Abstract
Intratumour heterogeneity complicates biomarker discovery and treatment personalization, and pervasive cancer evolution is a key mechanism leading to therapy failure and patient death. Thus, understanding subclonal heterogeneity architectures and cancer evolution processes is critical for the development of effective therapeutic approaches which can control or thwart cancer evolutionary plasticity. Current insights into heterogeneity are mainly limited to the macroheterogeneity level, established by cancer subclones that have undergone significant clonal expansion. Novel single cell sequencing and blood-based subclonal tracking technologies are enabling detailed insights into microheterogeneity and the dynamics of clonal evolution. We assess how this starts to delineate the rules governing cancer evolution and novel angles for more effective therapeutic intervention.
Current Opinion in Genetics & Development 2015, 30:1–6
This review comes from a themed issue on Cancer genomics
Edited by Christine A Iacobuzio-Donahue and Elaine A Ostrander
For a complete overview see the Issue
Available online 31st December 2014
http://dx.doi.org/10.1016/j.gde.2014.12.001
0959-437X/© 2014 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/).
Introduction
Cancer is a genetic and epigenetic disease arising from a single cell that has acquired the hallmarks of cancer. Although monoclonal in origin, the background mutation rate and genomic instability mechanisms which are operative in many cancers foster the generation of new mutations during the ensuing expansion of the cancer cell population [1, 2]. Although most new mutations are likely to be deleterious or have no impact on cellular fitness, the enormous number of mutations which can be generated during progression into an advanced cancer, harbouring up to hundreds of billions of malignant cells [3], likely generates a wealth of viable phenotypes. This subclonal diversity is the substrate which Darwinian selection can act upon, permitting the on-going evolutionary adaptation of cancer populations through the expansion of subclones harbouring beneficial aberrations [4].
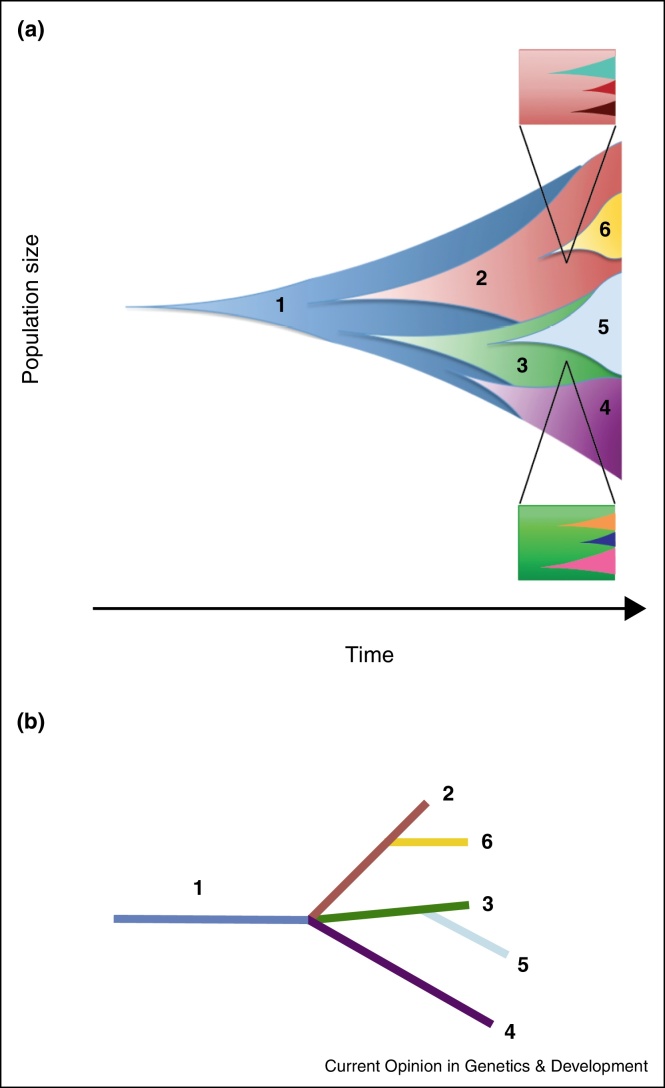
However, new mutations, bestowing genetic diversity, are initially confined to individual cells and to small subclones after subsequent rounds of cell division. These remain below the detection limit of standard exome or genome sequencing approaches, which have low sensitivity and high false positive rates when applied for the detection of mutations with allele frequencies below 10% in the DNA extracted from a tumour sample [5]. Thus, this microheterogeneity remains undetectable until a significant expansion of one or more subclones establishes macroheterogeneity (Figure 1a), which may have a branched evolutionary pattern (Figure 1b). Novel single cell [6, 7, 8•] and ultra-deep DNA sequencing technologies [9•] only recently started to permit investigations into intratumour micro-heterogeneity and macro-heterogeneity, whilst circulating tumour DNA (ctDNA) detection techniques [10, 11, 12, 13] and circulating tumour cell molecular analyses [14] provide insight into the dynamics of evolutionary adaptation. We review how these techniques reveal intratumour micro-heterogeneity and macro-heterogeneity, thereby unravelling the fundamental evolutionary nature of cancer and the central role of genetic intratumour heterogeneity for patient outcome.
