Abstract
Post-oligomerization synthesis is a useful technique for preparing site-specifically modified DNA oligomers. This approach involves site-specific incorporation of inherently reactive halogenated nucleobases into DNA strands using standard solid phase synthesis, followed by post-oligomerization nucleophilic aromatic substitution (SNAr) reactions with carcinogen-derived synthons. In these reactions, the inherent reactivities of DNA and carcinogen-derived species are reversed: the modified DNA nucleobase acts as an electrophile, while the carcinogen-derived species acts as a nucleophile. In the present protocol, we describe the use of the post-oligomerization approach to prepare DNA strands containing site- and stereospecific N6-adenine and N1, N6-adenine adducts induced by epoxide metabolites of the known human and animal carcinogen, 1,3-butadiene (BD). The resulting oligomers containing site specific, structurally defined DNA adducts can be used in structural and biological studies to reveal the roles of specific BD adducts in carcinogenesis and mutagenesis.
Keywords: post-oligomerization synthesis; 1,3-butadiene-induced DNA adducts; adenine; site- and stereospecific oligodeoxynucleotides
INTRODUCTION
1,3-Butadiene (BD) is a known human and animal carcinogen present in cigarette smoke, automobile exhaust, and occupational settings (Himmelstein et al., 1997). BD is metabolically activated to 3,4-epoxybut-1-ene (EB) and 1,2,3,4-diepoxybutane (DEB), which alkylate the adenine bases in DNA to give N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenine (N6-HB-dA), N6-(2-hydroxy-3,4-epoxybutan-1-yl)-2'-deoxyadenine (N6-HEB-dA), 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenine (1,N6-HMHP-dA) and N6,N6-(2,3-dihydroxybutan-1,4-diyl)-2'-deoxyadenine (N6,N6-DHB-dA) adducts (Tretyakova et al., 1997a)(Seneviratne et al., 2010a). The availability of DNA strands containing site- and stereospecific BD-DNA adducts is a prime requirement to establish the molecular mechanisms by which BD and its metabolites elicit their adverse biological effects.
This unit describes the use of post-oligomerization synthesis to prepare DNA oligodeoxy-nucleotides containing site- and stereospecific N6-HB-dA, N6-HEB-dA, 1,N6-HMHP-dA , and N6,N6-DHB-dA adducts. The post-oligomerization synthesis conducted on solid support has proven to be a useful strategy capable of generating highly pure synthetic DNA containing exocylic amine-functionalized adenine adducts of BD in good yields and with reduced number of HPLC purification steps. The resulting oligodeoxynucleotides were successfully used in a number of studies to evaluate the effects of BD-DNA adducts on DNA structure and replication and to identify their repair mechanisms.
Our overall strategy is to generate 6-chloropurine-containing DNA strands, which are subsequently reacted with the appropriate amino functionalized BD derivatives to generate the corresponding N6-alkyladenine adducts.
Basic Protocol 1 describes stereospecific synthesis of N-Fmoc-1-amino-3-butene-2-ol and N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane, the synthons required for the preparation of exocyclic adenine adducts induced by EB and DEB, respectively.
Basic Protocol 2 describes solid phase synthesis in combination with manual coupling to prepare DNA oligodeoxynucleotides containing site-specific 6-chloropurine (6-Cl-Pu).
Basic Protocol 3, 4 and 5 describe the synthesis of DNA oligomers containing site- and stereospecific N6-HB-dA, N6-HEB-dA, 1,N6-HMHP-dA , and N6,N6-DHB-dA adducts using the stereospecific synthons obtained from basic Protocol 1 and site-specific 6-Cl-Pu DNA obtained basic Protocol 2. These protocols allow for the synthesis of BD-adducted DNA oligomers on solid phase using controlled-pore glass (CPG) support or in solution. The use of post-oligomerization reactions on solid support dramatically reduces the number of HPLC purification steps required, decreasing the time required to complete the synthesis and improving the overall yield of the adducted DNA.
BASIC PROTOCOL 1: SYNTHESIS OF STEREOISOMERIC N-FMOC-1-AMINO-2-HYDROXY-3,4-EPOXYBUTANE
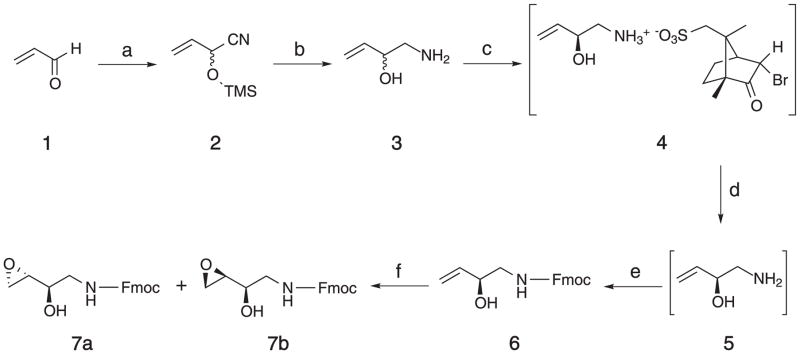
This protocol describes the synthesis of (2R,3S) and (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a and S.7b) (Seneviratne et al., 2010a), the amino-functionalized synthons derived from DEB (Figure 1). These stereoisomers are employed in the synthesis of oligodeoxynucleotides containing exocyclic BD-dA adducts, R,R- and R,S-1,N6-HMHP-dA. The synthesis involves five steps. (1) Addition of trimethylsilyl cyanide to acrolein (S.1) to prepare 2-[(trimethylsilyl)oxy]-3-butenenitrile (S.2). (2) Reduction of S.2 to racemic 1-amino-3-butene-2-ol (S.3). (3) Chiral resolution of the enantiomers of S.3 by reacting with [(1R)-(endo,anti)]-(+)-3-bromocamphor-8-sulfonic acid and crystallization of the S-isomer. (4) Protection of the amine functionality to yield (2S)-N-Fmoc-1-amino-3-butene-2-ol (S.6). This intermediate is used in the synthesis of the N6-BD-adenine adducts in basic protocol 3. (5) Epoxidation and resolution of the diastereomers of N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a and S.7b).
Figure 1.
Synthesis of (2R,3S) and (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane. a) TMSCN, ZnI2, reflux, 3 h; b) i. LiALH4, Et2O; ii. reflux, 75 min; iii. aqueous work up; c) ammonium salt of [(1R)-(endo,anti)]-(+)-3-bromocamphor-8-sulfonic acid, MeOH; d) excess aq. Ba(OH)2; e) Fmoc-Cl, aq. 1,4-dioxane, aq. Na2CO3, rt, 18 h; f) mCPBA, CH2Cl2, rt, 18 h.
NOTE: Anhydrous reaction conditions are required for several steps of this procedure. Anhydrous solvents can be purchased in Sure/Seal bottles (e.g. from Sigma-Aldrich), while glassware needs to be oven-dried. Several steps require flame drying of glassware.
CAUTION: Some of the chemicals and reagents used are highly toxic or flammable. Refer to material safety data sheets prior to use. All the reactions should be conducted in a well-ventilated fume hood. Use of personal protection equipment is highly recommended.
Materials
Acrolein
Zinc iodide (ZnI2)
Ice
Trimethylsilyl cyanide (TMSCN)
Lithium aluminum hydride (LiAlH4)
4 Å molecular sieves
N2 or Ar
Water
Triethanolamine
Anhydrous magnesium sulfate (MgSO4)
Diethyl ether
Ammonium salt of [(1R)-(endo,anti)]-(+)-3-bromocamphor-8-sulfonic acid
Methanol (MeOH)
Ethyl acetate (EtOAc)
Barium hydroxide (Ba(OH)2•8H2O)
Hydrochloric acid (HCl)
Sodium carbonate (Na2CO3)
9-fluorenylmethoxycarbonyl chloride (FmocCl)
Hexanes
Anhydrous dichloromethane (CH2Cl2)
meta-chloroperbenzoic acid (mCPBA)
Chloroform (CHCl3)
100, 250 and 500 mL round-bottom flasks
A 500 mL three-necked round-bottom flask
100 and 250 mL dropping funnels
Magnetic stir bars
Reflux condensers
A silicone/paraffin oil bath
A magnetic stirrer/hotplate
A reduced-pressure distillation setup
Rubber septa
A heating mantle
Syringe needles
A Buchner funnel setup
A 500 mL conical flask
A rotary evaporator equipped with a dry ice condenser and connected to an oil pump
Disposable Pasteur pipettes
A Whatman Partisil 10 (9.5 × 500 mm) column
Synthesis of 2-[(trimethylsilyl)oxy]-3-butenenitrile (S.2)
-
1
Weigh out 23 g (410 mmol) of freshly distilled acrolein (S.1) into a 500 mL round-bottom flask.
-
2
Add ~10 mg of ZnI2 and cool to 0 °C on an ice bath.
-
3
Weigh out 50 g (504 mmol) of ice-cold trimethylsilyl cyanide (TMSCN).
-
4
Transfer TMSCN into a 100 mL dropping funnel.
-
5
Fit the flask with a dropping funnel and add TMSCN to the mixture of acrolein (S.1) and ZnI2 dropwise over 1 hr.
-
6
Place a magnetic stir bar in the flask, fit the flask with a reflux condenser, and place the flask in a temperature-controlled oil bath on top of a magnetic stir plate.
-
7
Reflux the reaction mixture for 3 h, while running water through the reflux condenser, then allow to cool to room temperature.
