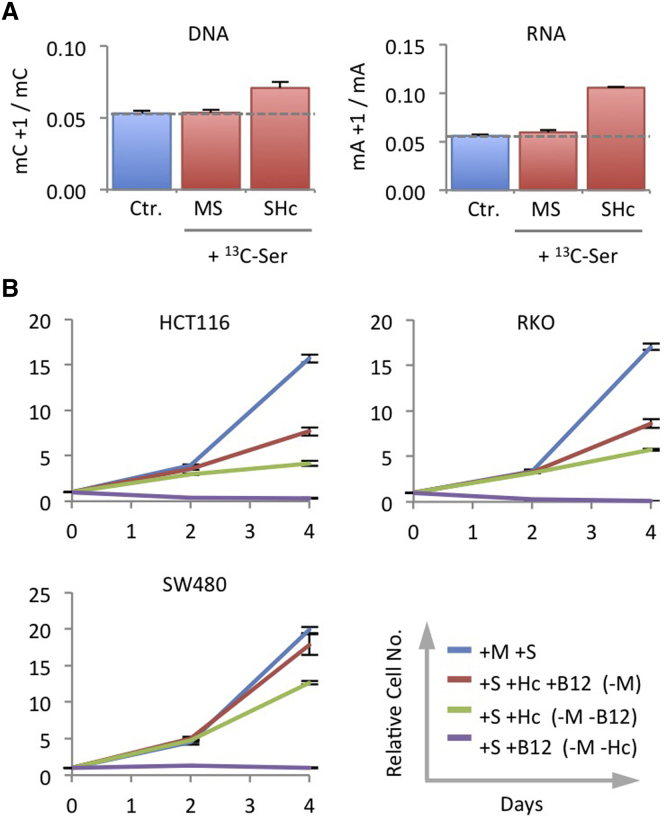
As we were unable to detect the use of serine-derived one-carbons for methionine re-methylation after 9 hr (Figures 1B and 1C), we considered that a longer period of methionine starvation could be necessary to promote detectable re-methylation. After 24 hr of methionine starvation (with supplementary vitamin B12 and homocysteine) with labeled serine for the final 3 hr, we were able to detect the transfer of serine-derived one-carbons to DNA and RNA (Figure 6A). Supplementing cells with homocysteine and vitamin B12 provided varying degrees of proliferative rescue (Figure 6B), most likely reflecting the efficiency with which these cells can recycle methionine from homocysteine. The small additional gain in rescue achieved by supplementary vitamin B12 suggests that background vitamin B12 levels are already present in the dialyzed serum in the protein bound form.
Figure 6.
Under Methionine Starvation, Homocysteine Is Re-Methylated Using One-Carbon Units from Serine
(A) RKO cells were grown with unlabeled methionine 0.1 mM and serine 0.8 mM (MS) or unlabeled serine plus homocysteine (SHc) for 21 hr, followed by matched medium containing 13C315N1-labeled serine for 3 hr. DNA and RNA were isolated an analyzed by LCMS. Broken lines indicate the background labeling due to natural 13C abundance, shown as control (Ctr.) blue bars. Data are averages of triplicate wells; error bars are SD.
(B) RKO, HCT116, and SW480 cells were grown in medium with serine 0.8 mM (S), with or without methionine 0.1 mM (M), homocysteine 0.8 mM (Hc), and vitamin B12 1uM (B12), and counted after 2 and 4 days. Data are averages of triplicate wells; error bars are SD.
