INTRODUCTION
Genetic instability is a common property of many human cancers[1], including those of HNPCC[2,3]. A novel form of genetic instability involving somatic alterations, such as deletions and insertions in simple repeated sequences[4], has been found. Microsatellites are relatively short runs of tandemly repeated sequences scattered throughout the genome[5,6]. Ubiquitous alterations in these sequences were initially detected by unbiased DNA fingerprinting in a subset of colorectal cancer[7,8], implying the presence of genome-wide genetic instability. Subsequently, amplification by polymerase chain reaction (PCR) of a few microsatellite loci should be used to reveal this microsatellites instability (MSI) in colorectal cancers[9,10] and other malignancies[11-14].
MSI was observed to be a common feature of HNPCC[15]. Physical mapping and finding of mutations in HNPCC patients revealed that a human homologues of the Escherichia coli DNA mismatch repair (MMR) enzyme MutS was one of the candidate genes for HNPCC and was named hMSH2[16]. Human homologues of the E. coli and yeast mismatch repair enzymes hMLH1, hPMS1 and hPMS2 have also been associated with HNPCC[17]. Defects of these mismatch repair genes have been reported to produce MSI in bacteria and yeast, and a high mutation rate of microsatellites has been observed in HNPCC patients and in cell lines established from HNPCC tumors[18]. Therefore, MSI in HNPCC is one of the results of mutations in these mismatch repair genes. In the present study, we analyzed MSI, expression of MMR genes and cell proliferation activity in colorectal cancer (CRC) patients with familial predisposition, to reveal the characteristics of these patients’ genetic defects and to afford the biological basis for screening high-risk relatives of CRC.
MATERIALS AND METHODS
Patients
Forty-six colorectal cancer patients who underwent surgical resection between 1993 and 1995 at Nanfang Hospital in Guangzhou, China, were analyzed. These included 26 patients with familial predisposition (Group A) by summarizing clinic archives and surveying their pedigrees, and 20 randomly selected colorectal cancer patients without familial predisposition (Group B). In Group A, 4 patients were eligible HNPCC according to the Amsterdam criteria[15].
PCR
Genomic DNA was extracted from formalin fixed, paraffin embedded tumor tissues and corresponding normal tissues, respectively, using a modification of the method reported previously[19-21]. About 100 ng of genomic DNA was used for PCR amplification of microsatellite sequences.
Microsatellite instability
Four microsatellite loci, D2S119, D2S123, D5S107 and D17S250 were analyzed in all patients, using silver staining polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) technique[22,23]. We classified patients as positive for MSI when the PCR product using tumor DNA revealed the presence of extra bands or shifting bands that were not visible in the PCR product of the corresponding normal tissue DNA, and these changes were found in at least two microsatellite loci, called MSI-H[24,25].
Immunohisochemical staining of hMLH1, hMSH2 and proliferation cell nucleus antigen (PCNA)
hMLH1, hMSH2 enzyme was detected using a rabbit polyclonal antibody, and PCNA, a monoclonal antibody (Dako Co). By streptavidin-peroxidase (SP) immunohisochemical method[26], we defined positive cases as those of hMLH1, hMSH2 that showed staining of tissues (tumor or normal) cell nuclei or cytoplasm in clusters[27], and PCNA that showed staining of tumor cell nuclei in clusters[28,29]. The label index (LI) of PCNA was analyzed.
Flow cytometry for DNA analysis
Using Flow Cytometry (Type of EPICS ELITE), the DNA index (DI), heteroploid rate, proliferation index (PI) and S-phase percentage were assayed in tumor cells. We defined heteroploid cells as that DI was not between 0.90 and 1.10.
Statistical analysis
Two-tailed Fisher’s and χ² exact tests were used to analyze the significance of differences between the groups. The significance level was set at P < 0.05.
RESULTS
All patients were detected for MSI in four microsatellite loci (Figure 1). MSI was detected in 20 (76.9%) out of 26 patients in Group A. The MSI-positive (MSI-H) proportion in Group A was 46.2% (12/26), significantly higher than 10% (2/20) in Group B (P < 0.05). Three of 4 HNPCC patients in Group A were MSI-positive.
Figure 1.