Figure 1.
Macro and microheterogeneity in cancer evolution. (a) Schematic illustrating clonal evolution. Multiple subclones evolve from the founding clone (blue) and undergo major clonal expansions, changing the composition of the tumour cell population. Subclones are detectable as macroheterogeneity by standard next generation sequencing approaches owing to their large population sizes. Magnifications of small proportions of the cancer cell population (insets) show the population structure at the microheterogeneity level. Newly generated mutations in single cells, which subsequently expand into small subclonal populations are below the detection limit of standard next-generation sequencing techniques and can only be detected through single cell or ultra-deep sequencing technologies. (b) Phylogenetic tree reconstructed from the macroheterogeneity data, depicting a branched evolutionary trajectory. The founding clone (blue) represents the trunk of the phylogenetic tree.
Evidence for intratumour macroheterogeneity
Intratumour macroheterogeneity has been observed across several solid tumour types. Exome sequencing of multiple tumour regions from ten clear cell renal cell carcinomas (ccRCC) demonstrated that, on average, over two thirds of driver somatic copy number aberrations (SCNAs) and of driver mutations were heterogeneous within individual tumours [15••]. Subclones were spatially demarcated within primary tumours and differed between primary tumours and metastatic sites within patients. Reconstructing the ancestral relationships of these subclones revealed branched evolutionary patterns with multiple subclones evolving simultaneously in each tumour but along distinct evolutionary paths [15••]. A characteristic found in all ten tumours was the presence of inactivating somatic alterations in the von Hippel Lindau (VHL) gene and loss of heterozygosity of chromosome 3p, harbouring the second copy of the VHL gene, on the trunk of the phylogenetic trees. Thus, these driver aberrations had been acquired early, most likely in the founding cell of each tumour. In contrast, other known ccRCC driver genes, including PI3K-mTOR pathway genes and those encoding epigenetic regulators were predominantly mutated in tumour subclones. SETD2, BAP1 and PBRM1 driver gene mutations were found in distinct subclones within the same tumour, defying that mutations in these genes define distinct molecular ccRCC subtypes [15••]. Studies into signalling pathway activity and prognostic and predictive biomarker expression demonstrated that genetic heterogeneity was associated with phenotypic diversity [15••, 16••, 17]. Multi-region exome sequencing of high-grade serous ovarian cancers also found macroheterogeneity and branched evolution, with early truncal TP53 mutations in five out of six patients, whereas driver genes such as PIK3CA, CTNNB1 and NF1 were mutated in subclones [18]. Multi-region SCNA profiling of nine glioblastomas demonstrated homogenous CDKN2A/B losses and EGFR amplifications, suggesting early acquisition on the trunk of the phylogenetic trees [19]. In contrast, SCNAs harbouring RB1, AKT3, and MDM4 were always found to be subclonal whereas those affecting CDK6, MET, PDGFRA, PTEN and TP53 were subclonal in some and truncal in other cases [19].