-
8
Perform a reduced-pressure distillation on the resulting light yellow reaction mixture.
The product S.2 is a clear oil and can be obtained in 60–90% yields. 1H NMR (400 MHz, C6D6) δ 0.00 (s, 9H), 4.39 (ddd, J = 1.6, 1.6, 5.2 Hz, 1H), 4.88 (ddd, J = 0.8, 1.6, 10.2 Hz, 1H), 5.25 (ddd, J = 0.8, 1.6, 16.8 Hz, 1H), 5.50 (ddd, J = 5.2, 10.2, 16.8 Hz, 1H). 13C NMR (100 MHz, C6D6) δ 0.4, 62.4, 118.1, 118.4, 133.4. ESI+-MS m/z 155.9 [M+H]+, 178.1 [M+Na]+.
Synthesis of racemic 1-amino-3-butene-2-ol (S.3)
-
9
Place a magnetic stir bar in a 500 mL three-necked round-bottom flask and fit the flask with a reflux condenser. Fit a calcium chloride drying tube atop the reflux condenser. Connect a 250 mL dropping funnel capped with a rubber septum to one of the side arms of the flask, and connect a rubber septum to the other sidearm of the flask.
-
10
Put the flask on a heating mantle, which is placed on top of a magnetic stir plate and flame dry the setup well.
-
11
Quickly transfer 4.96 g (131 mmol) of LiAlH4 into the flask by removing the septum from the sidearm, and reconnect the septum.
-
12
Add 200 mL of anhydrous diethyl ether into the flask with the aid of a syringe needle.
-
13
Transfer 8.9 g (57 mmol) of 2-[(trimethylsilyl)oxy]-3-butenenitrile (S.2) dried over 4 Å molecular sieves and 100 mL of anhydrous diethyl ether into the dropping funnel with the aid of a syringe needle.
-
14
Add the ether solution of 2-[(trimethylsilyl)oxy]-3-butenenitrile (S.2) dropwise into the flask under N2 or Ar atmosphere, while stirring.
-
15
Reflux the reaction mixture for 75 min, while running water through the reflux condenser, and allow to cool to room temperature.
-
16
Remove the dropping funnel and leave the water running through the condenser.
-
17
Carefully quench any excess LiAlH4 by the sequential addition of 15 mL (113 mmol) of triethanolamine over 60 min and 5 mL of water over 30 min.
-
18
Stir the resulting globular mass for 24 h.
The reaction mixture becomes sandy in consistency.
-
19
Vacuum filter the reaction mixture using a Buchner funnel setup or filter through Celite, and wash with 50 mL of diethyl ether.
-
20
Dry the filtrate over anhydrous MgSO4 for 3 h, vacuum filter the reaction mixture using a Buchner funnel setup and evaporate the solvent using a rotary evaporator equipped with a dry ice condenser and connected to an oil pump.
The product S.3 is a clear to pale yellow oil and can be obtained in ~90% yield. 1H NMR (500 MHz, DMSO) δ 2.48 (m, 2H), 3.83 (m, 1H), 5.02 (ddd, J = 1.6, 2.4, 10.4 Hz, 1H), 5.16 (ddd, J = 1.6, 2.4, 17.2 Hz, 1H), 5.80 (ddd, J = 5.2, 10.4, 17.2 Hz, 1H). ESI+-MS m/z 88.4 [M+H]+, MS2 m/z 88.4 → 70.3 [M+H-H2O]+.
Synthesis of (2S)-N-Fmoc-1-amino-3-butene-2-ol (S.6)
-
21
Weigh out 3.50 g (40 mmol) of racemic 1-amino-3-buten-2-ol (S.3) into a 250 mL round-bottom flask.
-
22
Add 13.13 g of the ammonium salt of [(1R)-(endo,anti)]-(+)-3-bromocamphor-8-sulfonic acid (BrCSA) and mix well for 5–10 min.
-
23
Completely dissolve the solids in 100 mL of hot MeOH, gently heating if necessary, and evaporate the solvent using a rotary evaporator equipped with a dry ice condenser and connected to an oil pump.
-
24
Completely dissolve the resulting yellow solid residue in 150 mL of hot EtOAc .
-
25
Add water dropwise with the aid of a disposable Pasteur pipette until cloudiness appears (~5 mL), and allow the BrCSA salt of (2S)-1-amino-3-butene-2-ol (S.4) to crystallize.
-
26
Vacuum filter the resulting white needle-like crystals using a Buchner funnel setup, wash with cold EtOAc, and dry under vacuum.
NOTE: Additional batch(es) of crystals may be obtained by repeating the recrystallization using the filtrate.
-
27
Transfer the dried crystals into a 250 mL round-bottom flask, and add 50 mL of saturated, aqueous Ba(OH)2.
-
28
Place a magnetic stir bar in the flask, fit the flask with a reflux condenser, and place the flask in a temperature-controlled oil bath on top of a magnetic stir plate.
-
29
Reflux the reaction mixture for 30 min, while running water through the reflux condenser, and allow to cool to room temperature.
-
30
Neutralize the solution with 1 M HCl (~12 mL).
-
31
Evaporate the solvent using a rotary evaporator equipped with a dry ice condenser and connected to an oil pump. Set the temperature of the water bath to 35–40 °C.
-
32
Dissolve the light yellow crude solid of (2S)-1-amino-3-butene-2-ol (S.5) in 50 mL of 10% aqueous Na2CO3, and cool the resulting solution to 0 °C on an ice bath.
-
33
Place a magnetic stir bar in the flask, fit the flask with a 100 mL dropping funnel, and place the flask on top of a magnetic stir plate.
-
34
Weigh out 2.0 g of 9-fluorenylmethoxycarbonyl chloride (FmocCl) into the dropping funnel, and dissolve in 50 mL of 1,4-dioxane.
-
35
Add the solution of FmocCl dropwise into the flask with stirring, and continue stirring for 18 h.
-
36
Evaporate the solvent using a rotary evaporator equipped with a dry ice condenser and connected to an oil pump, and purify the product S.6 by silica gel chromatography using hexane:EtOAc.
The product S.6 is obtained as a white solid in 30–50% yield. 1H NMR (400 MHz, DMSO) δ 3.00 (m, 2H), 4.00 (m, 1H), 4.21 (dd, J = 7.2, 14.4 Hz, 1H), 4.27 (dd, J = 2.8, 7.2 Hz, 1H), 4.98 (d, J = 2.8 Hz, 1H), 5.05 (ddd, J = 1.6, 1.6, 10.4 Hz, 1H), 5.20 (ddd, J = 1.6, 1.6, 17.0 Hz, 1H), 5.80 (ddd, J = 5.6, 10.4, 17.0 Hz, 1H), 7.26 (t, J = 6.0 Hz, 1H), 7.33 (dd, J = 7.2, 7.6 Hz, 2H), 7.41 (dd, J = 7.2, 7.6 Hz, 2H), 7.70 (d, J = 7.6 Hz, 2H), 7.89 (d, J = 7.2 Hz, 2H); 13C NMR (100 MHz, DMSO) δ 46.4, 46.7, 65.3, 70.2, 120.1-127.6, 139.9, 140.7, 143.9, 156.2; ESI+-MS m/z 310.0 [M+H]+, MS2 m/z 310.0 → 131.6 [M+H-C14H11]+.
Synthesis of (2R,3S) and (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a and S.7b)
-
37
Place 665 mg (2.15 mmol) of (2S)-N-Fmoc-1-aminobut-3-en-2-ol (S.6) in a 100 mL round-bottom flask, and dry under high vacuum overnight.
-
38
Cap the flask with a rubber septum, transfer 25 mL of anhydrous CH2Cl2, and cool the flask to 0 °C in an ice bath.
-
39
Quickly transfer 1.9 g (4 eq.) of meta-chloroperbenzoic acid (mCPBA, 77% max) to the flask and purge with Ar or N2 after recapping the flask with a septum.
-
40
Stir the reaction mixture under Ar or N2 at room temperature for 18 h.
-
41
Evaporate the solvent using a rotary evaporator equipped with a dry ice condenser and connected to an oil pump, and purify the product by silica gel chromatography using hexane:EtOAc.
-
42
Dissolve the solid obtained from silica gel chromatography in CHCl3.
-
43
Isolate the diastereomers S.7a and S.7b by normal phase HPLC using a Whatman Partisil 10 (9.5 × 500 mm) column eluted at a flow rate of 5 mL/min with 0.5% MeOH in CHCl3 (isocratic elution).
