Abstract
The cyanobacterium Synechococcus is a ubiquitous, important phytoplankter across the world’s oceans. A high degree of genetic diversity exists within the marine group, which likely contributes to its global success. Over 20 clades with different distribution patterns have been identified. However, we do not fully understand the environmental factors that control clade distributions. These factors are likely to change seasonally, especially in dynamic coastal systems. To investigate how coastal Synechococcus assemblages change temporally, we assessed the diversity of Synechococcus at the Martha’s Vineyard Coastal Observatory (MVCO) over three annual cycles with culture-dependent and independent approaches. We further investigated the abundance of both phycoerythrin (PE)-containing and phycocyanin (PC)-only Synechococcus with a flow cytometric setup that distinguishes PC-only Synechococcus from picoeukaryotes. We found that the Synechococcus assemblage at MVCO is diverse (13 different clades identified), but dominated by clade I representatives. Many clades were only isolated during late summer and fall, suggesting more favorable conditions for isolation at this time. PC-only strains from four different clades were isolated, but these cells were only detected by flow cytometry in a few samples over the time series, suggesting they are rare at this site. Within clade I, we identified four distinct subclades. The relative abundances of each subclade varied over the seasonal cycle, and the high Synechococcus cell concentration at MVCO may be maintained by the diversity found within this clade. This study highlights the need to understand how temporal aspects of the environment affect Synechococcus community structure and cell abundance.
Electronic Supplementary Material
The online version of this article (doi:10.1007/s00248-015-0644-1) contains supplementary material, which is available to authorized users.
Keywords: Cyanobacteria, Microdiversity, ntcA
Introduction
The marine Synechococcus group of cyanobacteria is a globally important primary producer in the world’s oceans. This picophytoplankter (∼1 µm diameter) is responsible for up to 20 % of carbon fixation in coastal systems [19, 25]. Thus, it is important that we understand the factors that affect Synechococcus abundance and enable it to be ecologically important across a wide range of environmental conditions. One of these key factors appears to be the high level of diversity contained within the marine Synechococcus lineage. Studies of molecular phylogeny have resolved isolated strains and environmental sequences into a total of 20 well-defined clades distributed over three main subclusters (5.1, 5.2, and 5.3) [9, 42]. These clades have been supported by phylogenies constructed from a variety of loci, including rpoC1 [35, 51], ITS [1, 17], narB [30, 32, 33], ntcA [38, 40], and petB [26]. Clade designation was recently shown to be congruent across these genetic markers [2] and multi-locus sequence analysis of core genes provides evidence that the clades are in fact distinct lineages [26].
This genetic diversity may be representative of physiological or ecological diversity, such that each clade (or closely related clades) corresponds to an ecotype that occupies a distinct niche [1]. This relationship between genetic diversity and ecological physiology has been well documented in the sister genus, Prochlorococcus, where genetically distinct clades show differences in light acclimation and nutrient utilization [27–29, 41]. Differences in clade physiology explain vertical distributions of Prochlorococcus in the water column, as well as clade biogeography across ocean basins [3, 20, 55, 59].
Similar relationships between genetic designation and physiological characteristics have been shown for some clades of Synechococcus. For example, strains belonging to clade III exhibit a motility that is unique to this clade [52, 54]. Clades also exhibit differences in nitrogen (N) utilization; some clades are unable to grow on or demonstrate reduced growth rate with nitrate (clades CRD2 and XV, respectively; [1, 28]), while others are able to utilize different N sources, such as urea and amino acids [28]. The response of growth rate to temperature can differ among clades as well as the response to temperature stress [39]. Marine Synechococcus also exhibit differences in light harvesting pigments [1, 7, 15, 47, 53]. Many members of subcluster 5.1 contain phycoerythrin (PE) in their pigment complements, and some of these clades can chromatically adapt to different light environments (clades I, III, XV, and XVI; [1, 36]). Many members of subcluster 5.2 contain only phycocyanin (PC) as their light harvesting phycobilipigment (type-I pigment, [42]). These pigment distinctions are not absolute, however, as some strains from subclusters 5.2 and 5.3 contain PE, and some PC-only strains have been found in subcluster 5.1 (clade VIII). Six et al. [47] suggest that phycobilisome rod genes have evolved independently from the rest of the core genome and are likely to have undergone horizontal gene transfer.
These physiological differences so far have not been sufficient to explain observed clade distributions in the ocean. Clades often co-occur [6, 11, 40], with reports of as many as six clades found at once [2]. Nonetheless, biogeographical and time series studies have begun to identify environmental factors that may shape some clade distributions. For instance, clades I and IV are typically found in colder, nutrient-rich, coastal waters at latitudes greater than 30° N and 30° S [59], and members of clades CB4 and CB5 are also found in coastal waters and in estuaries [5, 6, 17]. Other clades seem to prefer warmer and more oligotrophic environments; clades II and III are typically found in tropical waters, with clade II having a much wider distribution into subtropical areas [59].
A complex set of interacting factors likely determine clade distributions. Abundance will be governed by bottom-up conditions, such as light, nutrient availability, and temperature; top-down factors, such as grazing and viral lysis [4, 30, 60]; as well as sideways interactions, such as with heterotrophic bacteria [46]. These factors change over different time scales, such as across seasons and over water columns with variable mixing. The time scales of environmental changes may contribute to the ability of clades to simultaneously coexist. Consistent with this idea, time series studies of clade abundances have demonstrated shifting dominance. In California coastal waters, Tai and Palenik [51] found that clades I and IV were always dominant, but with changing relative abundance over the seasonal cycle, while clades II and III only appeared during autumn and even then at relatively low abundance. In the Gulf of Aqaba, Post et al. [40] observed a succession of clades across the transition from winter mixing to summer stratification, which led to insights of possible preferred nutrient environments for clades I, III, and V/XII. These studies highlight the need to understand how temporal aspects of the environment shape and maintain Synechococcus diversity. At present, there is little knowledge of how seasonal environmental changes affect clade abundances in North Atlantic coastal waters, including on the New England Shelf. To better understand how clade patterns may change over time, we used ntcA clone libraries and culture isolations to assess the diversity of Synechococcus at the Martha’s Vineyard Coastal Observatory (MVCO) over three annual cycles. We further investigated the abundance of both PE-containing and PC-only Synechococcus in these coastal waters by analyzing time series samples with a flow cytometric setup that separates PC-only Synechococcus from picoeukaryotes.
Materials and Methods
Sample Collection
Seawater samples were collected near the MVCO offshore tower (41° 19.500′ N, 70° 34.0′ W) or at the offshore node (41° 20.195′ N, 70° 33.3865′ W); ∼3 km from the south shore of Martha’s Vineyard, MA (Fig. 1) at roughly bimonthly-to-monthly intervals over a 3-year period from 2010–2012 (43 total samples). Seawater was collected at 2, 6, 10, and 15 m depth with Niskin bottles attached to a rosette sampler or at the surface via bucket sample. Glutaraldehyde was added to a 5-mL aliquot of the water sample to a final concentration of 0.1 % by volume for later flow cytometric analysis. These samples were incubated for 10 min at room temperature before being frozen in liquid N2. Samples for nutrient analysis were immediately filtered through a 0.2 µm Sterivex® filter into acid-washed vials and frozen at −20 °C. Samples were analyzed for phosphate, ammonium, silicate, and combined nitrate + nitrite by standard autoanalyzer techniques at the Woods Hole Oceanographic Institution Nutrient Analytical Facility (Woods Hole, MA). Only surface water samples were used for subsequent DNA extraction and culture enrichments (see below). Near continuous measurements of water temperature and salinity were obtained with a MicroCat CTD (SeaBird Electronics) deployed on the MVCO offshore tower at 4 m below mean water level. When there were short gaps in this data record, MicroCat data from the MVCO offshore node (12 m depth) were substituted. These records have been previously shown to be very similar [37].