Evidence for macroheterogeneity with significantly expanded subclones has also been identified within individual cancer samples. Deep sequencing of triple negative breast cancer biopsies revealed that most TP53, PIK3CA, and PTEN mutations had been acquired early during tumour evolution although they were subclonal in a small proportion of cases [20•]. In contrast, mutations in cytoskeletal, cell shape and motility proteins were predominantly subclonal, suggesting on-going evolutionary adaptation. Fluorescence in situ hybridisation of driver SCNAs identified genetically distinct subclonal populations in the majority of ETV6-RUNX1 positive acute lymphoblastic leukaemias (ALL) [21]. Twenty-four cases exhibited branched evolution and only six malignancies followed a linear evolutionary pattern [21]. Sequencing of single biopsies from non-small cell lung cancers (NSCLC) revealed subclonal heterogeneity in ten out of 17 cases [22]. Tumours harbouring KRAS or EGFR mutations had always acquired these in the founding clone, whereas putative driver mutations in HGF were subclonal. A large study investigating mutation concordance within NSCLC primary tumours and between primary tumours and metastases or recurrences found no macroheterogeneity of EGFR driver mutations, further supporting the notion that activating EGFR mutations are generally truncal [23]. This was also confirmed by two recent NSCLC multi-region exome sequencing studies. All identified activating EGFR mutations and indeed the majority of all other known NSCLC driver mutations and driver SCNAs were located on the trunks of the phylogenetic trees [24, 25]. Macroheterogeneity and branched phylogenetic patterns were nevertheless identified in each tumour. Although most heterogeneous aberrations may be passengers, the high mutation rate in NSCLCs impairs the ability to define the driver gene catalogue of these tumours [26] and subclonal drivers may have remained undetected as a consequence. Mutational signature analysis showed that a cell-endogenous mutational process caused by up-regulation of the APOBEC deaminase [24, 25] was the predominant mechanism of NSCLC subclonal mutation generation [27••], even in patients with on-going tobacco smoke exposure.
Insights into intratumour microheterogeneity
Novel sequencing technologies increasingly allow the investigation of microheterogeneity at the fundamental level of the single cell. The detection of SCNAs and point mutations in up to 60 individual cell nuclei from each of two breast cancers identified major subclones evolving in a branched evolutionary fashion in each tumour [8•], corroborating the conclusion from breast cancer macroheterogeneity studies [20•]. Single cell resolution further revealed relatively stable SCNA profiles across cancer cells within a tumour whereas point mutations differed between major subclonal populations but also within subclones. Thus, SCNAs had been acquired early during carcinogenesis and point mutation acquisition was continuously driving microheterogeneity generation. Mutation rate estimates based on this data revealed ∼8 new mutations per cell division in a triple negative cancer and 0.6–0.9 new mutations in an ER positive tumour, which is similar to the estimated 0.6 new mutations per division for normal cells. Importantly, single cell mutational heterogeneity allows insights into current mutation rates. In contrast, mutations observed at the macroheterogeneity level have been acquired many generations before clonal expansion made them detectable and only provides a historical record of the mutational processes that were operative in earlier tumour stages [28•].
Reconstruction of SCNA profiles from single cell RNA sequencing data from glioblastomas identified a monoclonal structure in four cases and two major subclones within one further case [29••]. Within the limits of the assay, which has a low sensitivity to detect small aberrations, SCNAs were similar between individual cells of a clone or subclone. Thus, the generation of new SCNAs may be a rare event and the observed profiles were likely acquired early during cancer evolution, similar to the results in breast cancers [8•]. Single cell RNA expression data further enabled the simultaneous assessment of gene expression profiles between single cells with similar SCNA profiles. This detected transcriptional signatures of different glioblastoma subtypes and variable degrees of stemness co-existing in different cells within a tumour. The simultaneous interrogation of genetic and non-genetic macroheterogeneity within a cancer cell population provides powerful opportunities to assess phenotypic consequences of subclonal genetic aberrations.