Both (2R,3S)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a) and (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7b) are white solids and can be obtained in 40–60% yield. S.7a 1H NMR (400 MHz, DMSO) δ 2.59 (dd, J = 2.8, 5.6 Hz, 1H), 2.64 (dd, J = 4.0, 5.6 Hz, 1H), 2.85 (ddd, J = 2.8, 4.0, 7.4 Hz, 1H), 3.07 (ddd, J = 6.4, 6.8, 13.6 Hz, 1H), 3.16 (ddd, J = 5.6, 11.2, 13.6 Hz, 1H), 3.37 (ddd, J = 5.2, 5.2, 11.2 Hz, 1H), 4.22 (dd, J = 7.2, 14.4 Hz, 1H), 4.28 (dd, J = 1.2, 7.2 Hz, 2H), 5.04 (d, J = 4.8 Hz, 1H), 7.30 (s, 1H), 7.33 (ddd, J = 0.8, 7.2, 7.6 Hz, 2H), 7.41 (dd, J = 7.2, 7.6 Hz, 2H), 7.71 (d, J = 7.6 Hz, 2H), 7.89 (d, J = 7.2 Hz, 2H); 13C NMR (100 MHz, DMSO) δ 43.8, 46.7, 52.7, 65.4, 69.0, 79.1, 120.1, 125.2, 127.1, 127.6, 140.7, 143.9, 156.3; ESI+-MS m/z 326.6 [M+H]+, MS2 m/z 326.6 → 147.9 [M+H-C14H11]+. S.7b 1H NMR (400 MHz, DMSO) δ 2.48 (m, 1H), 2.64 (dd, J = 4.4, 5.2 Hz, 1H), 2.83 (ddd, J = 2.8, 4.4, 6.2 Hz, 1H), 3.08 (m, 2H), 3.25 (ddd, J = 6.4, 6.4, 12.0 Hz, 1H), 4.21 (dd, J = 6.8, 6.8 Hz, 1H), 4.31 (m, 2H), 5.11 (d, J = 5.2 Hz, 1H), 7.29 (s, 1H), 7.33 (ddd, J = 0.8, 7.2, 7.6 Hz, 2H), 7.41 (dd, J = 7.2, 7.6 Hz, 2H), 7.71 (d, J = 7.6 Hz, 2H), 7.89 (d, J = 7.2 Hz, 2H); 13C NMR (100 MHz, DMSO) δ 43.6, 46.7, 53.7, 65.3, 70.1, 79.2, 120.1, 125.2, 127.0, 127.6, 140.7, 143.9, 156.2; ESI+-MS m/z 326.6 [M+H]+, MS2 m/z 326.6 → 147.9 [M+H-C14H11]+.
BASIC PROTOCOL 2: SYNTHESIS, PURIFICATION AND CHARACTERIZATION OF OLIGODEOXYNUCLEOTIDES CONTAINING 6-CHLOROPURINE
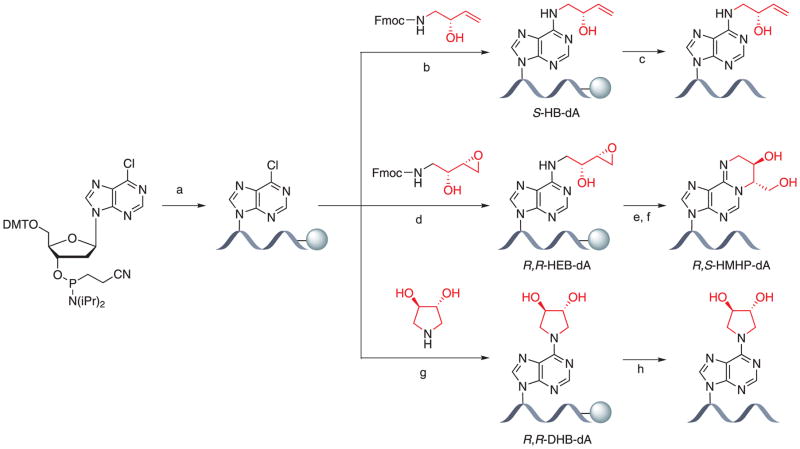
This protocol describes the synthesis of oligodeoxynucleotides containing 6-chloropurine (6-Cl-Pu) (Quirk Dorr et al., 2007; Seneviratne et al., 2010b), common DNA intermediates used in the syntheses of N6-functionalized adenine adducts (Figure 2). Solid-phase DNA synthesis uses phenoxyacetyl (PAC)-protected phosphoramidites and phenoxyacetic anhydride in THF/pyridine as the cap mix to ensure the compatibility of synthesis and deprotection of 6-Cl-Pu containing synthetic DNA.
Figure 2.
Synthesis of oligo-2'-deoxynucleotides containing 1,3-butadiene-induced deoxyadenosine lesions using a post-oligomerization approach.
The synthesis involves three major steps. (1) Automated solid-phase synthesis of the 3′-portion of the DNA strand of interest up to the location of the modified base on controlled-pore glass (CPG) support. (2) Manual coupling of the 6-Cl-Pu phosphoramidite. (3) Automated solid-phase synthesis of the 5′-portion of the DNA strand of interest to obtain the oligodeoxynucleotides containing a site-specific 6-Cl-Pu moiety. Post-oligomerization reactions to prepare site- and stereospecific DNA oligomers can be carried out on CPG support or in solution. This protocol further describes the deprotection and cleavage of the 6-Cl-Pu adducted DNA from the solid support using NaOH and HPLC purification to obtain oligodeoxynucleotides containing 6-Cl-Pu required for the post-oligomerization reactions in solution.
NOTE: Performing the post-oligomerization on solid support reduces the number of HPLC purification steps, which in turn reduces the time required for the oligodeoxynucleotide synthesis and improves the overall yields of the adducted DNA.
Materials
5′-O-(4,4′-dimethoxytrityl)-3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite of 6-chloropurine-2′-deoxyriboside (6-Cl-Pu phosphoramidite, ChemGenes Corp., Wilmington, MA)
-
5′-O-(4,4′-dimethoxytrityl)-3′-O-(2-cyanoethyl)-N,N-diisopropyl phosphoramidites (Glen Research, Sterling, VA):
Phenoxyacetyl-2′-deoxyadenosine (PAC-dA-CE) phosphoramidite
N-Acetyl-2′-deoxycytidine (Ac-dC-CE) phosphoramidite
N-p-isopropyl-phenoxyacetyl-2′-deoxyguanosine (p-iPr-PAC-dG-CE) phosphoramidite
Thymidine (dT-CE) phosphoramidite
-
1 μmol of DNA controlled-pore glass (CPG) supports (Glen Research, Sterling, VA):
PAC-dA-CPG ABI
Ac-dC-CPG ABI
p-iPr-PAC-dG-CPG ABI
dT-CPG ABI
-
Reagents for DNA synthesis (Glen Research, Sterling, VA):
Phenoxyacetic anhydride in THF/pyridine
16% 1-Methylimidazole in THF
0.02 M Iodine in THF/pyridine/water
1H-tetrazole in anhyd. ACN
3% Trichloroacetic acid in CH2Cl2
Anhydrous ACN
Anhydrous CH2Cl2
Anhydrous ACN
Concentrated ammonia
NaOH
AcOH
Triethylamine
3-hydroxypicolinic acid
10 mM Tris-HCl/5 mM MgCl2 (pH 7)
DNase I
Phosphodiesterase I (PDE I)
Phosphodiesterase II (PDE II)
Alkaline phosphatase
A glove box or a glove bag
A DNA synthesizer (e.g. Applied Biosystems Model 394)
1 mL syringes with needles
3 mL graduated V-vials (Wheaton, Millville, NJ)
Spin-X centrifuge tube filters (Corning Life Sciences, Wilkes-Barre, PA)
A Jupiter Proteo 90 Å C12 HPLC column (250 mm × 10.0 mm, Phenomenex, Torrance, CA)
A SpeedVac concentrator
Illustra NAP-25 columns (GE Healthcare, Pittsburgh, PA)
A heating block
Solid-phase synthesis of oligodeoxynucleotides containing site-specific 6-chloropurine
-
1
Weigh out 11 mg (15 μmol) of 5′-O-(4,4′-dimethoxytrityl)-3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite of 6-chloropurine-2′-deoxyriboside (6-Cl-Pu phosphoramidite) (ChemGenes Corp., Wilmington, MA) in an Ar atmosphere using a glove box or a glove bag.
-
2
Load a DNA synthesis column with 1 μmol of DNA controlled-pore glass (CPG) support corresponding to the first nucleotide at 3’-end of the desired DNA sequence onto the synthesis port of the DNA synthesizer (Applied Biosystems Model 394).
-
3
Perform the automated solid-phase oligonucleotide synthesis from the 3’-end up to the location of the modified base in ABI Begin, End Empty and DMT-off mode.
-
4
Dissolve the 6-Cl-Pu phosphoramidite in 100 μL of anhydrous ACN and withdraw into a 1 mL syringe.
-
5
Remove the CPG column from the DNA synthesizer. Attach a syringe containing the 6-Cl-Pu phosphoramidite solution to one side of the column and another syringe containing 400 μL of the activator to the other side of the column.
-
6
Pass the solutions through the CPG column for 15–20 min to couple the 6-Cl-Pu phosphoramidite to the 5’-end of the oligonucleotide.
-
7
Rinse the column using 1 mL anhydrous ACN, and put the column back onto the synthesis port on the DNA synthesizer.
-
8
Continue the automated solid-phase oligonucleotide synthesis from the modified base in Begin Extension, End Empty and DMT-off mode.
-
9
Remove the column from the synthesizer, dry it by a stream of Ar, and store the column at -20 °C.
NOTE: Post-oligomerization reactions can be carried out on CPG support or in solution.
Deprotection of the oligodeoxynucleotides (to perform the post-oligomerization reactions in solution)
-
10
Transfer the CPG support into a 3 mL graduated V-vial (Wheaton, Millville, NJ), add 1.5–2.0 mL of 0.1 M NaOH, and incubate at room temperature for 3 days in the dark.
NOTE: Concentrations of NaOH and incubation time should be followed strictly. Higher concentrations and longer incubation may results in degradation of the CPG support.
-
11
Neutralize the solution using 0.1 M AcOH and filter through a Spin-X centrifuge tube filter (Corning Life Sciences, Wilkes-Barre, PA).