Fig. 1.
Map showing location of the Martha’s Vineyard Coastal Observatory (indicated by red dot on inset map) and surrounding shelf waters. Bathymetric contours are shown for 100 m, and for 500 m to 3000 m in 500-m intervals
Flow Cytometry Analysis
A modified Epics V flow cytometer (FCM; Coulter Electronics Corp.) interfaced with a Cicero acquisition system (Cytomation, Inc.) was used to analyze preserved water samples. The instrument was equipped with a 5-W argon ion laser (Coherent Innova 90-5) and photomultipliers for three wavelengths of fluorescence detection and forward light scattering. Excitation was at 515 nm (300 mW) and a 540 long pass barrier filter was used to eliminate scattered laser light from the fluorescence detectors. Fluorescent emissions were split by successive dichroic mirrors and interference filters to measure wavelength bands of 562–588 nm (PE fluorescence), 610–660 nm (PC fluorescence), and 660–700 (chlorophyll fluorescence) (see Fig. S1 for schematic and filters used). Forward light scattering was measured at ∼3° − 19° above the axis of the laser beam. Samples were allowed to thaw in water before analysis and were injected into the sheath flow (MilliQ water, Millipore) by a peristaltic pump (Harvard Apparatus) at 0.1 mL min −1. Polystyrene microspheres (Polysciences, Inc.) of diameter 0.5 µm (polychromatic) and 1.0 µm (red-fluorescing) were measured as reference particles.
PE-containing Synechococcus were determined from characteristic PE fluorescence values and forward light scattering [31]. PC-only Synechococcus were determined from values of PC fluorescence, PC-to-chlorophyll fluorescence ratio, and forward light scattering. These features allowed separation and enumeration of PC-only Synechococcus from picoeukaryotes. Values of these parameters fell within a well-confined range for cultures (Fig. S2), and these values were used to guide analysis of field samples. Specifically, an event was designated as a PC-only Synechococcus if it did not show any PE fluorescence, had minimum values of 1.5·104 arbitrary fluorescence units for both PC and chlorophyll fluorescence, and had a PC:chlorophyll fluorescence ratio of >0.5 but <1.0. Cell concentration was determined from sample flow rate (pump rate) and analysis time.
Synechococcus Isolation
On selected days (see markers in Figs. 2 and 7 and dates in Fig. S4), surface seawater was prefiltered over a 20- µm Nitex® mesh. Filtrate was then gravity filtered through either 1- or 2- µm polycarbonate filters (Poretics) to exclude larger cells. The final filtrate was amended with nutrients at one third the concentrations described for SNAX media [53]. Cycloheximide was added at a final concentration of 50 µg mL−1 to prevent growth of eukaryotic phytoplankton and nanoflagellate grazers. Tubes were incubated near ambient seawater temperature with light levels of 30−80 µmol quanta m−2 s−1. After 4–6 weeks, enrichments were visually inspected before transfer into fresh SNAX media (full strength) and then routinely transferred approximately every 4 weeks. After 2–3 months, any enrichment incubating at or below 12 °C was moved to a higher temperature (15−18 °C). For selected tubes, 2- to 4-month old enrichments were plated out onto 0.8 % agar SNAX plates with 2 mM NaSO3; agar was cleaned as described by Waterbury et al. [53]. Plates were incubated for approximately 1 month until single colonies of Synechococcus could be identified either visually or by epifluorescence. Single colonies were picked and inoculated into liquid media. This processes was repeated for some isolates, but not all. For longer-term culture maintenance, isolates were grown in SN media [53] or a variation of SN (half nutrient concentrations of SN with ammonium at 5 × concentration in SNAX, termed “SNAV”). Of these single isolates, we selected 148 to be identified by genotyping of ntcA, an N-regulatory gene. Isolates were selected on the basis of different pigment colors (peach, orange, golden, green, etc.) and colony morphologies (size of colony, raised, sunken, circular or polygons, etc.). Isolates for which there was a sequence discrepancy in the clone replicates (see below) were not used and presumably not clonal.
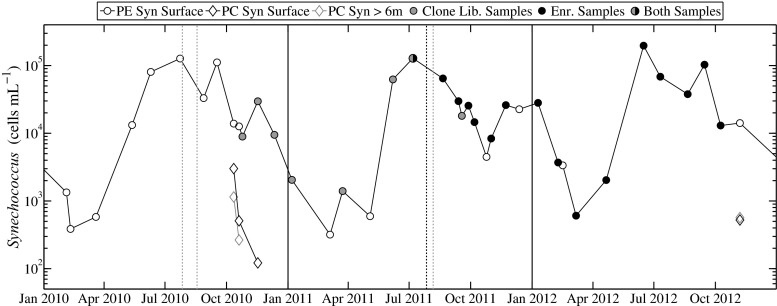
Fig. 2.
Cell concentration of PE-containing Synechococcus and PC-only Synechococcus for 2010–2012 at MVCO. PE-containing Synechococcus are represented by open circles and PC-Synechococcus by diamonds. Color of circle markers indicate samples used for either clone library construction (gray), culture enrichments (black), or both (half and half). Dotted vertical lines indicate dates when samples were taken for either clone libraries or culture enrichments (depending on respective color), but not for flow cytometric analysis. Solid black lines indicate start of years
Fig. 7.
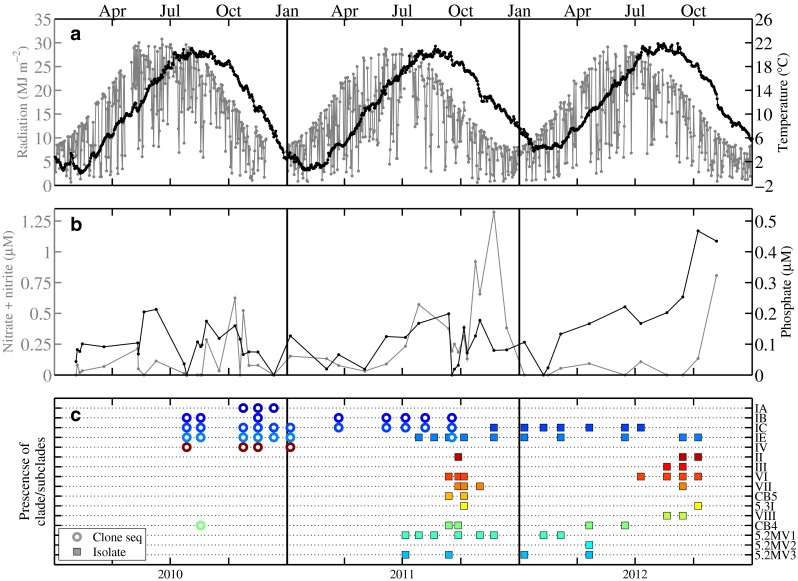
a Incident radiation (MJ m−2) (gray line) and water temperature (°C) at 4 m depth (black line) for 2010–2012. b Concentration of nitrate + nitrite (µM)(gray line) and inorganic phosphate (µM)(black line). c Occurrence of each clade in either clone library or enrichment for 2010–2012. Colors represent different clades or subclades, also separated on different lines. Clades detected by clone library sequence are designated by an open circle, while an isolate representative is designated by a filled square. Note that both clone library data and culture isolation data are only available for one sample
Spectral Analysis
To characterize pigment types of selected cultures, in vivo fluorescence excitation and emission spectra were obtained for cell suspensions with a SpectraMax M3 (Molecular Devices) spectrofluorometer. For the emission spectra, excitation was at 515 nm and emission measured from 530 to 700 nm at 2 nm increments, with a 530 long pass cutoff filter. Emission at 680 nm (Chl maximum) was measured from 400–660 nm at 2 nm increments with a 665 long pass cutoff filter. Cultures were determined to contain only PC (designated as type 1 [47]) if they did not show a representative PE peak (emission maximum between 530 and 580 nm), but rather a peak between 600 and 650 nm [56].