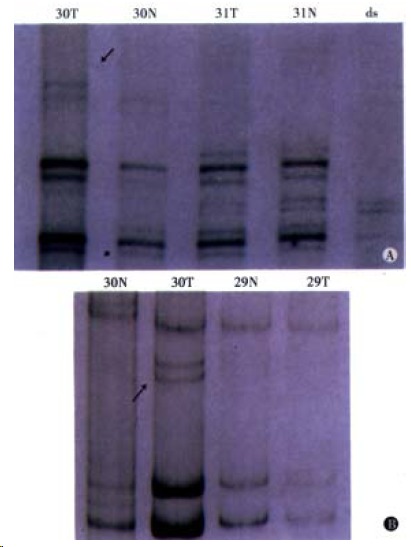
Patient of No.30 positive for MSI. A: Lane N: corresponding normal tissue, Lane T: tumor tissue, Lane ds: di-strand control. B: MSI is defined as showing the presence of extra or shifting bands in PCR products using tumor DNA that are not visible in the products from corresponding normal tissue. (Left) D17D250, (Right) D2S123.
We examined the association of MSI with clinicopathologic characteristics of CRC patients with familial predisposition, and elucidated the following features: ① An early age of onset (Table 1). Patients positive for MSI in Group A tended to have a younger mean age at onset than patients in Group B (P < 0.01) and patients negative for MSI in Group A. ② A preponderance of tumors in the proximal colon (Table 2). Proximal colon includes cecum, ascending and transverse colon[30]. Among patients positive for MSI, 9 (64.3%) of 14 tumors were located in the proximal colon. In contrast, among patients negative for MSI, 9 (28.1%) of 32 tumors were located in the proximal colon (P < 0.05). In MSI-negative patients, tumors in Group A also had a proclivity for the proximal colon (5/9) but tumors in Group B did not (4/14). ③ Correlation with a poorly differentiated phenotype (Table 2). Among patients positive for MSI, 11 (78.6%) of 14 tumors were poorly differentiated adenocarcinomas. It was significantly higher than that among MSI-negative patients (P < 0.05). Also in tumors of Group A, the poor differentiation rate positive MSI was higher than that of negative MSI. ④ Proclivity for extracolorectal malignancy. Among patients in Group A, 6 cancers were associated with cancers in other organs (2 in uterus, 2 in stomach, 1 in bladder, and 1 in bile duct), and 5 of these were positive for MSI. In contrast, there was no patient with cancers in other organs in Group B.
Table 1.
MSI and mean age at onset of colorectal cancer
| Group | MSI | n | Age (¯x ± s) |
| HNPCC | 4 | 41.8 ± 7.7b | |
| A | + | 12 | 45.4 ± 8.3b |
| - | 14 | 49.6 ± 9.7 | |
| B | + | 2 | 57.5 ± 9.2 |
| - | 18 | 58.9 ± 11.0 |
P < 0.01 vs Group B; Fisher’s test.
Table 2.
MSI and location, histologic differentiation of colorectal cancer
| Group | MSI |
Location n (%) |
Differentiation n (%) |
||
| Pr | Di | PD | WD | ||
| A | + | 8 (66.7) | 4 (33.3) | 9 (75.0)b | 3 (25.0) |
| - | 5 (35.7) | 9 (64.3) | 4 (28.6) | 10 (71.4) | |
| B | + | 1 (50.0) | 1 (50.0) | 2 | 0 |
| - | 4 (22.2) | 14 (77.8) | 5 (27.8) | 13 (72.2) | |
| Total | + | 9 (64.3)a | 5 (35.7) | 11 (78.6)a | 3 (21.4) |
| - | 9 (28.1) | 23 (71.9) | 9 (28.1) | 23 (71.9) | |
P < 0.05, vs patients negative for MSI; χ² test.
P < 0.05, vs patients negative for MSI in Group A; χ² test. Pr: proximal colon; Di: distal colon (descending, sigmoid colon and rectum); PD: poorly differentiated adenocarcinoma; WD: well and moderately differentiated adenocarcinoma.
Immunohisochemically, contrast with Group B, patients in Group A tended to be negative for hMLH1 protein staining in tumor tissues, and hMLH1, hMSH2 protein staining in normal tissues (Figure 2, Table 3). Among patients positive for MSI, 6 of 14 colorectal tumors were negative for hMLH1 together with hMSH2 protein, whereas 4 of 32 tumors negative for MSI (P < 0.05). The LI of PCNA staining (Figure 3) in tumors of Group A was 0.54 ± 0.10, which was lower than that of Group B (0.62 ± 0.07), P < 0.01, meanwhile in cancer tissues with MSI-positive, the LI was 0.53 ± 0.10, which was lower than that of negative MSI (0.59 ± 0.08), P < 0.05.