Macroheterogeneity of known driver mutations is rare between primary colorectal cancers (CRCs) and associated metastatic lesions. Mutations in KRAS, NRAS, BRAF and APC driver genes were always concordant and only low-level discordance was observed for mutations in TP53, PIK3CA and PTEN in a study of 69 primary CRC and metastasis pairs [30]. The absence of macroheterogeneity, for example for KRAS and NRAS mutations, suggests that these drivers were acquired on the trunk of the phylogenetic tree, in tumours in which they are detectable. The high concordance most likely explains the robust performance of KRAS and NRAS mutations as predictors of primary resistance to anti-EGFR therapy in CRCs [31, 32]. However, KRAS and NRAS mutations became detectable in the ctDNA from 23 out of 24 initially KRAS/NRAS wild-type CRCs at the time acquired resistance to anti-EGFR treatment had developed [33••]. Surprisingly, multiple distinct activating KRAS and NRAS mutations emerged in the ctDNA in 63% of patients, demonstrating that polyclonal resistance evolution was common. An analysis of the kinetics of KRAS mutation evolution in these patients further concluded that KRAS mutations had been present before anti-EGFR therapy initiation, in small subclones comprising ∼2000–3000 cancer cells [34]. Direct support for this microheterogeneity has been provided by the detection of low level KRAS mutations by sensitive digital PCR technology in patients found to be KRAS wild-type by standard detection techniques [41]. Thus, microheterogeneity of KRAS mutations and potentially also of other resistance driver mutations is likely to be present in many metastatic CRCs which are KRAS wild-type based on standard sequencing approaches. These mutations may evade detection owing to the small number of affected cells and through spatial segregation across metastatic sites but they eventually drive resistance evolution and therapy failure. The reliable evolution of one or multiple KRAS or NRAS mutant subclones in most patients during anti-EGFR therapy further suggest that the population size and mutation rates are sufficiently high to generate many beneficial driver mutations in any metastatic CRC. The presence of KRAS mutation microheterogeneity in many tumours which are KRAS wild-type by standard sequencing technologies together with the absence of KRAS mutational macroheterogeneity further indicated that these new subclones rarely undergo significant clonal expansion. Thus, KRAS mutations apparently have a low or no selective advantage unless they are acquired early during carcinogenesis or the tumour is treated with EGFR-targeted agents.
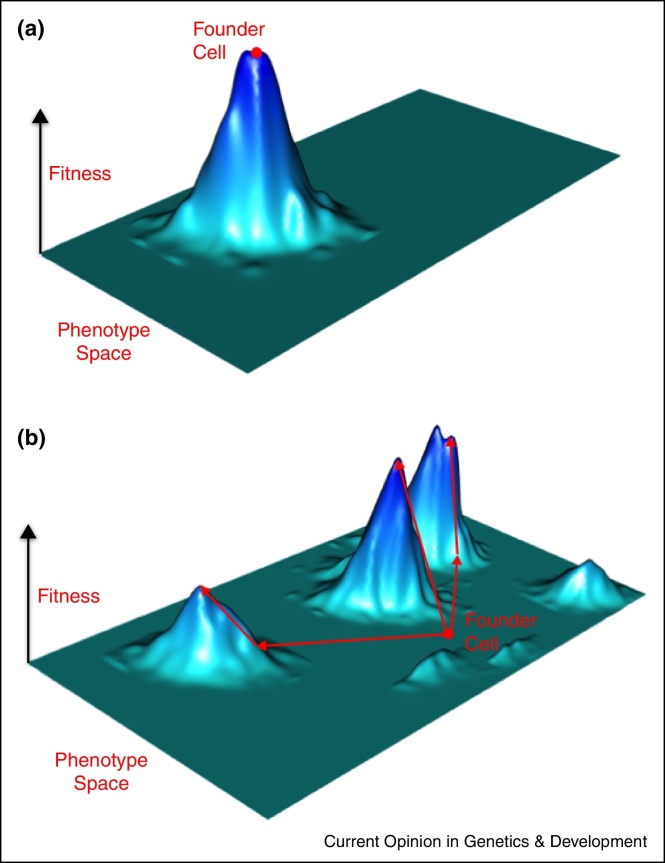
The presence of strong driver aberrations in the founding cell of most CRCs which leaves only limited opportunity to further optimize cancer cell fitness could be a parsimonious explanation for this paradox. In other words, many CRCs may already occupy a fitness peak on the fitness landscape at the time of cancer initiation (Figure 2a) precluding significant expansion and macroheterogeneity evolution of subclones harbouring additional drivers. This may be fundamentally different in the founding cell of a ccRCC (Figure 2b), which may only harbour a small number of weak drivers such as mutations in VHL and chromosome 3p loss. This is supported by studies in patients with germ-line VHL mutations which only showed a modest proliferative advantage of biallelic VHL inactivation in renal tubular cells [35] compared to the proliferative advantage conferred by biallelic inactivation of the APC tumour suppressor gene (which is altered in ∼80% of CRCs [36]) in colon cells. Thus, the founder clone of a typical ccRCC is likely to be located on a fitness landscape that permits significant further fitness increments through the acquisition of additional driver aberrations (Figure 2b). This would explain the frequent evolution of subclones harbouring additional driver genes and the detection of macroheterogeneity in these tumours.