To detect the presence of phycoerythrobilin (PEB) and phycobilins, PE emission at 585 nm was measured over the range 400-570 nm with a 570 nm long pass cutoff filter. Beginning at 550 nm, there was noticeable contribution due to stray light from the spectrofluorometer excitation source. This was between 2 and 16 % of the measured relative fluorescence value of isolate excitation spectra. To correct for this problem, excitation spectra of fresh media blanks (either SN or SNAV) were subtracted from these sample excitation spectra. Presence of PUB was determined if PE excitation spectra contained a peak or noticeable shoulder at ∼495 nm and if maximum Chl excitation fluorescence was between 540 and 550 nm. A PUB-lacking phenotype (PEB-only, designated as type 2) was characterized by no shoulder or peak at ∼495 nm and by shifted Chl excitation maximum towards 560–570 nm. PUB:PEB ratio was calculated as fluorescence excitation at 495 nm to that at 550 nm with corrected spectra. Correction to spectra did not influence pigment classification and ratio calculations varied less than 8 % before and after correction. Strains were designated as pigment type 3a if PUB:PEB ratio was low (∼0.4), and type 3b if PUB:PEB ratio was moderate (∼0.6–0.8) [10, 47].
Only a subset of the entire culture collection was characterized (∼ one strain representative per unique sequence). Strains that were not analyzed were assumed to have the same major phycobiliproteins as strains with the same culture color and clade type (i.e., if a strain color was peach and belonged to clade I, it was classified as containing PE, see Table S1).
Environmental Sample DNA Extraction and ntcA PCR Amplification
On selected days (dates marked on Figs. 2 and 7), 2–3 L of surface seawater was prefiltered through a 20- µm Nitex® mesh and then filtered onto 0.2- µm Sterivex® cartridge filters (Millipore) under vacuum pressure of no more than 40 kPa. Approximately 1.8 mL of DNA cell lysis buffer (Qiagen) was added to each cartridge before freezing and storage at -80 °C. For DNA extraction, samples were thawed on ice and, to break open cells, approximately 200 µL of 0.5-mm zirconia-silica beads (BioSpec Products) were added to the cartridges, which were then shaken vigorously at 2500 rpm for 10 min. Continued DNA extraction was carried out with Qiagen Purgene reagents but with the modified procedure described in Palacios et al. [34]. DNA concentration was determined with a NanoDrop 2000 Spectrophotometer (ThermoScientific). Depending on the sample, 120 to 1200 ng of DNA template was added to PCR reactions with the degenerate 1F/4R primer pair [38]. Final primer concentration was 2 µM in a total reaction volume of 50 µL with Qiagen Taq PCR Master Mix Kit reagents. BSA was added at 0.2 mg mL−1 final concentration. PCR reactions were performed on a GeneAmp PCR System 9700 thermocycler (Applied Biosystems), with an initial denaturation period of 4 min at 94 °C; followed by 40 cycles of 1 min at 94 °C, 1 min at 45 °C, 1 min at 72 °C; and then a final extension step at 72 °C for 7 min.
Culture Isolate DNA Extraction and ntcA PCR Amplification
Approximately 2 mL of dense culture isolate was centrifuged at 9300 ×g for 6 min to pellet cells, and DNA was extracted from this pellet with a Qiagen DNeasy Plant Kit, following manufacturer’s instructions with the exception of final elution volume (75 µL). Approximately 10–30 ng of DNA was added to PCR reactions with 1AF and 4AR primers (targeted Synechococcus primers, [38]). Final primer concentration was 2.5 µM in a total reaction volume of 50 µL with Qiagen Taq PCR Master Mix Kit reagents. BSA was added at 0.2 mg mL−1 final concentration. Reactions were preformed on a GeneAmp PCR System 9700 thermocycler (Applied Biosystems), with an initial denaturation period of 4 min at 94 °C; followed by 30 cycles of 1 min at 94 °C, 30 s at 55 °C, 30 s at 72 °C; and then a final extension step at 72 °C for 7 min.
ntcA Clone Libraries
All PCR products (an expected 449-bp fragment), from both the environmental and culture isolates, were gel purified with a Qiagen Qiaquick gel extraction kit. Cleaned products were cloned into TOPO vectors for sequencing (TOPO TA Kit, Invitrogen) and transformed into chemically competent E. coli TOP10 cells (Invitrogen) following manufacturer’s instructions. For isolates, at least five positive colonies (determined by blue/white selection on X-Gal, kanamycin LB plates) were picked. Plasmids were obtained via automated plasmid purification with a BiomekFX at the Josephine Bay Paul Center Keck Facility (Marine Biological Laboratory, Woods Hole, MA). Sequencing reactions used BigDye Terminator chemistry (Applied Biosystems) and contained at least 200 ng of purified plasmid and M13 reverse primer (15 µM, from TOPO kit). Sequencing was performed at the Josephine Bay Paul Center Keck Facility with a 3730 DNA Analyzer (Applied Biosystems). Primer and vector sequences were removed and resulting sequences were identified by BLAST search against ntcA accessions in NCBI Genbank. Sequences were manually evaluated and corrected if necessary from chromatograms. SynechococcusntcA sequences were deposited in NCBI GenBank with accession numbers KR360758–KR361175, KR364942 for environmental sequences, and KR361176–KR361318 for isolate sequences.
Phylogenetic Analysis
Sequences identified as ntcA were aligned with the ClustalW algorithm in BioEdit (version 7.2.0, [14]). Operational taxonomic unit (OTU) construction and rarefaction analysis was carried out in mothur v.1.23.1 [44] with the furthest neighbor clustering algorithm. Distinct OTUs were designated at a 10 % dissimilarity cutoff. Fourteen sequences were found that could cluster with either OTU IC or OTU IB (see below) based on distance, and these sequences were randomly assigned to either OTU (see Table S2). Phylogenetic reconstructions were carried out in the ARB software package (version 5.3, Ludwig et al. 2004) with a maximum likelihood approach using RAxML [48] and a GTR GAMMA rate substitution model. Bootstrap analysis for support of tree branches was also carried out in ARB with rapid bootstrap analysis and 500 sample trees.
Results
Environmental Conditions
Temperature at MVCO exhibited large seasonal fluctuations from a minimum of around 0 °C up to a maximum of 22 °C during the 3-year period of this study (Fig. 7a). Salinity was typically within the range of 31–32.5. The concentration of nitrate + nitrite was usually below 1 µM, with the majority of the samples below 0.5 µM and often at the limit of detection for the autoanalyzer technique (0.05 µM) (Fig. 7b). Higher nitrate + nitrite concentrations (0.75–1 µM) occasionally occurred during fall. The concentration of phosphate was also usually low (typically < 0.25 µM).
Flow Cytometry Analysis
Consistent with our previous reports for this site [18], cell abundance of PE-containing Synechococcus followed a repeatable seasonal pattern of low wintertime concentrations of a few hundred cells mL−1 to greater than 105 cells mL−1 in summertime (Fig. 2). Large changes in abundance (an order of magnitude) were observed during the late summer and fall. By contrast, only a few samples over three seasonal cycles appeared to contain signatures that matched our criteria for identification as a PC-only Synechococcus (see Fig. S2 for an example). The maximum observed concentration (∼3000 cells mL−1) was roughly fourfold less than that of the concurrent PE-containing Synechococcus (∼13,800 cells mL−1). The majority of small (<∼2 µm), non-PE-containing cells were classified as picoeukaryotes (Fig. S2).