Purification of the DNA oligodeoxynucleotides
-
12
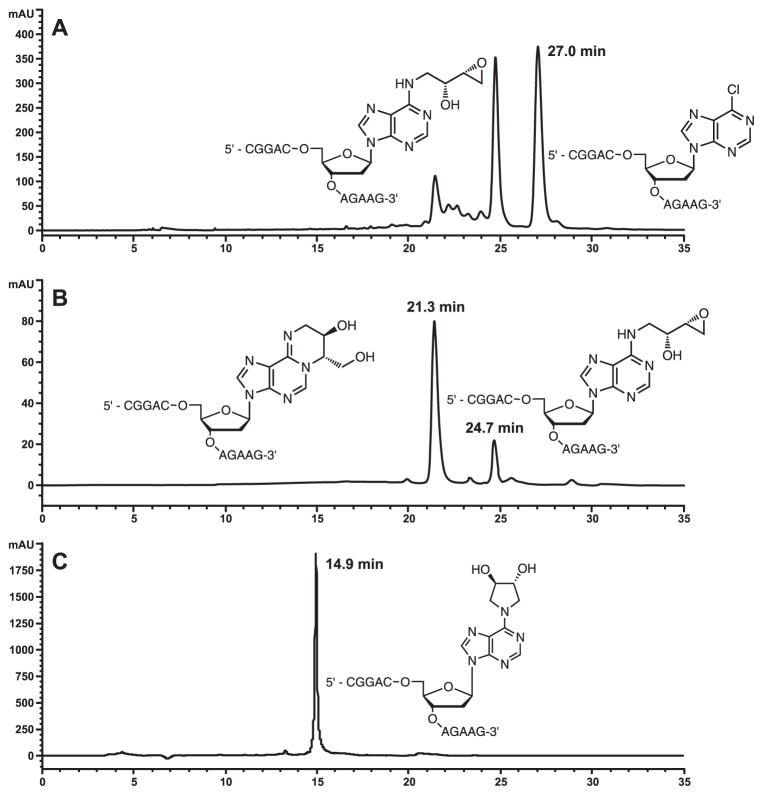
Purify the sample by reversed-phase high performance liquid chromatography (RP-HPLC) using a Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA) eluted at a flow rate of 2.5 mL/min with a linear gradient of acetonitrile (B) in 100 mM triethylammonium acetate (pH 7.0) (A). The solvent composition is changed from 3 to 9% B over 10 min, then to 11.7% B over 15 min, to 12.6% B over 7 min, to 13.0% B over 1 min, and finally to 60% B over 3 min (Figure 3).
-
13
Pool the fractions containing the 6-Cl-Pu oligodeoxynucleotide, concentrate the solution using a SpeedVac concentrator, and desalt by passing through an Illustra NAP-25 column (GE Healthcare, Pittsburgh, PA).
Figure 3.
HPLC chromatogram for the isolation of the 11-mer 5'-CGG ACX AGA AG-3' containing N6-HEB-dA (A), isolation of the 11-mer 5'-CGG ACX AGA AG-3' containing 1,N6-HMHP-dA (B), and purity check of 11-mer 5'-CGG ACX AGA AG-3' containing N6,N6-DHB-dA (C).
Characterization of synthetic oligodeoxynucleotides
-
14
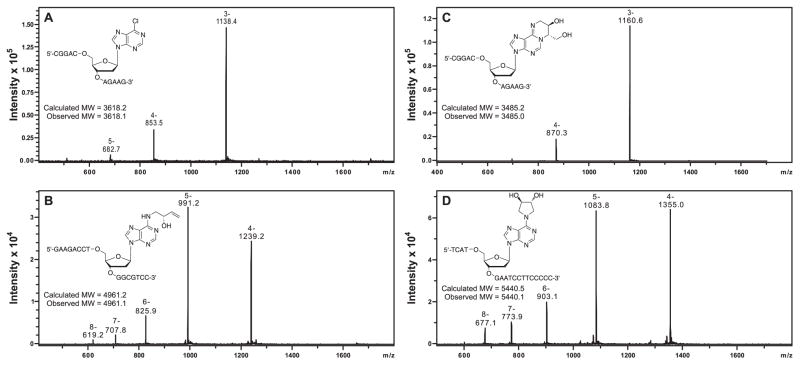
Characterize the oligodeoxynucleotide by capillary HPLC-ESI--MS using a Zorbax 300SB-C18 column (150 mm × 0.5 mm, 5 μm, Agilent Technologies, Inc., Wilmington, DE) eluted at a flow rate of 15 μL/min with a linear gradient of acetonitrile (B) in 15 mM ammonium acetate (A). The solvent composition is changed linearly from 2 to 20% B over 20 min (Figure 4A).
-
15
Dissolve ~1 nmol of the synthesized oligodeoxynucleotide in 10 mM Tris-HCl/5 mM MgCl2 (pH 7), add 35 U of DNase I, 120 mU of PDE I, 105 mU of PDE II, 22 U of alkaline phosphatase, and incubate at 37 °C for 18 h.
-
16
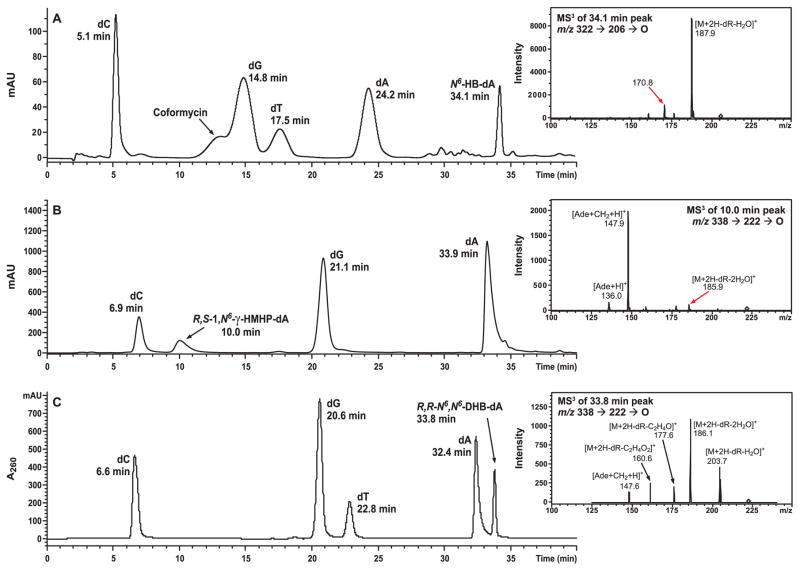
Analyze the enzymatic digests by capillary HPLC-ESI+-MSn using a Zorbax Extend-C18 column (150 mm × 0.5 mm, 5 μm, Agilent Technologies, Inc., Wilmington, DE) eluted at a flow rate of 15 μL/min with a linear gradient of acetonitrile (B) in 15 mM ammonium acetate (A). The solvent composition is kept at 0% B for 5 min, changed linearly from 0 to 2% B over 17 min, to 77% over 11 min, held at 7.7% for additional 8 min and increase to 50% B over 5 min (Figure 5).
-
17
Quantify the oligodeoxynucleotide by measuring UV absorbance at 260 nm, and store at −20 °C.
Figure 4.
Capillary HPLC-ESI--MS characterization of 11-mer 5'-CGG ACX AGA AG-3' containing 6-Cl-Pu (A), 16-mer 5'-GAA GAC CTX GGC GTC C-3' containing N6-HB-dA (B), 11-mer 5'-CGG ACX AGA AG-3' containing 1,N6-HMHP-dA (C) and 18-mer 5'-TCA TXG AAT CCT TCC CCC-3' containing N6,N6-DHB-dA (D).
Figure 5.
HPLC-UV analysis and ESI+-MS3 characterization of 18-mer 5'-TCA TXG AAT CCT TCC CCC-3' containing N6-HB-dA (A), 11-mer 5'-CGG ACX AGA AG-3' containing 1,N6-HPHP-dA (B) and 16-mer 5'-GAA GAC CTX GGC GTC C-3' containing N6,N6-DHB-dA (C).
BASIC PROTOCOL 3: SYNTHESIS OF OLIGODEOXYNUCLEOTIDES CONTAINING N6-(2-HYDROXY-3-BUTEN-1-YL)-ADENINE ADDUCTS ON CPG SUPPORT
This protocol describes site- and stereospecific synthesis, purification, and characterization of DNA oligomers containing S-N6-(2-hydroxy-3-buten-1-yl)-adenine (S-N6-HB-dA) on solid support (Figure 2b). S-N6-HB-dA is a DNA adduct induced by BD metabolite 3,4-epoxy-2-butene. Nucleophilic aromatic substitution reaction between 6-Cl-Pu adducted DNA and (2S)-N-Fmoc-1-amino-3-buten-2-ol (S.6) is carried out in DMSO in the presence of Hünig’s base, followed by deprotection and cleavage from the solid support and HPLC purification (Figure 2c).
Materials
(2S)-N-Fmoc-1-amino-3-buten-2-ol (S.6)
Anhydrous dimethylsulfoxide (DMSO)
Diisopropylethylamine (DIPEA)
ABI column containing the site-specific 6-Cl-Pu oligodeoxynucleotide on CPG support (from step 9 of basic protocol 2)
ACN
Water
NaOH
AcOH
Concentrated ammonia
Triethylamine
1.5 mL Microcentrifuge tubes
A benchtop vortex
A benchtop centrifuge
3 mL graduated V-vials (Wheaton, Millville, NJ)
Spin-X centrifuge tube filters (Corning Life Sciences, Wilkes-Barre, PA)
A Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA)
A SpeedVac concentrator
Illustra NAP-25 columns (GE Healthcare, Pittsburgh, PA)
Synthesis of (2S)-N6-(2-hydroxy-3-buten-1-yl)-adenine adducted DNA
-
1
Weigh out 7.8 mg of (2S)-N-Fmoc-1-amino-3-buten-2-ol (S.6) into a 1.5 mL microcentrifuge tube and dissolve in 300 °L of anhydrous DMSO.