Figure 2.
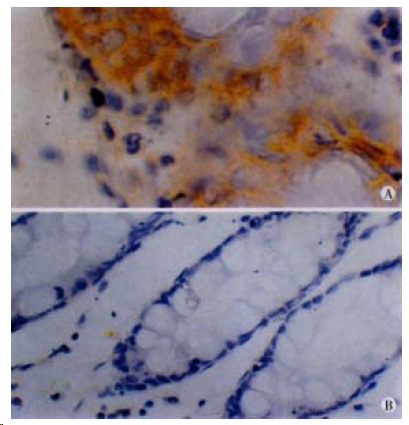
Immunohistochemistry of hMLH1. (Left) Patient positive for hMLH1 immunostaining. × 40. (Right) Patient negative for hMLH1 immunostaining, × 20.
Table 3.
Negative for hMLH1, hMSH2 protein staining in CRC patients’ tissues (n, %)
|
Group A (26 cases) |
Group B (20 cases) |
|||
| Tumor | Normal | Tumor | Normal | |
| hMLH1 - | 16 (61.5)a | 17 (65.4)b | 6 (30.0) | 5 (25.0) |
| hMSH2 - | 14 (53.9) | 16 (61.5)b | 9 (45.0) | 6 (30.0) |
P < 0.05, vs tumor tissues in Group B; χ² test.
P < 0.05, vs normal tissues in Group B; χ² test.
Figure 3.
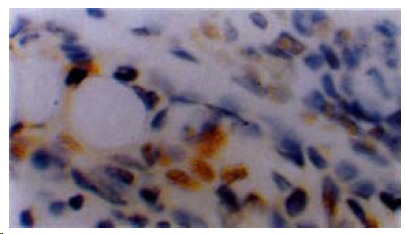
Immunostaining for PCNA in colorectal cancer tissue of patient with familial predisposition. × 40.
According to flow cytometry (Table 4), the heteroploid rate of DNA in tumors of Group A was 23.1%, PI and S-phase percentage in tumors with positive MSI were 14.34 ± 5.49 and 8.18 ± 2.55. In contrast with that of Group B and negative MSI respectively, they decreased obviously (P < 0.05).
Table 4.
MSI and DNA analysis in CRC tumor cells by flow cytometry (¯x ± s)
| Group | Cases | Heteroploid (n, %) | PI | S-phase |
| HNPCC | 4 | 1 (25.0) | 14.58 ± 3.12 | 8.70 ± 1.18 |
| A | 26 | 6 (23.1)a | 17.57 ± 6.51 | 9.47 ± 2.85 |
| B | 18 | 10 (55.6) | 17.94 ± 7.51 | 10.38 ± 3.89 |
| MSI + | 14 | 4 (28.6) | 14.34 ± 5.49b | 8.18 ± 2.55b |
| MSI - | 30 | 12 (40.0) | 19.30 ± 6.93 | 10.62 ± 3.36 |
P < 0.05, vs tumor cells in Group B; Fisher’s test.
P < 0.05, vs tumor cells negative for MSI; Fisher’s test.
DISCUSSION
MSI is the phenotype of a profound genomic instability caused by a mutator pheno type mechanism for cancer[31,32]. Four microsatellite loci (D2S119, D2S123, D5S107 and D17S250) used in our study resemble to the loci recommended by NCI workshop[25]. The ratio of MSI-positive patients in Group A (46.2%) was significantly higher than that among randomly selected patients in Group B (10%). These results showed that MSI was an important contributor in CRC with familial predisposition. Colorectal tumors with MSI exhibit several clinicopathologic characteristics. In this study, we confirmed that the age at onset of cancer in Group A was younger than that in Group B, especially in HNPCC patients and patients with MSI-positive in Group A. We also confirmed that the ratio of proximal colon tumors, complication by extracolorectal malignancies and poorly differentiated carcinomas in patients with positive MSI were higher than those with negative MSI, and these characteristics were significant in Group A. These results suggested that it was possible and effective to identify high-risk relatives of CRC by applying pedigree survey together with MSI detection.