Spectral Analysis
Isolates belonging to clades I, II, III, VI, VII, CB5, 5.2MV2, and 5.3I demonstrated excitation and emission spectra consistent with the presence of PE. Isolates belonging to clades VI, CB5, and 5.2MV2 appeared to lack PUB and demonstrated type 2 pigment spectra (Figs. 3 and S5). Emission maxima occurred at 574–578 nm and Chl excitation maxima at 554–566 nm. Interestingly, isolates of clade I were either of type 2 or appeared to contain PUB in low amounts (type 3a spectra) (Figs. 3 and S5). Isolates belonging to clades II, III, and 5.3I all appeared to contain PUB and were designated as either type 3a or 3b based on PUB:PEB ratio. Type 3 spectra had emission maxima from 564 to 574 nm and Chl excitation maxima from 542 to 548 nm. No seasonality was evident in the occurrence of PUB-containing or PUB-lacking strains from isolations (data not shown), and both types were found in almost all enrichments.
Fig. 3.
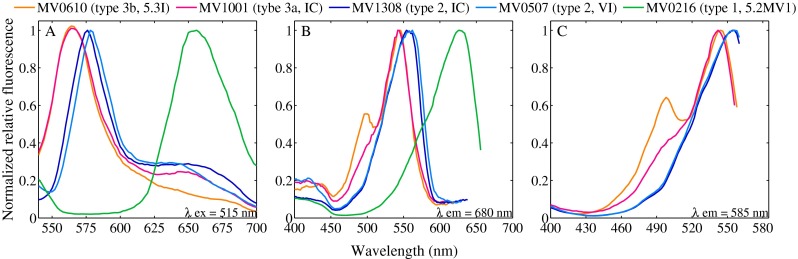
In vivo normalized excitation and emission spectra for 5 different isolates, representative of pigment types 1, 2, 3a, and 3b. a Normalized emission spectra (excitation at 515 nm), indicating presence or absence of PE (characteristic peak at 550–580 nm). b Normalized excitation spectra for chlorophyll emission at 680 nm for the same isolates, confirming presence or absence of PE. c Normalized excitation spectra for PE emission at 585 nm, illustrating presence or absence of PUB (peak or shoulder at ∼495 nm). Strain MV0610 is of pigment type 3b (medium PUB:PEB ratio), strain MV0801 of type 3a (low PUB:PEB), MV308 and MV0507 of type 2 (no PUB, PEB-only PE), and strain MV0216 of pigment type1 (PC-only). Strains belong to the following clades: IC for MV1001 and MV1308, VI for MV0507, 5.3I for MV0610, and 5.2MV1 for MV0216
Isolates belonging to clades VIII, CB4, 5.2MV1, and 5.2MV3 did not demonstrate characteristic excitation or emission peaks of PE and were designated as type 1 (only contained PC). Emissions peaks for these isolates were in the range of 648–658 nm and Chl excitation peaks within 622–630 nm. Despite similar pigment types, isolates exhibited a range of colors (Fig. 4). Synechococcus that contained only PEB appeared as brown, dark red, pink, or peach. Strains with PUB appeared as pink, peach, or orange. Synechococcus that contained only PC were dark bluish-green, green, or light yellow-green.
Fig. 4.
Cultures of representative strains for each clade and subclade isolated from MVCO. Two strains are shown for each of clades II and 5.2MV1 to demonstrate range of color differences. See Figs. 3 and S5 for representative excitation and emission spectra
Diversity of Environmental Sequences
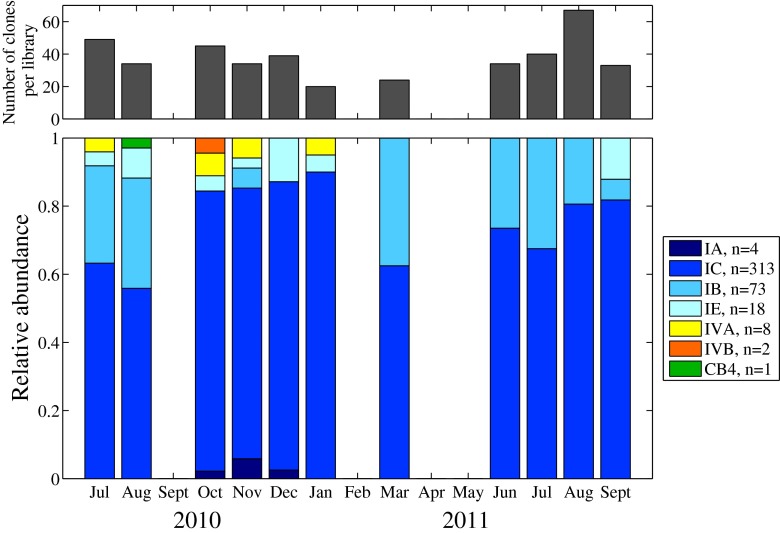
We obtained a total of 419 ntcA sequences from eleven amplicon libraries. Of these, 229 sequences were unique (i.e., different nucleotide sequences). The majority of the sequences (∼97 %; n = 408) belonged to clade I; ten sequences belonged to clade IV; and only 1 belonged to clade CB4 (Fig. 5). There was considerable diversity within the clade I sequences, and OTU clustering separated these into four distinct subclades (Figs. 6 and S3). Subclade IE grouped with reference strain WH8016, but the other subclades (IA, IB, and IC) did not group with reference strains WH8020 and CC9311 (for which ntcA sequences are available). These three subclades do not have previously known representatives in the ntcA tree and might be novel, but we cannot be certain they are not related to subclades previously observed with other markers. They could be related to the subclades observed by Tai and Palenik [51] in California coastal water or clade I strains analyzed by Mazard et al. [26]. Sequencing of the rpoC1 or petB regions for these strains would be needed to resolve this. Subclade IC sequences were found to dominate in all samples, ranging from 56 to 90 % of the sequences in each clone library (Fig. 6). The second most abundant subclade in the spring and summer libraries was IB, suggesting that this subclade made up more of the population at this time of the year. Subclade IE sequences appeared in samples taken in late summer through early winter, and subclade IA only appeared in late fall 2010. The most diverse Synechococcus populations were observed in the late summer and early fall, and they included sequences from clade IV and clade CB4, as well as from subclade IA. However, members of these clades and subclade (IV, CB4, IA) appeared to be relatively rare in the environment. Two subclades based on OTU clustering were also identified within clade IV (IVA and IVB).