Figure 2.
Influence of the fitness landscape on cancer evolutionary patterns. (a) Hypothetical cancer fitness landscape in which the founding cell (red dot) is already located at a fitness peak. Further evolutionary adaptation is only possible through a change in the fitness landscape, for example through a change in the environment or through drug therapy. Microheterogeneity can be extensive in this tumour but macroheterogeneity is absent. (b) Hypothetical fitness landscape where the founding cell is not located at a fitness peak. Tumour subclones can increase their relative fitness through the acquisition of further driver mutations which will lead to subclonal expansion. Increases in fitness are illustrated as arrows climbing up the fitness peaks. If multiple subclones acquire drivers that increase their relative fitness, branched evolution can occur. Multiple fitness peaks indicate multiple possible phenotypes which lead to increased cellular fitness.
Conclusions
Exome and genome sequencing studies of up to 500 cancer samples recently identified the most prevalent driver genes in many cancer types. In parallel, smaller studies started to portray the subclonal landscapes of many tumour types at the macroheterogeneity level through multi-region sequencing approaches or subclonal composition analysis of individual biopsies. This provided ample evidence for on-going evolutionary adaptation during cancer progression, frequently along complex branched trajectories, and started to delineate the spatial structures of subclonal architectures. The spatial segregation of functionally distinct subclones is a major hurdle for personalized cancer therapies as it complicates efforts to accurately assess the driver aberration landscapes of individual tumours. These results also question whether and how tumours harbouring subclones with different driver mutations can be optimally treated. The concept of a clinically dominant clone, which is not necessarily numerically dominant in a cancer but ultimately lethal for an individual patient [37•, 38], is emerging from this work and the development of strategies to detect, track and treat clinically dominant subclones is an important area of future research. At the same time, macroheterogeneity studies started to define cancer type specific ‘evolutionary rules’, such as the identification of driver genes which are commonly altered on the trunk of a specific tumour type, providing opportunities to prioritize the development of targeted therapeutics [39]. Most recently, new technologies enabled the study of microheterogeneity in exceedingly small subclones and even at the quantum level of the single cell. Combined with assessments of subclonal population dynamics through ctDNA or circulating tumour cell tracking [14], these tools start to unravel key mechanisms of cancer evolution at an unprecedented level of detail. For example, quantification of de novo mutation generation, the construction of genotype-phenotype maps and ultimately the mapping of dynamic fitness landscapes can now be accomplished. As on-going cancer evolution fosters cancer progression and therapy failure [40], a fundamental understanding of the rules governing cancer evolution may lead to novel therapeutic and preventive approaches to slow down or thwart evolution in order to improve clinical outcomes.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank Kamil Lipinski, Andrea Sottoriva and Chris Illingworth for critical reading of the manuscript. MG was funded by grants from Cancer Research UK, Prostate Cancer UK, the Prostate Cancer Foundation, the Royal Marsden NIHR Biomedical Research Centre for Cancer and the Wellcome Trust (Grant number: 105104/Z/14/Z). MD was funded by a grant from Cancer Research UK.