Leach et al[33] established the method for detecting the products of MMR gene mutation. If the MMR genes mutate, there may be a kind of methylation on promoter in transcription. And it brings the products in C-termination which block the normal expression of MMR genes. Therefore, negative for hMLH1, hMSH2 staining means potential mutation of MMR genes in tumor or normal tissues, and positive means no mutation[27,34]. In our study, we found that in Group A the incidence of negative hMLH1 staining in tumor tissues and hMLH1, hMSH2 staining in normal tissues was significantly high (Table 3). Among patients positive for MSI, ratio of negative hMLH1 together with hMSH2 staining in tumors was also higher than that among patients negative for MSI. It suggested that the rate of hMLH1, hMSH2 abnormality in CRC patients with familial predisposition increased in colorectal tumors and normal tissues. The colorectal epithelium of this group could have wide gene abnormality. And these gene changes had obviously been related with MSI.
According to the previous studies, the expressions of PCNA[35,36], DNA ploid and proliferation phases of cells[37,38] are the important markers for diagnosing and prognosticating cancers. We found that the LI of PCNA staining of tumor cells in Group A and MSI-positive group were lower than that in Group B and MSI-negative group respectively. According to Flow Cytometry, the heteroploid rate of DNA in tumor cells of Group A was lower than that of Group B. PI and S-phase percentage in tumor cells with positive MSI were also lower than those in negative MSI. It was reported that the metastasis and recurrence in colorectal cancer with MSI (due to replication error, RER) were less than in sporadic cancer. The low invasion of cancer could be explained that the mutation was too frequent and complicated to express the phenotype of metastasis and recurrence[18]. Our study showed that this low invasion was also associated with the decreased proliferation activity in colorectal cancer with familial predisposition and MSI.
Studies on MSI, MMR gene expression and proliferation kinetics showed the characteristics of colorectal cancer with famillial predisposition. According to this, the early diagnosis and warning system[39] of colorectal cancer can be established. It is important to screen high-risk relatives of colorectal cancer and diagnose colorectal cancer early[40].
Footnotes
Edited by You DY
Proofread by Ma JY
Project supported by the Natural Science Foundation of Guangdong Province, China, No.980120
References
- 1.Hong JY. Genetic polymorphism and human cancer. World J Gastroenterol. 1998;4(Suppl 2):37. [Google Scholar]
- 2.Li ZX, Jia YT, Ma ZQ. Hereditary nonpolypous colorectal cancer. Huaren Xiaohua Zazhi. 1998;6(Supp1 7):112–113. [Google Scholar]
- 3.Gao SK, Xu WH. Hereditary nonpolypous colorectal cancer. Huaren Xiaohua Zazhi. 1998;6:70–71. [Google Scholar]
- 4.Fang DC, Zhou XD. Advanced studies on microsatellite DNA instability in gastric-intestine tumor. Huaren Xiaohua Zazhi. 1998;6(Suppl 7):66–68. [Google Scholar]
- 5.Beckman JS, Weber JL. Survey of human and rat microsatellites. Genomics. 1992;12:627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- 6.Guo RF, Lu YY. Application of Microsatellite to human genome research and tumor related genes cloning. Huaren Xiaohua Zazhi. 1998;6:441–444. [Google Scholar]
- 7.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 8.Ji XL, Liu XB. Basis of molecular biology of gastrointestinal disease. Huaren Xiaohua Zazhi. 1998;6:905–910. [Google Scholar]
- 9.Zhang LL, Zhang ZS, Zhang YL, Wu BP, Guo W, Liu XX, Zhou DY. Microsatellite instability in multiple primary colorectalcancers. Shijie Huaren Xiaohua Zazhi. 1999;7:397–399. [Google Scholar]
- 10.Chen J, Gu HG, Lin WH, Luo YH. Study on microsatellite instability in 46 cases of sporadic colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:350–352. [Google Scholar]
- 11.Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 12.Zhou XD, Fang DC. Clinical implication of dinucleotide repeat sequence instability at D17S261 and D17S799 in gastric cancer. Huaren Xiaohua Zazhi. 1998;6:318–320. [Google Scholar]
- 13.Ji XL. Instability of microsatellites: new focus on gene studies. Shijie Huaren Xiaohua Zazhi. 1999;7:372–374. [Google Scholar]