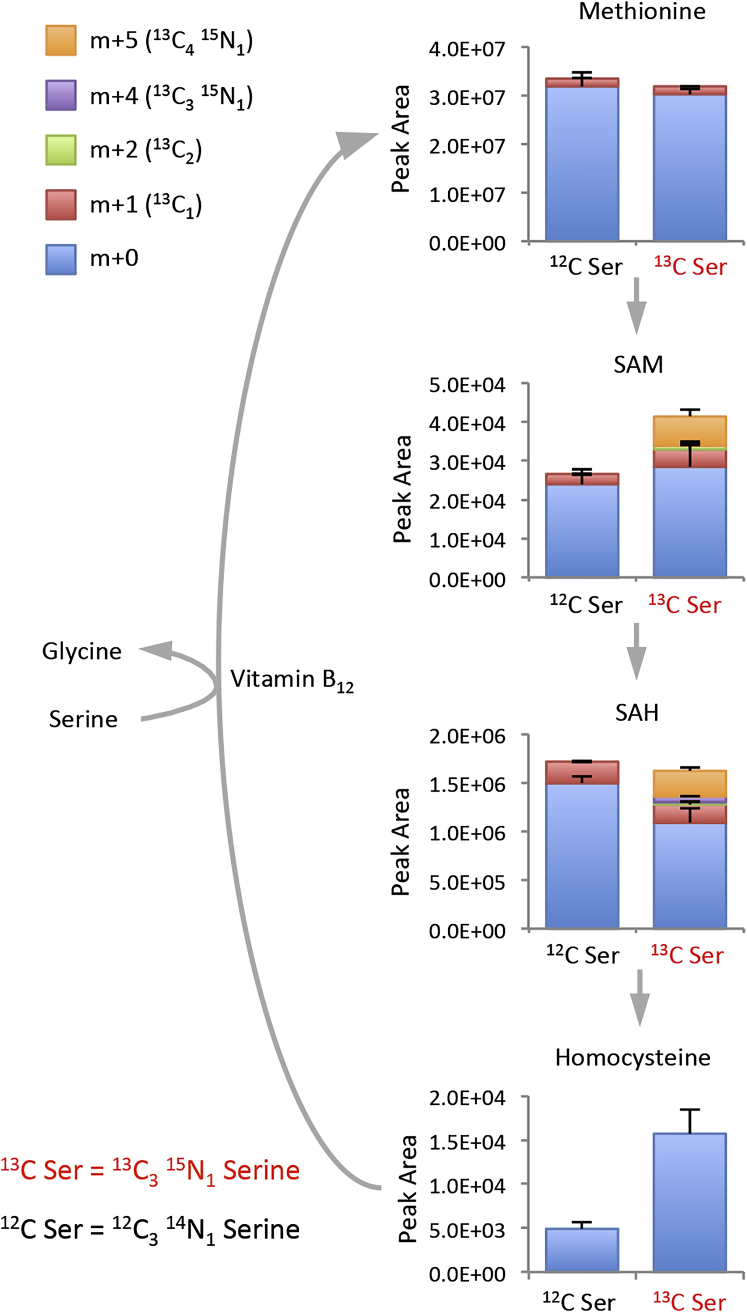
Serine Contributes to the SAM Cycle by Two Distinct Pathways that Can Operate Simultaneously
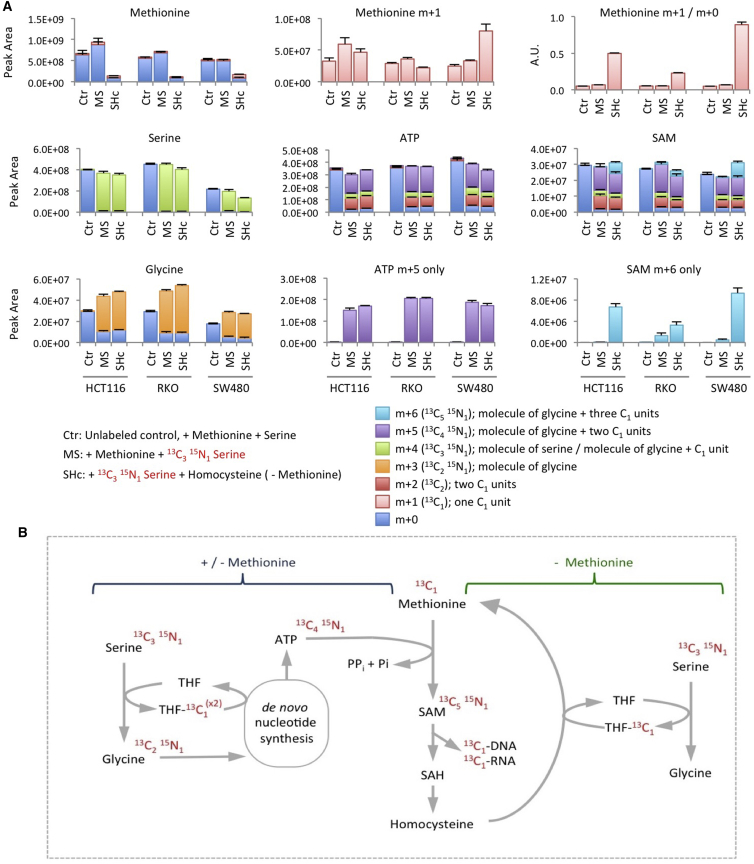
Our data suggest that serine can contribute to the methionine cycle both by providing one-carbon units for re-methylation of methionine and by supporting de novo ATP synthesis to allow the conversion of methionine to SAM. To assess the full extent of the contribution of serine to nucleotide synthesis and the methionine cycle, we cultured cells with labeled serine for 24 hr, either in the presence of methionine or without methionine plus homocysteine and vitamin B12. All three cell lines readily took up the exogenous serine, which was converted into glycine. One-carbons from serine and serine-derived glycine molecules entered de novo nucleotide synthesis so that almost the entire cellular pools of AMP, ADP, ATP GMP, GDP, and GTP were labeled after 24 hr (Figures 7A and S5). At this time point most NAD(H) was also labeled, although the proportion of the pool labeled was less than that seen for ATP and SAM (Figures 7A and S5).