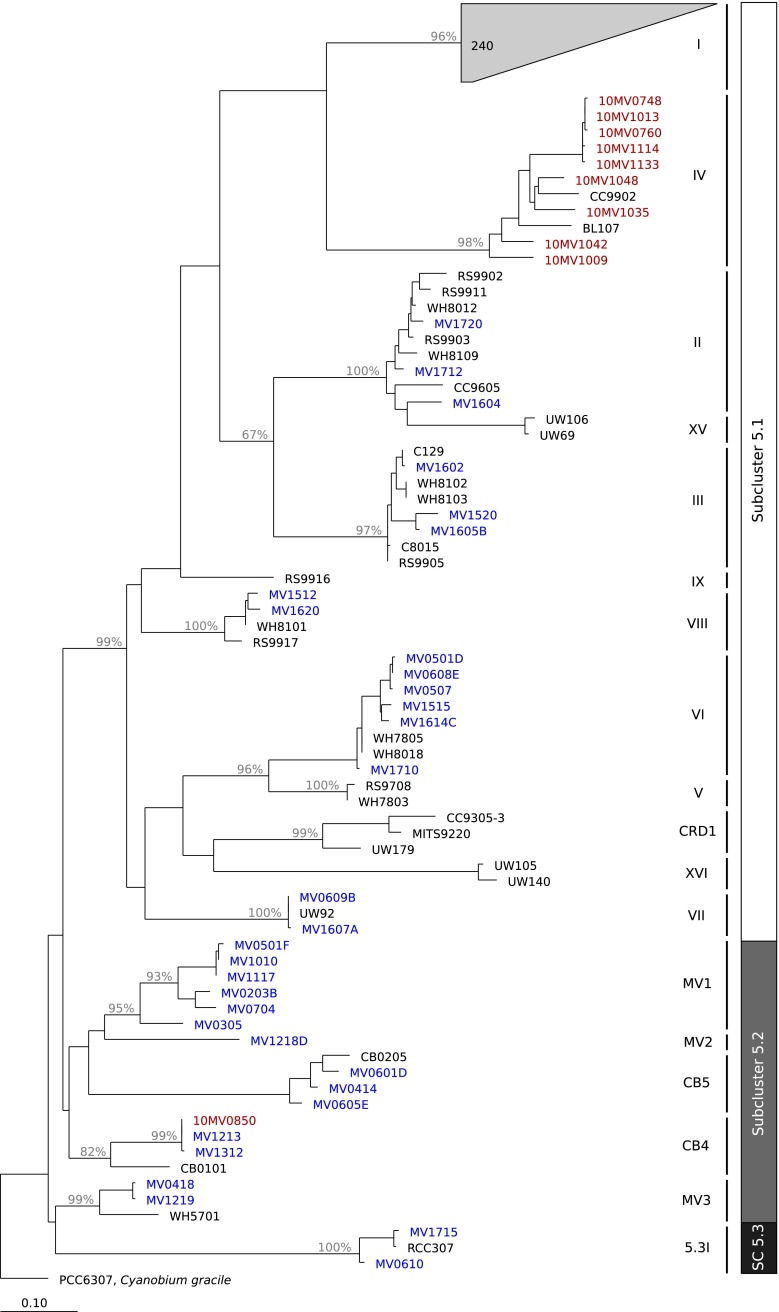
Fig. 5.
Phylogenetic tree constructed from ntcA sequences, illustrating the relationships between known clade representatives (in black), clone library sequences (red), and culture isolates (blue). Clade assignments for sequences were made by identifying the closest known clade representative or were assigned to possibly novel clades (designated as 5.2MV1 and 5.2MV2) if sequences did not cluster with known strains. Bootstrap values greater than 65 % are shown on branches
Fig. 6.
Relative abundance of ntcA clone library sequences belonging to 7 distinct OTUs for 11 samples taken between July 2010 and September 2011. Shades of blue indicate OTUs that belong to clade I, orange/yellow indicate OTUs that belong to clade IV, and green indicates the OTU that belongs to clade CB4. In the legend, n indicates total number of sequences retrieved for each OTU over the entire time series.The number of sequences retrieved for each library are indicated by the histogram in the top panel
Diversity of Isolates
From the 17 enrichments, 143 isolates were identified from their ntcA gene sequences. Of these, 50 unique strains were identified (i.e., different ntcA sequences). Isolates with identical ntcA sequences did occur not only within the same enrichment but also across different enrichments, with original sample collection dates sometimes separated by months (see Table S1). Analysis of ntcA phylogeny showed that the strains belonged to 12 different clades that spanned the three subclusters (Fig. 5). Isolates mapped to known clades I, II, III, VI, VII, and VIII of subcluster 5.1; three clades of subcluster 5.2, including CB4 and CB5; and one clade of subcluster 5.3 (5.3I/X). Strains also clustered into two other clades, belonging to subcluster 5.2 that did not match to other known strains. These clades have been labeled as 5.2MV1 and 5.2MV2 until confirmation of their status as either novel or known can be determined (strain representatives may exist for which phylogenetic markers other than ntcA have been sequenced). Strains of clade 5.2MV1 contained PC as their primary pigment, whereas the one strain of clade 5.2MV2, MV1218, contained PE (see Table S1). We have designated isolates that clustered closely with reference strain WH5701 as clade 5.2MV3.
Clade I isolates belonged to two subclades, IC and IE, observed from clone library sequences. Representatives of these subclades were consistently recovered in cultures from enrichments throughout the time series analysis (Figs. 7c and S4), which spanned a wide range of temperatures, light levels, and nutrient concentrations (Fig. 7a, b).
Surprisingly, we also isolated strains considered more common to subtropical and tropical waters (i.e., clades II, III, and VII [40, 59]). Isolates of such clade members only occurred in late summer and early fall. Clade II types were isolated during September (2011 and 2012) and October (2012) and clade III was isolated during August and September (2012). Clade VI isolates were also only obtained in late summer and early fall and clade VII was only found during the fall. In general, isolations of clades II, III, VI, VII, CB5, and 5.3I tended to occur when water temperature was relatively warm (17–20 °C) and nitrate + nitrite concentration was relatively low (<0.5 µM, Fig. 7). The frequency of isolation of these clades was much lower than for clade I representatives (Fig. S4).
We were able to culture PC-only pigment type strains from all but a few of the enrichments from MVCO (Fig. S4). Although not all of these isolates were sequenced (Fig. S4), those that were fell into clades VIII, CB4, 5.2MV1, and 5.2MV3 (Fig. 5). Isolation of these PC-only strains over the entire enrichment time series suggests that they were persistent members of the Synechococcus community with a year round presence at MVCO.
Discussion
Synechococcus Diversity at MVCO
From culture-dependent and culture-independent approaches, we find that the Synechococcus assemblage observed throughout the year at MVCO is diverse. Members of 13 different clades spanning all 3 known subclusters of marine Synechococcus were identified, but members of clade I dominate the Synechococcus population over the entire year. Below we discuss and highlight possible reasons for the prevalence and occurrence of these clades at MVCO.
Clade I and Subclade Diversity
This dominance of clade I is consistent with the known biogeography of clade I, which is primarily found in cooler, higher nutrient, coastal waters [7, 17, 51, 59], and has been shown to make up a majority of the Synechococcus assemblage in surrounding shelf areas [2]. While a preference for cooler water is likely to be a key factor explaining the presence of clade I at MVCO, the ability to survive colder winter water temperatures (∼0–4 °C) may also contribute to clade I dominance at this site. Recently, Pittera et al. [39] demonstrated that clade I strains were able to grow at temperatures lower (∼10–15 °C) than tolerated by strains from clades II and V. These authors also demonstrated that clade I strains were more tolerant of cold stress. At MVCO, strains of clade I were isolated from water at a range of temperatures (Fig. 7), but notably from water at ∼4 °C, indicating that cells were still viable during this time. The PE-containing Synechococcus population reaches a minimum cell concentration of a few hundred cells per mL−1 during winter (Fig. 2), and the ability to survive these colder temperatures may be an important factor that allows this population to “overwinter” until more favorable spring conditions.
Within clade I, we also found significant diversity at the subclade level, such that four different subclades could be resolved in the clone library sequences (Figs. 6 and S3). While not strictly quantitative, relative abundances of sequences in our clone libraries suggest that not all of these subclades are equally represented when they co-occur. While subclade IC appears to be dominant throughout the year, subclades IB and IE appeared to increase in relative abundance at different times of year (spring-summer, summer-early winter, respectively). This may indicate possible differences in environmental preference, but the small number of clone libraries in our study (n = 11), combined with the possible biases in this data type, preclude definitive investigation of relationships between environmental factors and subclade relative abundances. Given that clade I appears to be dominant, it will be important to understand how each subclade affects and contributes to this dominance as well as seasonal cell abundance patterns.
Interestingly, both pigment types 2 and 3a (no PUB and low PUB, respectively) were found among clade I isolates for both subclade IC and IE. To our knowledge, this is the first instance of PEB-only clade I strains. Other clade I representatives examined to date have contained PUB [1, 11, 53]. It is unknown whether this spectral phenotype results from a lack of PE-II subunit or associated genes [47] or can be achieved by chromatic adaption (CA), as clade I strains have been shown to exhibit type IV CA [36, 47]. The prevalence of low or no PUB phenotype among these isolates may reflect a strong pressure for efficient absorption of green light, which often predominates at coastal locations. It has long been appreciated that there are different geographical distributions of PUB-containing PEs, with higher PUB:PEB types dominating in the open ocean and low or no PUB types more prevalent nearshore [16, 31, 45, 57, 58]. This is thought to be an adaptation (or acclimation) to the in situ light environment, where PUB allows better absorption of blue, open water (PUB maximum absorption ∼495 nm), while PEB better absorbs greener, coastal water (maximum absorption ∼550 nm).