-
2
Add 100 μL of DIPEA, mix well by vortexing, and spin down by centrifugation.
-
3
Transfer 1/8th of the beads from the ABI column containing the 6-Cl-Pu oligomer (from basic protocol 2, assuming 80% yield) into a 1.5 mL microcentrifuge tube.
-
4
Add the reaction mixture from step 3 and incubate at 60 °C for 36 h in the dark.
Deprotection of the oligodeoxynucleotides
-
5
Remove the supernatant and wash the beads with 400–500 μL of ACN (3 times), followed by 400–500 μL of water (3 times).
-
6
Transfer the CPG support into a 3 mL graduated V-vial (Wheaton, Millville, NJ), add 1.5–2.0 mL of 0.1 M NaOH, and incubate at room temperature for 3 days in the dark.
-
7
Neutralize the solution using 0.1 M AcOH and filter through a Spin-X centrifuge tube filter (Corning Life Sciences, Wilkes-Barre, PA).
Purification of the oligodeoxynucleotides
-
8
Purify the sample by reversed-phase high performance liquid chromatography (RP-HPLC) using a Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA) eluted at a flow rate of 2.5 mL/min with a linear gradient of acetonitrile in 100 mM triethylammonium acetate (pH 7.0). The solvent composition is changed from 3 to 9% B over 10 min, then to 10.3% B over 21 min, and finally to 70% B over 2 min.
-
9
Pool the fractions containing the S-N6-HB-dA oligodeoxynucleotide, concentrate the solution using a SpeedVac concentrator, and desalt by passing through an Illustra NAP-25 column (GE Healthcare, Pittsburgh, PA).
-
10
Characterize and quantify the resulting oligodeoxynucleotide as described in steps 14–17 of basic protocol 2 (Figures 4B and 5A).
ALTERNATE PROTOCOL 3: SYNTHESIS OF OLIGODEOXYNUCLEOTIDES CONTAINING N6-(2-HYDROXY-3-BUTEN-1-YL)-ADENINE ADDUCTS IN SOLUTION
This protocol describes solution phase synthesis, purification, and characterization of DNA oligomers containing site- and stereospecific S-N6-HB-dA adducts (Quirk Dorr et al., 2007). Nucleophilic aromatic substitution reaction between 6-Cl-Pu adducted DNA and (2S)-N-Fmoc-1-amino-3-buten-2-ol (S.6) is carried out in DMSO in the presence of Hünig’s base, followed by HPLC purification generates the site- and stereospecifically modified DNA (Figure 2b,f).
Additional Materials
Synthetic oligodeoxynucleotides containing site-specific 6-Cl-Pu (from step 13 of Basic Protocol 2)
Synthesis of (2S)-N6-(2-hydroxy-3-buten-1-yl)-adenine adducted DNA
-
1
Weigh out 9.7 mg of (2S)-N-Fmoc-1-amino-3-buten-2-ol (S.6) into a 1.5 mL microcentrifuge tube, and dissolve the solid in 300 μL of anhydrous DMSO.
-
2
Add 100 μL of DIPEA, mix well by vortexing, and spin down by centrifugation.
-
3
Transfer 100 nmol of the 6-Cl-Pu containing oligodeoxynucleotide (from basic protocol 2) into a 1.5 mL microcentrifuge tube and completely dry using a SpeedVac concentrator.
-
4
Add the reaction mixture from step 3 into the tube containing the dry synthetic DNA and incubate at 60 °C for 36 h in the dark.
Purification of the oligodeoxynucleotide
-
5
Dilute the reaction mixture to 10% DMSO by adding water and separate by reversed-phase high performance liquid chromatography (RP-HPLC) using a Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA) eluted at a flow rate of 2.5 mL/min with a linear gradient of acetonitrile in 100 mM triethylammonium acetate (pH 7.0). The solvent composition is changed from 3 to 9% B over 10 min, then to 10.3% B over 21 min, and finally to 70% B over 2 min.
-
6
Pool the fractions containing the S-N6-HB-dA oligodeoxynucleotide, concentrate the solution using a SpeedVac concentrator, and desalt by passing through an Illustra NAP-25 column (GE Healthcare, Pittsburgh, PA).
-
7
Characterize and quantify the S-N6-HB-dA oligodeoxynucleotide as described in steps 14–17 of basic protocol 2.
BASIC PROTOCOL 4: SYNTHESIS OF OLIGODEOXYNUCLEOTIDES CONTAINING 1,N6-(2-HYDROXY-3-HYDROXYMETHYLPROPAN-2,3-DIYL)-ADENINE ADDUCTS ON CPG SUPPORT
This protocol describes site- and stereospecific synthesis, purification, and characterization of DNA oligomers containing R,R and R,S-1,N6-(2-hydroxy-3-hydroxymethylpropan-2,3-diyl)-adenine (HMHP-dA), adenine exocycles induced by R,S and R,R stereoisomers of 1,2,3,4-diepoxybutane (DEB), on solid support (Figure 2). This procedure involves two synthetic steps. (1) Nucleophilic aromatic substitution reaction between 6-Cl-Pu adducted DNA and (2R,3S) or (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a or S.7b) is carried out in DMSO in the presence of Hünig’s base to generate N6-(2-hydroxy-3,4-epoxybutan-1-yl)-adenine adducted DNA (Figure 2c). (2) Spontaneous cyclization of N6-(2-hydroxy-3,4-epoxybutan-1-yl)-adenine in water to generate 1,N6-(2-hydroxy-3-hydroxymethylpropan-2,3-diyl)-adenine adducted DNA. Deprotection and cleavage from the solid support using NaOH and HPLC purification generate the site- and stereospecifically modified DNA containing R,R and R,S-1,N6-(2-hydroxy-3-hydroxymethylpropan-2,3-diyl)-adenine (Figure 2e,f).
Materials
(2R,3S) or (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a or S.7b)
Anhydrous DMSO
DIPEA
ABI column containing the site-specific 6-Cl-Pu oligodeoxynucleotide on CPG support (from step 9 of Basic Protocol 2)
ACN
Water
NaOH
AcOH
Concentrated ammonia
Triethylamine
1.5 mL microcentrifuge tubes
A benchtop vortex
A benchtop centrifuge
3 mL graduated V-vials (Wheaton, Millville, NJ)
Spin-X centrifuge tube filters (Corning Life Sciences, Wilkes-Barre, PA)
A Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA)
A SpeedVac concentrator
Illustra NAP-25 columns (GE Healthcare, Pittsburgh, PA)
Synthesis of N6-(2-hydroxy-3,4-epoxybutan-1-yl)-adenine containing DNA
-
1
Weigh out 3.3 mg of (2R,3S) or (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (7a or 7b) into a 1.5 mL microcentrifuge tube, and dissolve the solid in 300 μL of anhydrous DMSO.
-
2
Add 100 μL of DIPEA, mix well by vortexing, and spin down by centrifugation.
-
3
Transfer 1/8th of the beads from the ABI column containing the 6-Cl-Pu oligomer (from basic protocol 2) into a 1.5 mL microcentrifuge tube.
-
4
Add the reaction mixture from step 3 and incubate at 37 °C for 24 h in the dark.
Synthesis of 1,N6-(2-hydroxy-3-hydroxymethylpropan-2,3-diyl)-adenine containing DNA
-
5
Remove the supernatant and wash the beads with 400–500 μL of ACN (3 times), followed by 400–500 μL of water (3 times).
-
6
Add 300 μL of water to the oligonucleotide solution, and incubate at 25 °C for 3 h in the dark.
NOTE: Leaving the reaction mixture longer than recommended incubation times may result in rearrangement products.
Deprotection, purification and characterization of 1,N6-HMHP-dA oligodeoxynucleotides
-
7
Cleave the 1,N6-HMHP-dA containing DNA from the CPG support as described in steps 5–7 of Basic Protocol 3.
-
8
Purify the desired oligomer by reversed-phase high performance liquid chromatography (RP-HPLC) using a Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA) eluted at a flow rate of 2.5 mL/min with a linear gradient of acetonitrile (B) in 100 mM triethylammonium acetate (pH 7.0) (A). Solvent composition is changed linearly from 3 to 9% B over 10 min, further to 10.3% B over 21 min, and finally to 70% B over 2 min.
-
9
Characterize and quantify the 1,N6-HMHP-dA oligodeoxynucleotides as described in steps 14–17 of Basic Protocol 2 (Figures 4C, 5B).