Figure 7.
Serine Contributes to the SAM Cycle by Two Distinct Pathways, which Can Act Simultaneously
(A) HCT116, RKO, and SW480 cells were grown with unlabeled methionine 0.1 mM plus 13C315N1-labeled serine 0.8 mM (MS) or without methionine, with homocysteine 0.8 mM, vitamin B12 1 μM, plus 13C315N1-labeled serine (SHc) for 24 hr; glycine was not included in any of the media. Metabolites were extracted and analyzed by LCMS. The major isotopomers resulting from serine labeling are shown. Data are averages of triplicate wells; error bars are SD.
(B) Schematic diagram depicting the contribution of serine/glycine metabolism to the SAM cycle under methionine-fed and methionine-starved conditions. The contribution of labeled carbons and nitrogens from exogenous serine are shown in red text. See also Figure S5.
The major ATP isotopomer contained four labeled carbons and one labeled nitrogen (m+5), indicating incorporation of serine-derived glycine (two carbons, one nitrogen) and two serine-derived one-carbon units (Figures 7A and 7B). While total methionine levels were lower in the methionine-starved cells, an increase in the proportion of m+1 labeled methionine was observed. Hence the ratio of labeled to unlabeled methionine was dramatically increased in the methionine-starved cells. Labeling of SAM generally mirrored ATP labeling; however, an additional m+6 isotopomer was also observed in SAM, which was increased during methionine starvation. This m+6 isotopomer is a result of SAM labeling via re-methylated methionine (one carbon from serine) plus serine-labeled ATP (four carbons and one nitrogen) (Figures 7A and 7B). This labeling pattern indicates that serine can contribute simultaneously to two independent pathways that support the SAM cycle.
For technical clarity, we have provided examples of chromatogram peaks for adenine, methyl-adenine, cytosine, and methyl-cytosine with theoretical and measured masses (Figure S6), as well as a breakdown of our data analysis and presentation for 13C methyl transfer (Figures S7A–S7C) and total RNA/DNA methylation analysis (Figures S7D–S7F).
Discussion
One of the most important cellular uses for serine is to provide one-carbon units to support the THF-cycle (Tibbetts and Appling, 2010). While the cleavage of glycine could also provide one-carbons, we showed previously that glycine cannot substitute for serine, and increasing glycine in cells starved of serine further depletes the one-carbon pool as cells convert the glycine to serine (Labuschagne et al., 2014). Serine-dependent one-carbon metabolism is necessary for purine synthesis, but it can also contribute to several other metabolic pathways. The biochemical capacity for transfer of one-carbons from the THF cycle to homocysteine re-methylation is well known (Field et al., 2015, Herbig et al., 2002, Shlomi et al., 2014, Tang et al., 2000). However, the activity of this pathway in cancer cells is poorly characterized. In the present study, we show that during methionine starvation, serine can provide one-carbon units to recycle homocysteine to methionine and so support the methionine cycle. Intriguingly, however, we also found that serine contributes to the SAM cycle and subsequent DNA and RNA methylation, even in methionine-fed cells, although this was not a reflection of methionine recycling. Instead, we found that serine-dependent de novo ATP synthesis is needed to support the conversion of methionine to SAM and that limitation of this ATP pool can impact the rate of SAM generation and new methylation of DNA and RNA.