References
- 1.Greaves M., Maley C.C. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burrell R.A., McGranahan N., Bartek J., Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 3.Del Monte U. Does the cell number 10(9) still really fit one gram of tumor tissue? Cell Cycle. 2009;8:505–506. doi: 10.4161/cc.8.3.7608. [DOI] [PubMed] [Google Scholar]
- 4.Gerlinger M., McGranahan N., Dewhurst S.M., Burrell R.A., Tomlinson I., Swanton C. Cancer: evolution within a lifetime. Annu Rev Genet. 2014;48:215–236. doi: 10.1146/annurev-genet-120213-092314. [DOI] [PubMed] [Google Scholar]
- 5.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voet T., Kumar P., Van Loo P., Cooke S.L., Marshall J., Lin M.L., Zamani Esteki M., Van der Aa N., Mateiu L., McBride D.J. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 2013;41:6119–6138. doi: 10.1093/nar/gkt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni X., Zhuo M., Su Z., Duan J., Gao Y., Wang Z., Zong C., Bai H., Chapman A.R., Zhao J. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Wang Y., Waters J., Leung M.L., Unruh A., Roh W., Shi X., Chen K., Scheet P., Vattathil S., Liang H. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]; A novel single cell sequencing method that utilizes G2/M nuclei to reduce the problem of allelic dropout which occurrs during whole genome amplification of single cell genomes. Application to ER+ and triple-negative breast cancer cells showed intratumour microheterogeneity and macroheterogeneity. Chromosomal rearrangements tended to occur early in tumorigenesis whereas point mutations continued to evolve, generating clonal diversity.
- 9•.Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe a novel next generation sequencing error correction technology which improves the sensitivity and specificity for ultra-rare subclonal mutation detection. This has been used subsequently by Wang et al. to validate mutations identified by single cell sequencing.
- 10.Diaz L.A., Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson S.J., Tsui D.W., Murtaza M., Biggs H., Rueda O.M., Chin S.F., Dunning M.J., Gale D., Forshew T., Mahler-Araujo B. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 12.De Mattos-Arruda L., Weigelt B., Cortes J., Won H.H., Ng C.K., Nuciforo P., Bidard F.C., Aura C., Saura C., Peg V. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heitzer E., Ulz P., Belic J., Gutschi S., Quehenberger F., Fischereder K., Benezeder T., Auer M., Pischler C., Mannweiler S. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:p30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mateo J., Gerlinger M., Rodrigues D., de Bono J.S. The promise of circulating tumor cell analysis in cancer management. Genome Biol. 2014;15:448. doi: 10.1186/s13059-014-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Gerlinger M., Horswell S., Larkin J., Rowan A.J., Salm M.P., Varela I., Fisher R., McGranahan N., Matthews N., Santos C.R. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used multi-region exome sequencing to analyze intratumour heterogeneity of chromosomal aberrations and point mutations in ten primary ccRCCs and their metastases. This is one of the first studies to define the subclonal architecture and the evolutionary anatomy of a solid cancer. This is also one of the first studies demonstrating temporal heterogeneity of mutational processes.
- 16••.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first study demonstrating branched evolutionary trajectories through exome sequencing in a solid tumour. Extensive driver mutation heterogeneity and heterogeneity of predictive and prognostic biomarkers was also identified.
- 17.Gulati S., Martinez P., Joshi T., Birkbak N.J., Santos C.R., Rowan A.J., Pickering L., Gore M., Larkin J., Szallasi Z. Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers. Eur Urol. 2014;66:936–948. doi: 10.1016/j.eururo.2014.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashashati A., Ha G., Tone A., Ding J., Prentice L.M., Roth A., Rosner J., Shumansky K., Kalloger S., Senz J. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231:21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sottoriva A., Spiteri I., Piccirillo S.G., Touloumis A., Collins V.P., Marioni J.C., Curtis C., Watts C., Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Shah S.P., Roth A., Goya R., Oloumi A., Ha G., Zhao Y., Turashvili G., Ding J., Tse K., Haffari G. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study pioneered the use of variant allelel frequencies to define the subclonal compositions of individual biopsies from 104 triple negative breast cancers.