ALTERNATE PROTOCOL 4: SYNTHESIS OF OLIGODEOXYNUCLEOTIDES CONTAINING 1,N6-(2-HYDROXY-3-HYDROXYMETHYLPROPAN-2,3-DIYL)-ADENINE ADDUCTS (1,N6-HMHP-dA ) IN SOLUTION
This protocol describes site- and stereospecific synthesis, purification, and characterization of DNA oligomers containing R,R and R,S-1,N6-HMHP-dA in solution (Seneviratne et al., 2010b). This procedure involves two synthetic steps. (1) Nucleophilic aromatic substitution between 6-Cl-Pu containing DNA and (2R,3S) or (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a or S.7b) is carried out in DMSO in the presence of Hünig’s base to generate (R,S) or (R,R)-N6-(2-hydroxy-3,4-epoxybutan-1-yl)-adenine adducted DNA, which are isolated by reversed-phase HPLC (Figure 2d). (2) Spontaneous cyclization of N6-(2-hydroxy-3,4-epoxybutan-1-yl)-adenine in water to generate 1,N6-(2-hydroxy-3-hydroxymethylpropan-2,3-diyl)-adenine adducted DNA followed by HPLC purification generate the site- and stereospecifically modified DNA containing R,R and R,S-1,N6-(2-hydroxy-3-hydroxymethylpropan-2,3-diyl)-adenine (Figure 2e).
Additional Materials
Synthetic oligodeoxynucleotides containing site-specific 6-Cl-Pu (from step 13 of Basic Protocol 2)
Synthesis of N6-(2-hydroxy-3,4-epoxybutan-1-yl)-adenine adducted DNA
-
1
Weigh out 3.3 mg of (2R,3S) or (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane (S.7a or S.7b) into a 1.5 mL microcentrifuge tube and dissolve the solid in 300 μL of anhydrous DMSO.
-
2
Add 100 μL of DIPEA, mix well by vortexing, and spin down by centrifugation.
-
3
Transfer 100 nmol of the 6-Cl-Pu- containing oligodeoxynucleotide into a 1.5 mL microcentrifuge tube and completely dry using a SpeedVac concentrator.
-
4
Add the solution from step 2 into the tube containing the dried synthetic DNA and incubate at 37 °C for 24 h in the dark.
-
5
Dilute the reaction mixture to 10% DMSO by adding water and filter through a Spin-X centrifuge tube filter.
-
6
Purify and characterize the 1,N6-HMHP-dA oligodeoxynucleotides as described in steps 8–9 of Basic Protocol 4 (Figures 3 and 4).
Purification and characterization of 1,N6-HMHP-dA adducted DNA
-
7
Concentrate the desalted 1,N6-HMHP-dA containing DNA using a SpeedVac concentrator.
-
8
Add water to make the final volume of the oligonucleotide solution to 300 μL, and incubate at 25 °C for 3 h in the dark.
-
9
Purify and characterize the resulting oligodeoxynucleotide containing 1,N6-(2-hydroxy-3-hydroxymethylpropan-2,3-diyl)-adenine adducts as described in steps 8–9 of Basic Protocol 4 (Figure 3B).
BASIC PROTOCOL 5: SYNTHESIS OF OLIGODEOXYNUCLEOTIDES CONTAINING N6,N6-(2,3-DIHYDROXYBUTAN-1,4-DIYL)-ADENINE ADDUCTS ON CPG SUPPORT
This protocol describes site- and stereospecific synthesis, purification, and characterization of DNA oligomers containing R,R-N6,N6-(2,3-dihydroxybutan-1,4-diyl)-adenine (N6,N6-DHB-dA), exocyclic N6,N6-dA adducts induced by DEB, on solid support (Figure 2). Nucleophilic aromatic substitution between 6-Cl-Pu containing DNA and (3R,4R)-pyrrolidine-3,4-diol (S.9) is carried out in DMSO in the presence of Hünig’s base, followed by deprotection and cleavage from the solid support using NaOH. The resulting site- and stereospecifically modified DNA containing N6,N6-DHB-dA are purified by reversed-phase HPLC. The corresponding S,S streoisomer can be prepared analogously starting with (3S,4S)-pyrrolidine-3,4-diol.
Materials
(3R,4R)-Pyrrolidine-3,4-diol (S.9, see Support Protocol 5)
Anhydrous DMSO
DIPEA
ABI column containing the site-specific 6-Cl-Pu oligodeoxynucleotide on CPG support (from step 9 of Basic Protocol 2)
ACN
Water
NaOH
AcOH
Concentrated ammonia
Triethylamine
1.5 mL Microcentrifuge tubes
A benchtop vortex
A benchtop centrifuge
3 mL graduated V-vials (Wheaton, Millville, NJ)
Spin-X centrifuge tube filters (Corning Life Sciences, Wilkes-Barre, PA)
A Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA)
A SpeedVac concentrator
Illustra NAP-25 columns (GE Healthcare, Pittsburgh, PA)
Synthesis of (2R,3R)-N6,N6-(2,3-dihydroxybutan-1,4-diyl)-adenine containing DNA
-
1
Weigh out 5.0 mg of (3R,4R)-pyrrolidine-3,4-diol into a 1.5 mL microcentrifuge tube and dissolve the solid in 300 μL of anhydrous DMSO.
-
2
Add 100 μL of DIPEA, mix well by vortexing, and spin down by centrifugation.
-
3
Transfer 1/8th of the beads from the ABI column containing the 6-Cl-Pu oligomer (from basic protocol 2) into a 1.5 mL microcentrifuge tube.
-
4
Add the reaction mixture from step 3 and incubate at 37 °C for 72 h in the dark.
Deprotection, purification and characterization of the oligodeoxynucleotides
-
5
Cleave the N6,N6-DHB-dA containing DNA from CPG support as described in steps 5–7 of Basic Protocol 3.
-
6
Purify the resulting synthetic oligomers by reversed-phase HPLC using a Jupiter Proteo 90 Å C12 column (250 mm × 10.0 mm, Phenomenex, Torrance, CA) eluted at a flow rate of 2.5 mL/min with a linear gradient of acetonitrile (B) in 100 mM triethylammonium acetate (pH 7.0) (A). The solvent composition is changed from 3 to 11% B over 10 min, then to 11.7% B over 21 min, and finally to 50% B over 2 min (Figure 3C).
-
7
Characterize and quantify the N6,N6-DHB-dA oligodeoxynucleotide by HPLC-ESI-MS as described in steps 14–17 of Basic Protocol 2 (Figures 4D and 5C).
ALTERNATE PROTOCOL 5: SYNTHESIS OF OLIGODEOXYNUCLEOTIDES CONTAINING N6,N6-(2,3-DIHYDROXY-BUTAN-1,4-DIYL)-ADENINE ADDUCTS IN SOLUTION
This protocol describes solution-state synthesis, purification, and characterization of DNA oligomers containing site- and stereospecific R,R-N6,N6-DHB-dA (Seneviratne et al., 2010b). Nucleophilic aromatic substitution reaction between 6-Cl-Pu adducted DNA and (3R,4R)-pyrrolidine-3,4-diol (S.9) is carried out in DMSO in the presence of Hünig’s base, followed by HPLC purification generates the site- and stereospecifically modified DNA containing R,R-N6,N6-DHB-dA. The corresponding S,S stereoisomer can also be prepared by using (3S,4S)-pyrrolidine-3,4-diol.
Additional Materials
Synthetic oligodeoxynucleotides containing site-specific 6-Cl-Pu (from step 13 of Basic Protocol 2)
Synthesis of R,R-N6,N6-DHB-dA adducted DNA
Weigh out 5.0 mg of (3R,4R)-pyrrolidine-3,4-diol into a 1.5 mL microcentrifuge tube, and dissolve the solid in 300 μL of anhydrous DMSO.
Add 100 μL of DIPEA, mix well by vortexing, and spin down by centrifugation.
Transfer 100 nmol of the oligodeoxynucleotide containing 6-Cl-Pu (from basic protocol 2) into a 1.5 mL microcentrifuge tube and completely dry using a SpeedVac concentrator.
Add the reaction mixture from step 3 into the tube containing the dried synthetic DNA, and incubate at 37 °C for 72 h in the dark.
Dilute the reaction mixture to 10% DMSO by adding water and filter through a Spin-X centrifuge tube filter.
Purify and characterize the oligodeoxynucleotide containing N6,N6-(2,3-dihydroxybutan-1,4-diyl)-adenine as described in steps 6–7 of basic protocol 5.
SUPPORT PROTOCOL 5: SYNTHESIS OF (3R,4R)-PYRROLIDINE-3,4-DIOL (S.9)
This procedure describes the synthesis of (3R,4R)-pyrrolidine-3,4-diol (S.9) by hydrogenolysis of commercial(3R,4R)-(-)-1-benzyl-3,4-pyrrolidinediol. Similarly, (3S,4S)-pyrrolidine-3,4-diol can be obtained by the hydrogenolysis of (3S,4S)-(+)-1-benzyl-3,4-pyrrolidinediol.
Materials
(3R,4R)-(-)-1-Benzyl-3,4-pyrrolidinediol (Sigma Aldrich, St. Louis, MO)
10% Pd/C
Anhydrous MeOH
H2 gas
Celite
A small magnetic stir bar
A 50 mL round-bottom flask
A heating mantle
A magnetic stir plate
A rubber septum
Syringe needles
A glass fritted Buchner funnel setup
A rotary evaporator equipped with a dry ice condenser and connected to an oil pump
Place a small magnetic stir bar in an oven-dried 50 mL round-bottom flask.
Place the flask on a heating mantle, which is placed on top of a magnetic stir plate.
Weigh out 1.0 g of (3R,4R)-(−)-1-benzyl-3,4-pyrrolidinediol (S.8) and 150 mg of 10% Pd/C into the flask, and mix the solids after capping the flask with a rubber septum.
-
Add 5 mL of anhydrous MeOH dropwise into the solid mixture using a syringe needle.
CAUTION: Allow the flask to cool to ambient temperature, and carefully add MeOH in a well-ventilated fume hood to prevent ignition of the solvent.