The method we have used to track transfer of methyl units from 13C-labeled methionine and serine has potential utility to evaluate methylation dynamics of DNA and RNA in other contexts. There is a growing realization that methylation of DNA and RNA is more dynamic than previously appreciated. The ability to analyze these changes with a temporal dimension (e.g., by adapting the method to include multiple time points) may allow this technique to be used to achieve this goal. In addition, the analysis method we employed for total cytosine/adenine methylation offers a robust method for global methylation analysis. The advantages of this LCMS method compared to other measures of global methylation (e.g., immuno-detection of 5 mC) are that it is direct and properly normalized for total cytosine/adenine content. However, the present method is not sensitive enough to detect small changes (e.g., less than ∼2%) in methylation or to provide information on where in the genome these changes may occur.
Serine has been shown to play an important role in supporting the proliferation of cancer cells. Although not an essential amino acid, serine starvation promotes the induction of a program of metabolic adaptation including the de novo serine synthesis pathway, allowing cells to generate serine from glycolytic intermediates. In some cancers, amplification of the SSP enzymes renders the cells less dependent on exogenous serine. However, most cancer cells respond to serine starvation with an abrupt depletion of intracellular serine. Successful adaptation to serine starvation depends on the ability to partition the depleted serine pool into pathways important for cell survival (such as the generation of glutathione to control ROS) while limiting flow of serine-derived one-carbon units into nucleotide synthesis. While this strategy is consistent with meeting the needs of cells that have undergone a transient cell-cycle arrest (in response to serine depletion) and so have lower need for nucleic acid synthesis, our data indicate that a broader view of the contribution of purines—especially ATP—to cell growth and survival needs to be considered.
Numerous studies demonstrate that cells monitor and sense energetic stress through AMPK, which detects elevated AMP/ADP levels, signifying an increase in the AMP-to-ATP ratio (Hardie et al., 2012, Xiao et al., 2013). AMPK activation causes a host of changes that promote ATP regeneration and inhibit ATP consumption, so allowing the cell to balance ATP demand with ATP supply (Hardie et al., 2012, Hochachka and McClelland, 1997). The intracellular ATP level is frequently considered to be a function of changes in ATP turnover (i.e., the regeneration of ATP by phosphorylation of AMP/ADP) (Hardie et al., 2012, Hochachka and McClelland, 1997, Xiao et al., 2013), and clearly, the changing ratios of these metabolites in relation to ATP are signals for the activation of energetic stress responses like AMPK (Xiao et al., 2013). Less explored, however, is the contribution of de novo ATP synthesis (i.e., ATP generated by de novo purine synthesis, a biosynthetic pathway that requires glucose and amino acids). Cancer cells show enhanced rates of de novo purine synthesis, which has been interpreted as contributing to increased demand for nucleic acid (primarily RNA and DNA) synthesis. However, ATP generated by this pathway has the potential to be used in other metabolic reactions, including functions in energy transfer, as a phosphate donor, or as an adenosine donor—as in SAM and NAD(H) synthesis. It is possible that these alternative functions for the de novo synthesized purines (both ATP and GTP) could help explain why cancer cells devote so many resources to this pathway. Indeed, guanine nucleotides (GMP, GDP, and GTP) are critical for supporting the Ras pathway and a range of other signaling and metabolic processes important in cancer (Grewal et al., 2011, Ostrem et al., 2013).
Given that we observed almost complete labeling of the ATP pool within 24 hr, our data also suggest that just as other pools of cellular components (such as DNA and proteins) need to be replicated to support cell division, so do pools of “free nucleotides” such as ATP and GTP. Since AMPK is seemingly insensitive to concurrent decreases in AMP and ATP, it is tempting to speculate that other mechanisms exist to detect absolute cellular ATP (and GTP) content. Given the dependence of the methionine cycle on ATP and its ability to influence gene expression and protein activity by methylation, it is conceivable that the methionine cycle has a role in sensing absolute ATP levels.