- 21.Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 22.Govindan R., Ding L., Griffith M., Subramanian J., Dees N.D., Kanchi K.L., Maher C.A., Fulton R., Fulton L., Wallis J. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatabe Y., Matsuo K., Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 24.de Bruin E.C., McGranahan N., Mitter R., Salm M., Wedge D.C., Yates L., Jamal-Hanjani M., Shafi S., Murugaesu N., Rowan A.J. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Fujimoto J., Zhang J., Wedge D.C., Song X., Zhang J., Seth S., Chow C.W., Cao Y., Gumbs C. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imielinski M., Berger A.H., Hammerman P.S., Hernandez B., Pugh T.J., Hodis E., Cho J., Suh J., Capelletti M., Sivachenko A. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; Whole genome sequencing of 21 breast cancers identified clustered mutations which were thought to occurr in a punctuated fashion and which the authors termed kataegis. The mutational signature of this novel genomig instability mechanisms suggested the involvement of APOBEC deaminases in this process.
- 28•.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg Å., Børresen-Dale A.-L. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of over 20 different mutational signatures which can be operative in human cancers. The data suggests that multiple mutational processes can contribute to mutation generation in a cancer type and provides a novel tool to detect these in individual cancers.
- 29••.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors pioneered the use of single-cell RNA sequencing to determine and transcriptomic heterogeneity in parallel from the same single cells.
- 30.Brannon A., Vakiani E., Sylvester B.E., Scott S.N., McDermott G., Shah R.H., Kania K., Viale A., Oschwald D.M., Vacic V. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:p454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lievre A., Bachet J.B., Le Corre D., Boige V., Landi B., Emile J.F., Cote J.F., Tomasic G., Penna C., Ducreux M. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 32.Douillard J.-Y., Oliner K.S., Siena S., Tabernero J., Burkes R., Barugel M., Humblet Y., Bodoky G., Cunningham D., Jassem J. Panitumumab — FOLFOX4 Treatment and RASMutations in Colorectal Cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 33••.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study applied ctDNA detection to interrogate mechanisms of anti-EGFR therapy resistance in colorectal cancer patients. The identification of polyclonal resistance in a large proportion of these patient shows the enormous evolutionary plasticity of cancer cell populations.
- 34.Diaz L.A., Jr., Williams R.T., Wu J., Kinde I., Hecht J.R., Berlin J., Allen B., Bozic I., Reiter J.G., Nowak M.A. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandriota S.J., Turner K.J., Davies D.R., Murray P.G., Morgan N.V., Sowter H.M., Wykoff C.C., Maher E.R., Harris A.L., Ratcliffe P.J. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Haffner M.C., Mosbruger T., Esopi D.M., Fedor H., Heaphy C.M., Walker D.A., Adejola N., Gurel M., Hicks J., Meeker A.K. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study found that a small low grade subclone nested within a predominantly high grade primary prostate cancer had seeded the metastases which were eventually lethal for this patient. This case study illustrates how small subclones within the primary tumour can dominate clinical outcomes and the difficululties this causes for personalized cancer medicine.
- 38.Gerlinger M., Catto J.W., Orntoft T.F., Real F.X., Zwarthoff E.C., Swanton C. Intratumour heterogeneity in urologic cancers: from molecular evidence to clinical implications. Eur Urol. 2014 doi: 10.1016/j.eururo.2014.04.014. (epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 39.Yap T.A., Gerlinger M., Futreal P.A., Pusztai L., Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 40.Gerlinger M., Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurent-Puig P., Pekin D., Normand C., Kotsopoulos S.K., Nizard P., Perez Toralla K., Rowell R., Olson J., Srinivasan P., Le Corre D. Clinical relevance of KRAS-mutated sub-clones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin. Can. Res. 2014 doi: 10.1158/1078-0432.CCR-14-0983. [DOI] [PubMed] [Google Scholar]