Slowly and carefully purge the flask with H2 gas using syringe needles to remove any air and to fill the flask with H2 gas.
Fit the flask with a balloon filled with H2 gas using a needle.
Stir the reaction mixture at 55 °C for 24 h and allow the reaction mixture to cool to room temperature.
Add celite (1–1.5 cm layer) onto a glass-fritted Buchner funnel setup, and wash the celite with MeOH.
Filter the reaction mixture through the celite bed, and wash the celite bed with 5–10 mL of MeOH.
-
Evaporate the solvent using a rotary evaporator equipped with a dry ice condenser and connected to an oil pump.
(3R,4R)-pyrrolidine-3,4-diol (S.9) is a yellowinsh-white solid and can be obtained in 90–98% yield. 1H NMR (400 MHz, DMSO) δ 2.45 (s, 1H), 2.90 (dd, J = 5.6, 12.0 Hz, 4H), 3.78 (d, J = 5.6 Hz); 13C NMR (100 MHz, DMSO) δ 53.2, 77.5; ESI+-MS m/z 104.1 [M+H]+, MS2 m/z 104.1 → 86.2 [M+H-H2O]+, 104.1 → 68.2 [M+H-2H2O]+.
REAGENTS AND SOLUTIONS
100 mM triethylammonium acetate (pH 7.0)
-
Dissolve 14 mL of triethylamine and 5.5 mL of glacial acetic acid in 950 mL of deionized, distilled water. Adjust the pH using acetic acid to pH 7, and add water to make to final volume to 1 L.
The buffer can be stored at ambient temperature for 1–2 weeks.
COMMENTARY
Background Information
Structural modification of DNA bases by carcinogens or their reactive metabolites is a critical step in chemical carcinogenesis. The resulting structurally altered nucleobases can be misread by DNA polymerases, leading to heritable mutations and cancer (Geacintov and Broyde, S.2010). A complex mixture of DNA lesions can be formed following exposure, making it difficult to attribute the observed biological effects to specific type of carcinogen-DNA adducts. For example, human and animal carcinogen 1,3-butadiene is metabolically activated to the corresponding mono and diepoxides, which react with DNA to produce a range of nucleobase monoadducts, exocyclic lesions, inter- and intrastrand DNA-DNA cross-links, and DNA-protein crosslinks. In order to understand the exact mechanisms of carcinogen-induced mutations, it is necessary to characterize each adduct’s effects of DNA structure, its miscoding potential, and its ability to be recognized and removed by DNA repair proteins. This requires the preparation of DNA oligodeoxynucleotides containing site- and stereospecific DNA adducts of defined structure (Geacintov and Broyde, S.2010).
Three general strategies have been developed for synthesis of DNA oligodeoxynucleotides containing site-specific adducts. In the direct adduction approach, synthetic oligomers are reacted with metabolically activated form of the carcinogen, and the adducted DNA molecules are isolated by high performance liquid chromatography (Delaney and Essigmann, J. M. 2008; Harris and Harris, C. M. 2001). This strategy have been useful for carcinogens that exhibit a high degree of chemoselectivity, e.g. benzo[a]pyrene diolepoxide, aflatoxin B1, and N-acetyl-2-aminofluorene (Harris and Harris, C. M. 2001). However, direct adduction approach has serious limitations. First, strongly electrophilic carcinogens may react with several nucleobases in the oligomer and modify multiple nucleophilic sites to generate a mixture of products that are difficult to resolve. Secondly, the target oligonucleotide cannot contain more than 1–3 target bases, limiting the applicability of this strategy. Finally, the reaction can lead to a mixture of stereoisomers if the activated carcinogen contains a chiral center.
The phosphoramidite approach involves the use of structurally modified nucleoside phosphoramidites as monomers in solid phase synthesis of DNA (Delaney and Essigmann, J. M. 2008; Harris and Harris, C. M.2001; Iwai 2006; Takamura-enya et al., 2006). This route allows for a complete control of site specificity and stereochemistry of adduction and enables the synthesis of DNA strands of any sequence. However, nucleoside phosphoramidite synthesis requires multiple synthetic steps which can be low yielding and labor intensive. Any carcinogen-derived functionalities present on the DNA adduct of interest must be compatible with synthetic steps involved in phosphoramidite chemistry and solid phase synthesis and in many case require protective groups. Furthermore, the adduct of interest much be stable enough to survive harsh conditions of oligonucleotide deprotection and cleavage form solid support.
The post-oligomerization approach originally reported by Matteucci and Webb and further developed by Benkovic, Verdine, and Harris (Harris and Harris, C. M.2001) uses solid phase synthesis to prepare DNA molecules containing site-specific an intrinsically reactive electrophilic nucleoside (e.g. 6-chloropurine or 2-fluoro-dI) which serves as a key intermediate for post-oligomerization reactions to introduce the carcinogen moiety via nucleophilic aromatic substitution (SNAr) reactions (Harris and Harris, C. M.2001). Reactions of the electrophilic equivalent of the nucleobase on DNA with the nucleophilic equivalent of the carcinogen affords the adducted oligomer of interest in a regio- and stereospecific manner (Delaney and Essigmann, J. M.2008; Harris and Harris, C. M.2001; Omumi et al., 2011; Stone et al., 2008; Wang et al., 2006). In this approach,
The post-oligomerization strategy has been proven to be a useful strategy for synthesis of DNA molecules containing a range of exocylic amine-functionalized nucleobase adducts including N6-BPDE-dA, N2-BPDE-dG, ethanocytosine, ethanoadenosine and 1,N2-hydroxypropano-deoxyguanosine (Harris and Harris, C. M.2001). This approach overcomes many of the limitations of the phosphoramidite and direct adduction approaches described above by employing synthetic oligonucleotides bearing a unique reactive site for in situ modifications which generate the desired adducted DNA strands. The resulting adduct-containing DNA strands can be used for structural studies by NMR, X-ray crystallography, UV melting, and circular dichroism spectroscopic studies to evaluate the effects of DNA lesions on DNA structure and stability. Furthermore, these modified oligodeoxynucleotides can be used to characterize adduct repair, polymerase bypass, and their effects on DNA replication and transcription.
However, several considerations should be kept in mind when assembling DNA molecules containing halogenated intermediates for postoligomerization reactions. Because of their intrinsic reactivity towards nucleophiles, standard protection-deprotection chemistry may not be appropriate. Instead of traditional acetyl or dimethylformamide protected phosphoramidites, phenoxyacetyl (PAC) protected phosphoramidites are typically used, which enable ultra-mild deprotection conditions (incubation at room temp. in 0.1 M NaOH instead of heating in conc. ammonia for typical solid phase synthesis). Furthermore, the postoligomerization approach is only applicable to exocyclic amino adducts.
In the present Protocol, we describe a post-oligomerization approach for the preparation of site-specific oligodeoxynucleotides containing structurally defined 2’-deoxyadenosine adducts of 1,3-butadiene (BD). BD is an industrial chemical widely used in synthetic polymer industry (White2007), and also found in automobile exhaust and in cigarette smoke (Hecht 1999; Hoffmann et al., 2001). Metabolites of BD like 3,4-epoxybut-1-ene (EB), 3,4-epoxybutan-1,2-diol (EBD) and 1,2,3,4-diepoxybutane (DEB) contain highly reactive epoxide groups. EB and DEB form a range of DNA adducts by alkylating multiple nucleophilic sites on DNA, specifically N7 position of guanine, and N1, N3 and N6 positions of adenine (Carmical et al., 2000; Tretyakova et al., 1997b; Tretyakova et al., 1997c). Since BD and DEB induce a large number of A→T transversions, it has been proposed that BD-adenine adducts are critically important for its mutagenic activity. Thus, it is essential to prepare synthetic oligonucleotides containing stereo- and regiospecific BD-dA adducts so that their biological consequences can be examined.
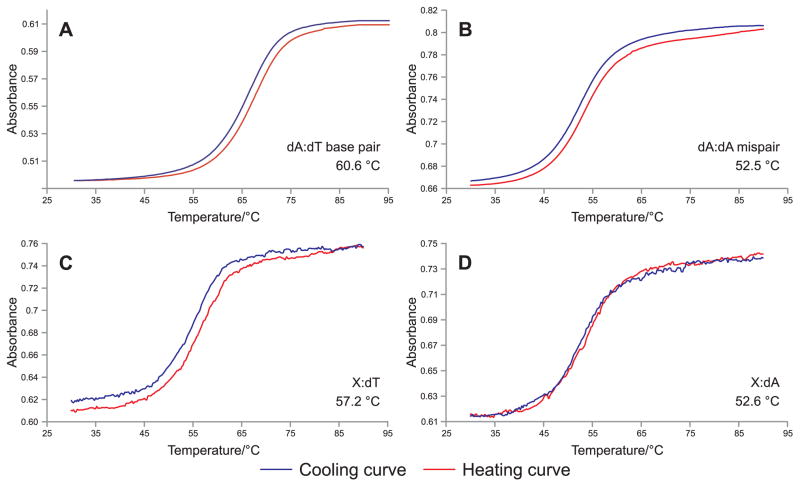
The post-oligomerization strategies developed in our laboratory and described in this protocol have been successfully employed to prepare a range of DNA molecules containing site and stereospecific N6-HB-dA, N6-HEB-dA, 1,N6-HMHP-dA, and N6,N6-DHB-dA adducts, which were subsequently employed in structural studies by NMR, thermodynamic duplex stability experiments (Figure 6), polymerase bypass assays, site-specific mutagenesis, and in vitro repair studies. Our studies exemplify the power of the postoligomerization approach, which has the advantage of not requiring high carcinogen chemoselectivity as in direct adduction approach or chemical compatibility as in phosphoramidite approach. This methodology can be exploited in various ways to synthesize a broad spectrum of DNA adducts, which can be useful for a range of critical experiments.