In the present work, we observe the contribution of newly synthesized ATP to the SAM cycle and a requirement for serine in supporting transfer of methyl groups to DNA and RNA through ATP synthesis. A wealth of literature supports the role of serine-derived one-carbon in methionine re-methylation through the synthesis of 5-methyl THF, and polymorphisms in one-carbon metabolism enzymes can influence chromatin methylation and ultimately disease risk (Stover, 2011). However, we did not observe the expected serine-dependent re-methylation in cancer cell lines when methionine was present. We show that non-energetic metabolic stress can have a dramatic and rapid effect on total ATP levels, causing an even greater decrease than glucose starvation. However, this drop in ATP is not clearly detected by the classic energetic stress sensor AMPK, likely because lower de novo ATP synthesis is accompanied by lower levels of AMP. We see that despite dramatic decreases in ATP levels, cells survive and maintain proliferative potential. These are important observations given the widely held belief—based on studies involving energetic stress—that cells maintain constant ATP levels at all costs and that ATP concentration is “universally homeostatic” (Hochachka and McClelland, 1997). The term “ATP synthesis” is very commonly used (Pubmed returns over 600 papers with this term in the title) to describe the generation of ATP by adding phosphate(s) to AMP or ADP. However, this form of ATP generation can more accurately be referred to as ATP turnover or regeneration, as the adenosine nucleotide is conserved during such reactions. As the present study demonstrates, true (i.e., de novo) ATP synthesis—achieved by the assembly of glucose and amino acids through multi-step de novo purine synthesis—is also a major contributor to the functional ATP pool in cancer cells.
Experimental Procedures
Cell Culture
HCT116, RKO, and SW480 cells were obtained from ATCC and subsequently authenticated using the Promega GenePrint 10 system according to the manufacturer’s instructions. Unless otherwise stated, cell culture media were purchased from GIBCO, product numbers are shown in parenthesis. Stock HCT116 and RKO cells were grown in McCoys 5A Medium (26600) supplemented with 10% FBS and penicillin-streptomycin. SW480 cells were grown in DMEM (21969) supplemented with 2 mM L-glutamine 10% FBS. All stock cell culture and experiments were conducted in 37°C, 5% CO2 incubators.
Nutrient Deprivation
For experiments where nutrient levels were manipulated, cells received formulated medium containing MEM vitamins (11120), dialysed FBS (Hyclone, Thermo Scientific), penicillin-streptomycin, 17 mM D-glucose (Sigma), sodium bicarbonate (Sigma) lacking methionine, and serine and glycine (but containing all other amino acids), with varying concentrations of these nutrients added back as specified in each experiment. For re-methylation experiments (where cells were deprived of methionine) 0.8 mM homocysteine (Sigma) and 1 μM vitamin B12 (methylcobalamin, Sigma) were added. Azacytidine (Sigma) was used at 0.5 μM. Labeled amino acids 13C315N1-Serine and 13C515N1-methionine (Cambridge Isotope Laboratories Inc. USA, via CK Gas Ltd, UK) were used at concentrations specified in each experiment.
Methylation Analysis
Cells were seeded in McCoys/DMEM in 6-well plates and left for 16–24 hr; initial seeding density was 2 to 8 × 105 cells per well and was calculated so that cells were approximately 80% confluent at the time of harvest. Cells were washed once with PBS and received formulated assay medium as described above under “Nutrient Deprivation.” For each experiment, cells were initially grown with unlabeled nutrients for various time periods (stated in each experiment) followed by a fixed period of 3 hr with labeled 13C315N1-Serine or 13C515N1-Methionine. The relative contribution of these labeled metabolites to DNA/RNA methylation over the 3 hr period was then assessed as follows: cells were removed from wells with trypsin, and each well was split into two cell pellets that were frozen on dry ice/−80°C. One pellet was used for DNA extraction using QIAamp DNA mini kit (QIAGEN, 51304) with RNase treatment, RNA was extracted from the second pellet using RNeasy mini kit (QIAGEN, 74104) with DNase treatment according to manufacturer’s instructions.