Isolation and Detection of Other Clades
We were able to culture representatives of clades that have typically been found in either warmer or more oligotrophic environments (e.g., clades II, III, VII, 5.3I), but only in late summer and early fall. Ahlgren and Rocap [2] found clades II and 5.3I at an outer shelf location (304 km south of MVCO) and detected clade III in samples from the Gulf Stream. One possibility for these clade occurrences at MVCO is that they are advected onto the inner New England shelf from more offshore locations and can only survive at this location during late summer and fall. The general circulation on the New England Shelf is part of the larger shelf circulation of the Middle Atlantic Bight, which is characterized by a southwestward along-shelf flow of relatively fresh water, with across-shelf offshore currents at the surface and bottom and onshore currents in the middle of the water column [24]. Shelf water is separated from saltier slope water by a shelf-slope front, but exchanges between these water types can occur due to frontal instabilities [13], eddies [12], warm-core ring shelf interactions [8, 21], and saline intrusions at the seasonal pycnocline [23]. Locally, there is a counterclockwise recirculation just south of MVCO, which is strongest in the summer months [22]. Slope water intrusions plus this recirculation feature could make it possible for clades growing in warmer, saltier water to be advected and then retained near MVCO. During summer, water temperature is warm for this location (∼20 °C) and nitrate levels are typically low or undetectable (Fig. 7b), such that conditions may allow persistence of clades that would not typically thrive in coastal waters at other times of the year.
Clades VI, VIII, and CB5 were also only isolated during late summer and fall. Noticeably, this is when water temperature was relatively warm. This is most apparent for clade VI, which was cultured from seven different enrichments, but only when water temperature at time of sampling was greater than 16 °C. The global distribution of clade VI is ambiguous (previous studies utilized probes that could not separate V, VI, and VII from one another [11]). In general, clade VI representatives have been isolated from coastal environments (Woods Hole Harbor, [53]; East Sea and East China Sea, [7]), suggesting tolerance or preference of coastal conditions. Clade CB5 has also been isolated from estuarine or coastal locations [6, 7]. Representatives of both VI and CB5 were of pigment type 2, which as stated above, can allow better absorption of green light that is prevalent in coastal waters, and may contribute to their presence at MVCO.
Interestingly, clade IV, which has been reported to co-occur with clade I in other coastal waters [59], may have lower relative abundance at MVCO. Strain representatives were not able to be isolated and only ∼2 % of the clone library sequences belonged to this clade. While we cannot be sure that biases in clone libraries and culture isolations are not a factor, it may be that this clade is very rare at MVCO. This is consistent with observations of Ahlgren and Rocap [2] who found clade IV to be much less abundant than clade I at outer shelf locations (80–304 km south of MVCO). This is different from other coastal locations for which the abundance of clade IV usually matches or exceeds that of clade I [51]. The potentially low abundance of clade IV at MVCO raises interesting questions as to the physiological differences between these two clades and why, for this coastal system, clade I is much more abundant. As highlighted by Ahlgren and Rocap [2], these questions are relevant for the wider shelf region of the northern Middle Atlantic Bight.
PC-Only Synechococcus
Strains of Synechococcus that only have PC as their light-harvesting pigment have been isolated previously from either estuarine or nearshore coastal waters [6, 7, 11, 15, 53], and sequences that match these strain representatives have been found in similar regions [5, 6, 17]. PC-only Synechococcus are well suited to absorb the quality of light found in these more turbid waters [49, 50]. Given the nearshore location of MVCO, it is not surprising then that PC-only Synechococcus strains were isolated. However, consistently low to undetectable cell concentrations from flow cytometry analysis suggest that these strains may not be ecologically relevant at MVCO. These strains may have been transported from more estuarine sites and then grow poorly in the environmental conditions at this location. Although the site is exposed to the open shelf, it is located on the inner shelf only 3 km from the south shore of the island of Martha’s Vineyard. Another hypothesis is that these organisms are found in the benthos at MVCO (water column 15 m deep), such that storms or other mixing events could transport them to the surface. The sudden appearance and then rapid decline of PC-only Synechococcus in samples from October 2010 (Fig. 2, separated only by a week) is consistent with a hypothesis of a population advected to the site (either from shore or the bottom) but not able to thrive.
The PC-only Synechococcus strains appeared to flourish, however, in the culture conditions, and in fact dominated many enrichments (Fig. S4). The media recipe used to culture and maintain the strains contained only 75 % seawater. Many members of subcluster 5.2 are halotolerant, such that they do not require elevated salt requirements for growth [43]. If these strains grow better in lower salinity, then the enrichment conditions may have selected for these representatives. Anecdotal observations from our enrichments also suggest that these strains may persist at background levels for months in a low-nutrient enrichment that is dominated by another pigment type (as judged by color of the culture). Once supplied with higher nutrient concentrations (i.e., when we switched to SN media), these PC-only strains were able to quickly out compete PE-containing strains that appeared to dominate originally. This highlights important questions about the factors that either allow clades to coexist or certain groups to dominate. In particular, little is known about the ability of certain strains to survive unfavorable conditions and how variation in this ability can impact clade distributions in nature.
Comparison of Diversity from Clone Libraries and Culture Isolations
Some isolated clade representatives did not appear in environmental clone libraries and vice versa. Members of clades II, III, VI, VII, VIII, CB5, 5.2MVI, 5.2MV2, 5.2MV3, and 5.3I were isolated into culture but did not show up in the clone library sequences. The reverse occurred for clade IV and subclades IA and IB, for which sequences were found in the clone libraries, but not in the isolated and sequenced strains. Ahlgren and Rocap [1] found a similar mismatch of diversity recovered from simultaneous culture isolations and construction of clone libraries for samples from the Sargasso Sea. This is perhaps not surprising given the potential biases in each method. Isolation procedures are likely to favor growth of certain clades over others, and we can surmise that subclades IA and IB and clade IV cells do not grow well or were outcompeted by other strains in our enrichment conditions. The strength of possible culturing biases is clearly demonstrated with the isolations of PC-only Synechococcus strains. These were numerous in the culture collection, but almost absent in both the clone libraries and flow cytometry record. For the clone libraries, there may be primer biases that selectively favor the amplification of certain clades. The overwhelming dominance of clade I cells would also make rarer clades difficult to detect in the clone libraries. Even with these biases, rarefaction analysis suggests that for all but a few samples, further sampling with either method would have yielded more diversity, as most curves do not reach a plateau (Fig. 8). In combination, though, culture isolation and clone library techniques have captured a more complete picture of total diversity than either one alone (Fig. 7c).
Fig. 8.