Figure 6.
UV melting curves for DNA duplexes containing N6-HB-dA (X) opposite dT or dA.
Critical Parameters and Troubleshooting
Careful attention to details of basic organic synthesis procedures and sold phase DNA synthesis is advised as some of the procedures involve flammable and toxic materials. Further, familiarity with routine techniques for organic laboratory operation such as solvent distillation, evaporation, extraction, TLC analysis, column chromatography, and air free techniques is recommended. Anhydrous solvents and reaction conditions are required for several procedures in this unit. It is recommended to purchase anhydrous solvents in Sure/Seal bottles in the smallest size to fit the requirement for the synthetic procedure. The use of anhydrous or freshly distilled solvents and thoroughly dried starting materials is essential. All glassware needs to be oven-dried, and several steps require flame drying of glassware.
Adherence to good laboratory safety practices is required. Some of the chemicals and reagents used are highly toxic, corrosive or flammable. Refer to material safety data sheets and safety information prior to use. All reactions should be conducted in a well-ventilated fume hood. Use of personal protection equipment including safety goggles, a laboratory coat and gloves is required.
The post-oligomerization reactions described above can be carried out on solid support or in solution. Synthesis on solid support reduces the number of HPLC purification steps, reducing the synthesis time improving the overall yields of adducted DNA. The procedures are optimized to carry out the syntheses of specific BD-dA adducts described in this unit. If applied to other dA adducts, they may require additional optimization to achieve optimum yields. Conditions described for the deprotection and cleavage of synthetic DNA from solid support are also optimized for the syntheses outlined in this unit. NaOH concentration and incubation times should be followed strictly since higher concentrations of the base and longer incubations may result in degradation of DNA and the CPG support.
Anticipated Results
The procedures described in this unit produce highly pure site- and stereospecifically modified oligodeoxynucleotides in good to moderate yields. While the yields can vary depending on specific modification and the length of the oligonucleotide sequence, significantly higher amounts of target oligomers are obtained if the post-oligomerization is carried out on solid support.
Time Considerations
With prior experience in basic organic techniques, (2R,3S) or (2R,3R)-N-Fmoc-1-amino-2-hydroxy-3,4-epoxybutane can be prepared in two to three weeks. Oligonucleotides containing 6-Cl-Pu can be prepared in a day. Deprotection and HPLC purification can be performed in approximately 5 days (since deprotection requires 3 days), if the post-oligomerization is to be carried out in solution. Pure oligonucleotides containing BD-induced deoxyadenosine adducts can be obtained in approximately a week. It is recommended that all intermediates and oligodeoxynucleotides on solid support are completely dried and stored at or below −20 °C under Ar or N2. Conditions such as air and moisture can significantly affect the shelf life. It is also recommended that the pure modified oligonucleotides to be stored in dried, solid form under inert conditions, if long-term storage is required.
Considerable variation in these times is to be expected based on the time required for drying and/or distillation of reagents and synthetic compounds, recrystallization, chromatographic separation and analysis, as well as on the level of expertise of the experimenter.
Acknowledgments
This research was supported by grants from the NIH (R01-CA-100670, R01-CA-095039). We thank Danae Quirk Dorr, Sergey Antsypovich, and Uthpala Seneveratne for their great contributions to synthetic methodology development and Carmelo Rizzo, Michael Stone, and Thomas Harris (Vanderbilt University) and Pramod Upadhyaya (University of Minnesota) for helpful discussions.
Contributor Information
Susith Wickramaratne, Email: wickr011@umn.edu, Department of Medicinal Chemistry and the Masonic Cancer Center, University of Minnesota, Cancer and Cardiovascular Research Building, 2231 6thStreet SE - Room 2-220, Minneapolis, MN 55455, 612-624-0638.
Christopher L. Seiler, Email: seile053@umn.edu, Department of Medicinal Chemistry and the Masonic Cancer Center, University of Minnesota, Cancer and Cardiovascular Research Building, 2231 6thStreet SE - Room 2-220, Minneapolis, MN 55455, 612-624-0638
Natalia Y. Tretyakova, Email: trety001@umn.edu, Department of Medicinal Chemistry and the Masonic Cancer, Center University of Minnesota, Cancer and Cardiovascular Research Building, 2231 6thStreet SE - Room 2-147, Minneapolis, MN 55455, 612-626-3432
LITERATURE CITED
- Carmical JR, Nechev LV, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of adenine N6 adducts of monoepoxide and diolepoxide derivatives of butadiene. Environ Mol Mutagen. 2000;35:48–56. [PubMed] [Google Scholar]
- Delaney JC, Essigmann JM. Biological properties of single chemical-DNA adducts: a twenty year perspective. Chem Res Toxicol. 2008;21:232–252. doi: 10.1021/tx700292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geacintov NE, Broyde S. The chemical biology of DNA damage 2010 [Google Scholar]
- Harris TM, Harris CM. Synthesis of oligodeoxynucleotides bearing structurally defined adducts of mutagens. ARKIVOC. 2001;vi:16–29. doi: 10.1021/tx9501795. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- Himmelstein MW, Acquavella JF, Recio L, Medinsky MA, Bond JA. Toxicology and epidemiology of 1,3-butadiene. Crit Rev Toxicol. 1997;27:1–108. doi: 10.3109/10408449709037482. [DOI] [PubMed] [Google Scholar]
- Hoffmann D, Hoffmann I, el Bayoumy K. The less harmful cigarette: a controversial issue a tribute to Ernst L Wynder. Chem Res Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- Iwai S. Chemical synthesis of oligonucleotides containing damaged bases for biological studies. Nucleosides, Nucleotides & Nucleic Acids. 2006;25:561–582. doi: 10.1080/15257770600685826. [DOI] [PubMed] [Google Scholar]
- Omumi A, Beach DG, Baker M, Gabryelski W, Manderville R. Postsynthetic guanine arylation of DNA by Suzuki-Miyaura cross-coupling. J Am Chem Soc. 2011;133:42–50. doi: 10.1021/ja106158b. [DOI] [PubMed] [Google Scholar]
- Quirk Dorr D, Murphy K, Tretyakova N, Dorr DQ. Synthesis of DNA oligodeoxynucleotides containing structurally defined N 6 -(2-hydroxy-3,4-buten-1-yl)-adenine adducts of 3,4-epoxy-1-butene. Chem Biol Interact. 2007;166:104–111. doi: 10.1016/j.cbi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Seneviratne U, Antsypovich S, Goggin M, Dorr DQ, Guza R, Moser A, Thompson C, York DM, Tretyakova N. Exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, structural elucidation, and mechanistic studies. Chem Res Toxicol. 2010a;23:118–133. doi: 10.1021/tx900312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne U, Antsypovich S, Quirk-Dorr D, Dissanayake T, Kotapati S, Tretyakova NY. DNA oligomers containing site-specific and stereospecific exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, characterization, and effects on DNA structure. Chem Res Toxicol. 2010b;23:1556–1567. doi: 10.1021/tx100146v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MP, Cho Yj, Huang HAI, Kim Hy, Kozekov ID, Kozekova A, Wang HAO, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA Crosslinks Induced by α,β,-Unsaturated Aldehydes Derived from Lipid Peroxidation and Environmental Sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura-enya T, Enomoto S, Wakabayashi K. Palladium-catalyzed direct N-arylation of nucleosides, nucleotides, and oligonucleotides for efficient preparation of dG-N2 adducts with carcinogenic amino-nitroarenes. J Org Chem. 2006;71:5599–5606. doi: 10.1021/jo0605243. [DOI] [PubMed] [Google Scholar]
- Tretyakova N, Lin Y, Sangaiah R, Upton PB, Swenberg JA. Identification and quantitation of DNA adducts from calf thymus DNA exposed to 3,4-epoxy-1-butene. Carcinogenesis. 1997a;18:137–147. doi: 10.1093/carcin/18.1.137. [DOI] [PubMed] [Google Scholar]
- Tretyakova N, Sangaiah R, Yen TY, Gold A, Swenberg JA. Adenine adducts with diepoxybutane: isolation and analysis in exposed calf thymus DNA. Chem Res Toxicol. 1997b;10:1171–1179. doi: 10.1021/tx9700681. [DOI] [PubMed] [Google Scholar]
- Tretyakova NY, Sangaiah R, Yen TY, Swenberg JA. Synthesis, characterization, and in vitro quantitation of N7-guanine adducts of diepoxybutane. Chem Res Toxicol. 1997c;10:779–785. doi: 10.1021/tx970004q. [DOI] [PubMed] [Google Scholar]
- Wang H, Kozekov ID, Kozekova A, Tamura PJ, Marnett LJ, Harris TM, Rizzo CJ. Site-specific synthesis of oligonucleotides containing malondialdehyde adducts of deoxyguanosine and deoxyadenosine via a postsynthetic modification strategy. Chemical Research in Toxicology. 2006;19:1467–1474. doi: 10.1021/tx060137o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White WC. Butadiene production process overview. Chem Biol Interact. 2007;166:10–14. doi: 10.1016/j.cbi.2007.01.009. [DOI] [PubMed] [Google Scholar]