The protocol for DNA/RNA acid hydrolysis was developed from methods presented by Kok & colleagues (Kok et al., 2007). 1ug DNA or 3ug RNA was placed in a 1.5ml tube and dried at 40°C under nitrogen gas. 100ul of formic acid (Sigma) was added to the dry pellets and incubated at 130°C for 3.5 hr. After cooling, the acid was dried off at 40°C under nitrogen gas, dry pellets were re-suspended in 25ul LCMS-grade water for 20 min at room temperature. 100ul of ice-cold LCMS-grade methanol (62.5%) acetonitrile (37.5%) solution was added to each sample. Samples were vortexed and spun at 4°C for 15 min, supernatant were transferred to LCMS vials.
Bases from hydrolyzed DNA and RNA were analyzed on a Dionex Ultimate 3000 LC system coupled to a Q Exactive mass spectrometer (Thermo Scientific). Chromatographic separation was achieved using a Sequant ZIC-pHILIC column (2.1 × 150 mm, 5 μm) (Merck) with elution buffers (A) and (B) consisting of 20 mM (NH4)2CO3, 0.1% NH4OH in H2O) and acetonitrile, respectively. The LC system was programmed to maintain a flow rate of 200 μl/min with the staring condition at 80% (A), which linearly decreased to 20% (A) over 10 min followed by washing and re-equilibration steps (20%–80% [A]) over 7 min. Ionization of the analytes occurred in a heated electrospray ionization (HESI) probe fitted to the mass spectrometer that operated in negative ion mode over a mass range between 75 and 200 m/z at a resolution of 70,000. Thermo LCquan software was used to identify and analyze the nucleotides. To quantify the contribution of labeled serine/methionine to DNA/RNA methylation the peak area for m+1,methyl-cytosine/m+1,methyl-adenine was divided by m+0,methyl-cytosine/m+0,methyl-adenine. For quantification of total DNA/RNA methylation, only unlabeled metabolites were used, and the peak area for methyl-cytosine/methyl-adenine was divided by peak area for cytosine/adenine, respectively. Examples of chromatogram peaks and a graphical representation of the data analysis and presentation are included in Figures S6 and S7.
Metabolite Analysis
Assays were performed as described previously (Labuschagne et al., 2014). Briefly, cells were plated in 6-well plates and cultured in McCoys 5A medium or DMEM for 24 hr before replacing the medium with fresh medium containing labeled amino acids and incubated for the indicated times. Cells were washed with PBS followed by metabolite extraction with ice-cold extraction buffer consisting of methanol, acetonitrile, and H2O (50:30:20). Extracts were analyzed by LCMS using a Dionex Ultimate 3000 LC system coupled to a Q Exactive mass spectrometer (Thermo Scientific). A Sequant ZIC-pHILIC column (2.1 × 150 mm, 5 μm) (Merck) was used to separate the metabolites using the same elution buffers ([A] and [B]) as described above. A gradient program starting at 80% (A) and linearly decreasing to 20% (A) over 17 min was used followed by washing and re-equilibration steps. The total run time of the method was 23.5 min. The Q Exactive was operated in full scan mode over a mass range of 75–1,000 m/z at a resolution of 35,000 with polarity switching. Ionization occurred in the HESI probe. Metabolites were identified and analyzed using Thermo LCquan software.
Western Blot
Whole-cell protein lysates were prepared in RIPA-buffer supplemented with complete protease inhibitors (Roche), sodium orthovanadate, and sodium fluoride (both Sigma). Lysates were separated using precast NuPAGE gels (Invitrogen, Life Technologies) and transferred to nitrocellulose membranes. Proteins were detected and quantified using a LI-COR Odyssey Infrared scanner and software (LI-COR Biosciences). Primary antibodies used were as follows: Anti-AMPK-a (2532) and Anti-phospho-AMPK-a (2535) both from Cell Signaling Technology. Secondary antibodies for the relevant species were IRDye680LT and IRDye800CW conjugated (LI-COR Biosciences).
Growth Curves
HCT116, RKO (2 to 3 × 104 cells per well in McCoys medium) and SW480 (5 × 104 cell per well in DMEM) were seeded in 24-well plates and allowed to adhere overnight. Adherent cells were washed with PBS and were fed formulated assay medium supplemented with the stated nutrients (see “Nutrient Deprivation” above). A separate “time-zero” counting plate was used to record starting cell number. Media were changed every 24 hr, and plates were counted after 2 and 4 days. Relative cell number was calculated by comparison to cell number at “time-zero.” For counting cells were trypsinized, re-suspended in PBS-EDTA, and counted with a CASY Model TT Cell Counter (Innovatis, Roche Applied Science).