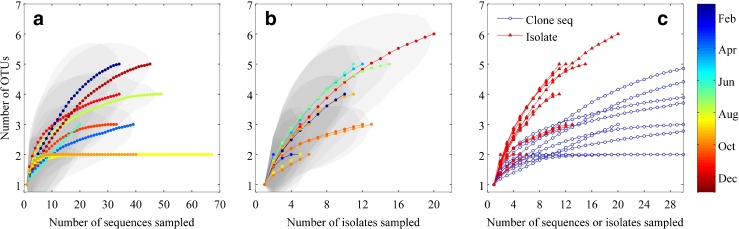
Rarefaction analysis of ntcA sequences for clone library and culture isolate sequences. In each panel, dotted lines are the average number of OTUs observed for 1000 sampling iterations for each number of sequences sampled. a Rarefaction curves for each clone library sample. b Rarefaction curves of culture isolate sequences for each enrichment sample. Color indicates time of year and gray shaded areas indicate 95 % confidence intervals. c Comparison of clone library (blue circles) and culture isolate (red triangles) rarefaction curves (same data as plotted in panels a and b) for sampling up to 30 sequences. OTU designation was at 10 % dissimilarity level
It is noteworthy that the occurrence of certain clades differed among enrichments that were separated only by a few weeks in time. For example, the samples for enrichments 15–17 came from similar nutrient and temperature conditions, but each of these yielded a very different array of clade representatives (Fig. S4). While there are many biases in culturing, these differences between enrichments may hint at fast changing dynamics either in the field or in the enrichment culture (probably during the first few weeks). This illustrates some of the challenges faced when attempting to isolate and culture novel strains of Synechococcus or other organisms. We do not yet understand all the factors that determine how an organism will grow in isolation under laboratory conditions, and caution is needed in extending findings to natural dynamics.
Future Considerations
Many questions remain not only concerning how environmental (temperature, light, nutrients) and ecological (grazers, viruses, heterotrophic bacteria) factors govern clade distributions but also how changes in these factors over a seasonal cycle affects the abundance of different clades. We emphasize the importance of investigating the temporal aspects of diversity patterns, especially in dynamic coastal systems. On the New England Shelf, Synechococcus cell concentration undergoes a dramatic (3 orders of magnitude) seasonal cycle, and it is likely that some of the abundance patterns are determined by which clades are favored under different conditions. With isolates in culture from this location, we are poised to begin exploration into some of the differences between clade and subclade types. Ultimately, high frequency monitoring of clade diversity, coupled with physiological and ecological knowledge of representative strains, will allow a greater insight into how diversity of this genus is maintained and how that diversity is linked to overall population dynamics.
Electronic supplementary material
Acknowledgments
The authors are indebted to Rob Olson and Alexi Shalapyonok for assistance with the flow cytometry setup. We are also grateful to John Waterbury for training and assistance in culture isolations and to Kasia Hammar, Erin Banning, and Heather Landry for assistance with clone library construction and troubleshooting. We thank Taylor Crockford, Janet Fredericks and the MVCO Operations Team, captain and mate of the R/V Tioga, and Paul Henderson of the WHOI Nutrient Analytical Facility. We would also like to thank Jennie C. Hunter-Cevera and Athena Aicher for culture collection assistance and Michael G. Neubert and Penny Chisholm for helpful discussions. We further thank the anonymous reviewers, whose comments and suggestions improved this manuscript. This work was supported in part by NSF Chemical Oceanography award 1155566 to AFP; Gordon and Betty Moore Foundation grant 934, NASA Ocean Biology and Biogeochemistry Program Grant NNX11AF07G, NSF OCE grant 0530830 to HMS; and a WHOI Ocean Ventures Fund award, a WHOI Coastal Ocean Institute award, private donation, and a National Defense Science and Engineering Graduate Fellowship to KRHC.
References
- 1.Ahlgren NA, Rocap G. Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl Environ Microbiol. 2006;72(11):7193–7204. doi: 10.1128/AEM.00358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlgren NA, Rocap G (2012) Diversity and distribution of marine Synechococcus: multiple gene phylogenies for consensus classification and development of qPCR assays for sensitive measurement of clades in the ocean. Front Microbiol 3(213) [DOI] [PMC free article] [PubMed]
- 3.Ahlgren NA, Rocap G, Chisholm SW. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol. 2006;8(3):441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- 4.Apple JK, Strom SL, Palenik B, Brahamsha B. Variability in protist grazing and growth on different marine Synechococcus isolates. Appl Environ Microbiol. 2011;77(9):3074–3084. doi: 10.1128/AEM.02241-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai H, Wang K, Huang S, Jiao N, Chen F. Distinct patterns of picocyanobacterial communities in winter and summer in the Chesapeake Bay. Appl Environ Microbiol. 2010;76(9):2955–2960. doi: 10.1128/AEM.02868-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F, Wang K, Kan J, Suzuki MT, Wommack KE. Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S–23S rRNA internal transcribed spacer sequences. Appl Environ Microbiol. 2006;72(3):2239–2243. doi: 10.1128/AEM.72.3.2239-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi DH, Noh JH. Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol Ecol. 2009;69(3):439–448. doi: 10.1111/j.1574-6941.2009.00729.x. [DOI] [PubMed] [Google Scholar]
- 8.Churchill JH, Cornillon PC, Milkowski GW. A cyclonic eddy and shelf-slope water exchange associated with a Gulf Stream warm-core ring. J Geophys Res Oceans (1978–2012) 1986;91(C8):9615–9623. doi: 10.1029/JC091iC08p09615. [DOI] [Google Scholar]
- 9.Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, Paulsen IT, de Marsac NT, Wincker P, Dossat C, et al. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;9(5):R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everroad CR, Wood MA. Comparative molecular evolution of newly discovered picocyanobacterial strains reveals a phylogenetically informative variable region of ß-phycoerythrin1. J Phycol. 2006;42(6):1300–1311. doi: 10.1111/j.1529-8817.2006.00282.x. [DOI] [Google Scholar]
- 11.Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol. 2003;69(5):2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gawarkiewicz G, Bahr F, Beardsley RC, Brink KH. Interaction of a slope eddy with the shelfbreak front in the Middle Atlantic Bight. J Phys Oceanogr. 2001;31(9):2783–2796. doi: 10.1175/1520-0485(2001)031<2783:IOASEW>2.0.CO;2. [DOI] [Google Scholar]
- 13.Gawarkiewicz G, Brink KH, Bahr F, Beardsley RC, Caruso M, Lynch JF, Chiu CS (2004) A large-amplitude meander of the shelfbreak front during summer south of New England: observations from the shelfbreak PRIMER experiment. J Geophys Res Oceans (1978–2012) 109(C3)
- 14.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 15.Haverkamp TH, Schouten D, Doeleman M, Wollenzien U, Huisman J, Stal LJ. Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the Baltic Sea. ISME J. 2009;3(4):397–408. doi: 10.1038/ismej.2008.118. [DOI] [PubMed] [Google Scholar]
- 16.Hoge FE, Wright CW, Kana TM, Swift RN, Yungel JK. Spatial variability of oceanic phycoerythrin spectral types derived from airborne laser-induced fluorescence emissions. Appl Opt. 1998;37(21):4744–4749. doi: 10.1364/AO.37.004744. [DOI] [PubMed] [Google Scholar]
- 17.Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 2012;6(2):285–297. doi: 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter-Cevera KR, Neubert MG, Solow AR, Olson RJ, Shalapyonok A, Sosik HM. Diel size distributions reveal seasonal growth dynamics of a coastal phytoplankter. Proc Natl Acad Sci. 2014;111(27):9852–9857. doi: 10.1073/pnas.1321421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardillier L, Zubkov MV, Pearman J, Scanlan DJ. Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 2010;4(9):1180–1192. doi: 10.1038/ismej.2010.36. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311(5768):1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 21.Joyce TM, Bishop JK, Brown OB. Observations of offshore shelf-water transport induced by a warm-core ring. Deep Sea Res Part A. 1992;39:S97—S113. [Google Scholar]