Acknowledgments
This work was funded by Cancer Research UK grant C596/A10419 and ERC Grant 322842-METABOp53. We thank Gillian Mackay and Niels van den Broek for assistance with metabolomics.
Published: January 7, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.12.014.
Supplemental Information
References
- Bachman M., Uribe-Lewis S., Yang X., Williams M., Murrell A., Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerle M.R., Schwalm E.L., Booker S.J. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J. Biol. Chem. 2015;290:3995–4002. doi: 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N., Burns D.M., Blau H.M. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun H.M., Nordio F., Coull B.A., Tarantini L., Hou L., Bonzini M., Apostoli P., Bertazzi P.A., Baccarelli A. Temporal stability of epigenetic markers: sequence characteristics and predictors of short-term DNA methylation variations. PLoS ONE. 2012;7:e39220. doi: 10.1371/journal.pone.0039220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni G.L. The Nature of the Active Methyl Donor Formed Enzymatically from L-Methionine and Adenosinetriphosphate. J. Am. Chem. Soc. 1952;74:2942–2943. [Google Scholar]
- Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Christensen B.C., Smith A.A., Zheng S., Koestler D.C., Houseman E.A., Marsit C.J., Wiemels J.L., Nelson H.H., Karagas M.R., Wrensch M.R. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J. Natl. Cancer Inst. 2011;103:143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Li T., Wang X., Zhao E., Choi J.H., Yang L., Zha Y., Dong Z., Huang S., Asara J.M. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013;18:896–907. doi: 10.1016/j.cmet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy D.C. Epigenetic gene regulation: early environmental exposures. Pharmacogenomics. 2007;8:5–10. doi: 10.2217/14622416.8.1.5. [DOI] [PubMed] [Google Scholar]
- Dolinoy D.C., Weidman J.R., Jirtle R.L. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod. Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Fan J., Ye J., Kamphorst J.J., Shlomi T., Thompson C.B., Rabinowitz J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann A., Ivanek R., Murr R., Gaidatzis D., Burger L., Schübeler D. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 2013;9:e1003994. doi: 10.1371/journal.pgen.1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M.S., Kamynina E., Watkins D., Rosenblatt D.S., Stover P.J. Human mutations in methylenetetrahydrofolate dehydrogenase 1 impair nuclear de novo thymidylate biosynthesis. Proc. Natl. Acad. Sci. USA. 2015;112:400–405. doi: 10.1073/pnas.1414555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Gilsbach R., Preissl S., Grüning B.A., Schnick T., Burger L., Benes V., Würch A., Bönisch U., Günther S., Backofen R. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014;5:5288. doi: 10.1038/ncomms6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal T., Koese M., Tebar F., Enrich C. Differential Regulation of RasGAPs in Cancer. Genes Cancer. 2011;2:288–297. doi: 10.1177/1947601911407330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Li X., Liang D., Li T., Zhu P., Guo H., Wu X., Wen L., Gu T.P., Hu B. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15:447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Gut P., Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502:489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig K., Chiang E.P., Lee L.R., Hills J., Shane B., Stover P.J. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J. Biol. Chem. 2002;277:38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- Hino S., Nagaoka K., Nakao M. Metabolism-epigenome crosstalk in physiology and diseases. J. Hum. Genet. 2013;58:410–415. doi: 10.1038/jhg.2013.57. [DOI] [PubMed] [Google Scholar]
- Hochachka P.W., McClelland G.B. Cellular metabolic homeostasis during large-scale change in ATP turnover rates in muscles. J. Exp. Biol. 1997;200:381–386. doi: 10.1242/jeb.200.2.381. [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S., Stride B., Métivier R., Polycarpou-Schwarz M., Ibberson D., Carmouche R.P., Benes V., Gannon F., Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kohli R.M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok R.M., Smith D.E., Barto R., Spijkerman A.M., Teerlink T., Gellekink H.J., Jakobs C., Smulders Y.M. Global DNA methylation measured by liquid chromatography-tandem mass spectrometry: analytical technique, reference values and determinants in healthy subjects. Clin. Chem. Lab. Med. 2007;45:903–911. doi: 10.1515/CCLM.2007.137. [DOI] [PubMed] [Google Scholar]
- Labuschagne C.F., van den Broek N.J., Mackay G.M., Vousden K.H., Maddocks O.D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- Locasale J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale J.W., Cantley L.C. Genetic selection for enhanced serine metabolism in cancer development. Cell Cycle. 2011;10:3812–3813. doi: 10.4161/cc.10.22.18224. [DOI] [PubMed] [Google Scholar]
- Lu S.C., Mato J.M. S-adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane A.J., Liu X., Perry C.A., Flodby P., Allen R.H., Stabler S.P., Stover P.J. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J. Biol. Chem. 2008;283:25846–25853. doi: 10.1074/jbc.M802671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane A.J., Perry C.A., Girnary H.H., Gao D., Allen R.H., Stabler S.P., Shane B., Stover P.J. Mthfd1 is an essential gene in mice and alters biomarkers of impaired one-carbon metabolism. J. Biol. Chem. 2009;284:1533–1539. doi: 10.1074/jbc.M808281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane A.J., Perry C.A., McEntee M.F., Lin D.M., Stover P.J. Shmt1 heterozygosity impairs folate-dependent thymidylate synthesis capacity and modifies risk of Apc(min)-mediated intestinal cancer risk. Cancer Res. 2011;71:2098–2107. doi: 10.1158/0008-5472.CAN-10-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D., Berkers C.R., Mason S.M., Zheng L., Blyth K., Gottlieb E., Vousden K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier R., Gallais R., Tiffoche C., Le Péron C., Jurkowska R.Z., Carmouche R.P., Ibberson D., Barath P., Demay F., Reid G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Nordgren K.K., Skildum A.J. The deep end of the metabolite pool: influences on epigenetic regulatory mechanisms in cancer. Eur. J. Clin. Invest. 2015;45(Suppl 1):9–15. doi: 10.1111/eci.12361. [DOI] [PubMed] [Google Scholar]
- Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.K., Jang H.G., Jha A.K. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- Shen L., Inoue A., He J., Liu Y., Lu F., Zhang Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15:459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T., Fan J., Tang B., Kruger W.D., Rabinowitz J.D. Quantitation of cellular metabolic fluxes of methionine. Anal. Chem. 2014;86:1583–1591. doi: 10.1021/ac4032093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Locasale J.W., Lyssiotis C.A., Zheng Y., Teo R.Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J.J., Zhu H. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z.D., Chan M.M., Humm K.C., Karnik R., Mekhoubad S., Regev A., Eggan K., Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K. Enzymes of serine metabolism in normal and neoplastic rat tissues. Biochim. Biophys. Acta. 1985;843:276–281. doi: 10.1016/0304-4165(85)90149-7. [DOI] [PubMed] [Google Scholar]
- Snell K., Natsumeda Y., Weber G. The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem. J. 1987;245:609–612. doi: 10.1042/bj2450609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover P.J. Physiology of folate and vitamin B12 in health and disease. Nutr. Rev. 2004;62:S3–S12. doi: 10.1111/j.1753-4887.2004.tb00070.x. discussion S13. [DOI] [PubMed] [Google Scholar]
- Stover P.J. Polymorphisms in 1-carbon metabolism, epigenetics and folate-related pathologies. J. Nutrigenet. Nutrigenomics. 2011;4:293–305. doi: 10.1159/000334586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Tang B., Li Y.N., Kruger W.D. Defects in methylthioadenosine phosphorylase are associated with but not responsible for methionine-dependent tumor cell growth. Cancer Res. 2000;60:5543–5547. [PubMed] [Google Scholar]
- Tibbetts A.S., Appling D.R. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- Turcan S., Rohle D., Goenka A., Walsh L.A., Fang F., Yilmaz E., Campos C., Fabius A.W., Lu C., Ward P.S. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Sanders M.J., Carmena D., Bright N.J., Haire L.F., Underwood E., Patel B.R., Heath R.B., Walker P.A., Hallen S. Structural basis of AMPK regulation by small molecule activators. Nat. Commun. 2013;4:3017. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.