- 22.Kirincich AR, Lentz SJ, Farrar JT, Ganju NK (2013) The spatial structure of tidal and mean circulation over the inner shelf south of Martha’s Vineyard, Massachusetts. J Phys Oceanogr 43(9)
- 23.Lentz S (2003) A climatology of salty intrusions over the continental shelf from Georges Bank to Cape Hatteras. J Geophys Res Oceans (1978–2012) 108(C10)
- 24.Lentz SJ (2008) Observations and a model of the mean circulation over the Middle Atlantic Bight continental shelf. J Phys Oceanogr 38(6)
- 25.Li WK. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. doi: 10.4319/lo.1994.39.1.0169. [DOI] [Google Scholar]
- 26.Mazard S, Ostrowski M, Partensky F, Scanlan DJ. Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ Microbiol. 2012;14(2):372–386. doi: 10.1111/j.1462-2920.2011.02514.x. [DOI] [PubMed] [Google Scholar]
- 27.Moore LR, Chisholm SW. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol Oceanogr. 1999;44(3):628–638. doi: 10.4319/lo.1999.44.3.0628. [DOI] [Google Scholar]
- 28.Moore LR, Post AF, Rocap G, Chisholm SW. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr. 2002;47(4):989–996. doi: 10.4319/lo.2002.47.4.0989. [DOI] [Google Scholar]
- 29.Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393(6684):464–467. doi: 10.1038/30861. [DOI] [PubMed] [Google Scholar]
- 30.Mühling M, Fuller NJ, Millard A, Somerfield PJ, Marie D, Wilson WH, Scanlan DJ, Post AF, Joint I, Mann NH. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ Microbiol. 2005;7(4):499–508. doi: 10.1111/j.1462-2920.2005.00713.x. [DOI] [PubMed] [Google Scholar]
- 31.Olson RJ, Chisholm SW, Zettler ER, Armbrust EV. Pigments, size, and distribution of Synechococcus in the North Atlantic and Pacific Oceans. Limnol Oceanogr. 1990;35:45–58. doi: 10.4319/lo.1990.35.1.0045. [DOI] [Google Scholar]
- 32.Paerl RW, Foster RA, Jenkins BD, Montoya JP, Zehr JP. Phylogenetic diversity of cyanobacterial narB genes from various marine habitats. Environ Microbiol. 2008;10(12):3377–3387. doi: 10.1111/j.1462-2920.2008.01741.x. [DOI] [PubMed] [Google Scholar]
- 33.Paerl RW, Johnson KS, Welsh RM, Worden AZ, Chavez FP, Zehr JP (2011) Differential distributions of Synechococcus subgroups across the California current system. Front Microbiol Microbiol 2(59) [DOI] [PMC free article] [PubMed]
- 34.Palacios C, Zettler E, Amils R, Amaral-Zettler L. Contrasting microbial community assembly hypotheses: a reconciling tale from the Río Tinto. PLoS ONE. 2008;3(12):e3853. doi: 10.1371/journal.pone.0003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palenik B. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl Environ Microbiol. 1994;60(9):3212–3219. doi: 10.1128/aem.60.9.3212-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palenik B. Chromatic adaptation in marine Synechococcus strains. Appl Environ Microbiol. 2001;67(2):991–994. doi: 10.1128/AEM.67.2.991-994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peacock EE, Olson RJ, Sosik HM. Parasitic infection of the diatom Guinardia delicatula, a recurrent and ecologically important phenomenon on the New England Shelf. Mar Ecol Prog Ser. 2014;503:1–10. doi: 10.3354/meps10784. [DOI] [Google Scholar]
- 38.Penno S, Lindell D, Post AF. Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N-regulatory gene ntcA. Environ Microbiol. 2006;8(7):1200–1211. doi: 10.1111/j.1462-2920.2006.01010.x. [DOI] [PubMed] [Google Scholar]
- 39.Pittera J, Humily F, Thorel M, Grulois D, Garczarek L, Six C. Connecting thermal physiology and latitudinal niche partitioning in marine Synechococcus. ISME J. 2014;8(6):1221–1236. doi: 10.1038/ismej.2013.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Post AF, Penno S, Zandbank K, Paytan A, Huse SM, Welch DM (2011) Long term seasonal dynamics of Synechococcus population structure in the Gulf of Aqaba, Northern Red Sea. Front Microbiol 2(131) [DOI] [PMC free article] [PubMed]
- 41.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, Johnson ZI, Land M, Lindell D, Post AF, Regala W, Shah M, Shaw SL, Steglich C, Sullivan MB, Ting CS, Tolonen A, Webb EA, Zinser ER, Chisholm SW. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424(6952):1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 42.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73(2):249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scanlan DJ, West NJ. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol. 2002;40(1):1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 44.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shalapenok L, Shalapenok A. Heterogeneous pigment composition of phycoerythrin-containing picocyanobacteria Synechococcus spp. in the black sea. Microbiology. 1997;66(1):80–84. [Google Scholar]
- 46.Sher D, Thompson JW, Kashtan N, Croal L, Chisholm SW. Response of Prochlorococcus ecotypes to co-culture with diverse marine bacteria. ISME J. 2011;5(7):1125–1132. doi: 10.1038/ismej.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Six C, Thomas JC, Garczarek L, Ostrowski M, Dufresne A, Blot N, Scanlan DJ, Partensky F, et al. Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol. 2007;8(12):R259. doi: 10.1186/gb-2007-8-12-r259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 49.Stomp M, Huisman J, Stal LJ, Matthijs HC. Colorful niches of phototrophic microorganisms shaped by vibrations of the water molecule. ISME J. 2007;1(4):271–282. doi: 10.1038/ismej.2007.59. [DOI] [PubMed] [Google Scholar]
- 50.Stomp M, Huisman J, Vörös L, Pick FR, Laamanen M, Haverkamp T, Stal LJ. Colourful coexistence of red and green picocyanobacteria in lakes and seas. Ecol Lett. 2007;10(4):290–298. doi: 10.1111/j.1461-0248.2007.01026.x. [DOI] [PubMed] [Google Scholar]
- 51.Tai V, Palenik B. Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J. 2009;3(8):903–915. doi: 10.1038/ismej.2009.35. [DOI] [PubMed] [Google Scholar]
- 52.Toledo G, Palenik B, Brahamsha B. Swimming marine Synechococcus strains with widely different photosynthetic pigment ratios form a monophyletic group. Appl Environ Microbiol. 1999;65(12):5247–5251. doi: 10.1128/aem.65.12.5247-5251.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterbury JB, Watson SW, Valois FW, Franks DG. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can Bull Fish Aquat Sci. 1986;214:71–120. [Google Scholar]
- 54.Waterbury JB, Willey JM, Franks DG, Valois FW, Watson SW. A cyanobacterium capable of swimming motility. Science. 1985;230(4721):74–76. doi: 10.1126/science.230.4721.74. [DOI] [PubMed] [Google Scholar]
- 55.West NJ, Scanlan DJ. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl Environ Microbiol. 1999;65(6):2585–2591. doi: 10.1128/aem.65.6.2585-2591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood AM. Discrimination between types of pigments in marine Synechococcus spp. by scanning spectroscopy, epifluorescnece microscopy, and flow cytometry. Limnol Oceanogr. 1985;30(6):1303–1315. doi: 10.4319/lo.1985.30.6.1303. [DOI] [Google Scholar]
- 57.Wood AM, Lipsen M, Coble P. Fluorescence-based characterization of phycoerythrin-containing cyanobacterial communities in the Arabian sea during the northeast and early southwest monsoon (1994–1995) Deep-Sea Res II Top Stud Oceanogr. 1999;46(8):1769–1790. doi: 10.1016/S0967-0645(99)00043-0. [DOI] [Google Scholar]
- 58.Wood AM, Phinney DA, Yentsch CS. Water column transparency and the distribution of spectrally distinct forms of phycoerythrin-containing organisms. Mar Ecol Prog Ser. 1998;162:25–31. doi: 10.3354/meps162025. [DOI] [Google Scholar]
- 59.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O, Scanlan DJ. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10(1):147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 60.Zwirglmaier K, Spence E, Zubkov MV, Scanlan DJ, Mann NH. Differential grazing of two heterotrophic nanoflagellates on marine Synechococcus strains. Environ Microbiol. 2009;11(7):1767–1776. doi: 10.1111/j.1462-2920.2009.01902.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.