- 14.Fang DC, Zhou XD, Luo YH, Wang DX, Lu R, Yang SM, Liu WW. Microsatellite instability and loss of heterozygosity ofsuppressor gene in gastric cancer. Shijie Huaren Xiaohua Zazhi. 1999;7:479–481. [Google Scholar]
- 15.Park JG. Genetic diagnosis and management of hereditary nonpolyposis colorectal cancer. World J Gastroenterol. 1998;4(Suppl 2):55. [Google Scholar]
- 16.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA, Nyström-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 17.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 18.Shibata D, Peinado MA, Ionov Y, Malkhosyan S, Perucho M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 19.Frank TS, Svoboda-Newman SM, Hsi ED. Comparison of methods for extracting DNA from formalin-fixed paraffin sections for nonisotopic PCR. Diagn Mol Pathol. 1996;5:220–224. doi: 10.1097/00019606-199609000-00012. [DOI] [PubMed] [Google Scholar]
- 20.Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 21.Li ZX, Liu PY, Xu WX, Cong B, Ma ZX, Li Y. p53 gene mutations in primary gastric cancer. China Natl J New Gastroenterol. 1996;2:41–43. [Google Scholar]
- 22.Ji CY, Smith DR, Goh HS. The role and prognostic significance of p53 mutation in colorectal carcinomas. World J Gastroenterol. 2000;6(Suppl 3):78. [Google Scholar]
- 23.Orita M, Suzuki Y, Sekiya T, Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 24.Burks RT, Kessis TD, Cho KR, Hedrick L. Microsatellite instability in endometrial carcinoma. Oncogene. 1994;9:1163–1166. [PubMed] [Google Scholar]
- 25.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 26.Xu QW, Li YS, Zhu HG. Relationship between expression p53 protein, PCNA and CEA in colorectal cancer and lymph node metastasis. World J Gastroenterol. 1998;4:218. [Google Scholar]
- 27.Thibodeau SN, French AJ, Roche PC, Cunningham JM, Tester DJ, Lindor NM, Moslein G, Baker SM, Liskay RM, Burgart LJ, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res. 1996;56:4836–4840. [PubMed] [Google Scholar]
- 28.Jia L, Chen TX, Sun JW, Na ZM, Zhang HH. Relationship between microvessel density and proliferating cell nuclear antigen and prognosis in colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2000;8:74–76. [Google Scholar]
- 29.Wang YK, Ji XL, Gu YG, Zhang SC, Xiao JH. p53 and PCNA expression in glandular dilatation of gastric mucosa. China Natl J New Gastroenterol. 1996;2:106–108. [Google Scholar]
- 30.Muta H, Noguchi M, Perucho M, Ushio K, Sugihara K, Ochiai A, Nawata H, Hirohashi S. Clinical implications of microsatellite instability in colorectal cancers. Cancer. 1996;77:265–270. doi: 10.1002/(SICI)1097-0142(19960115)77:2<265::AID-CNCR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 31.Peltomäki P, Lothe RA, Aaltonen LA, Pylkkänen L, Nyström-Lahti M, Seruca R, David L, Holm R, Ryberg D, Haugen A. Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res. 1993;53:5853–5855. [PubMed] [Google Scholar]
- 32.Xu CT, Pan BR. Gene changes in colonic cancer. Huaren Xiaohua Zazhi. 1998;6:58–60. [Google Scholar]
- 33.Leach FS, Polyak K, Burrell M, Johnson KA, Hill D, Dunlop MG, Wyllie AH, Peltomaki P, de la Chapelle A, Hamilton SR, et al. Expression of the human mismatch repair gene hMSH2 in normal and neoplastic tissues. Cancer Res. 1996;56:235–240. [PubMed] [Google Scholar]
- 34.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 35.Feng S, Song JD, Tian XR. Significance of proliferating cell nuclear antigen expression in colorectal carcinomas. Huaren Xiaohua Zazhi. 1998;6:146–147. [Google Scholar]
- 36.Zhang YL, Zhou DY. Evaluation of proliferation kinetics in large intestinal neoplasms: Compared study the expression of proliferating cell nuclear antigen (PCNA) and AgNOR counting. J Med Coll PLA. 1994;9:116–118. [Google Scholar]
- 37.Shankey TV, Rabinovitch PS, Bagwell B, Bauer KD, Duque RE, Hedley DW, Mayall BH, Wheeless L, Cox C. Guidelines for implementation of clinical DNA cytometry. International Society for Analytical Cytology. Cytometry. 1993;14:472–477. doi: 10.1002/cyto.990140503. [DOI] [PubMed] [Google Scholar]
- 38.Bauer KD, Bagwell CB, Giaretti W, Melamed M, Zarbo RJ, Witzig TE, Rabinovitch PS. Consensus review of the clinical utility of DNA flow cytometry in colorectal cancer. Cytometry. 1993;14:486–491. doi: 10.1002/cyto.990140506. [DOI] [PubMed] [Google Scholar]
- 39.Yan XJ. Establishing early diagnosis and warning system of gastric carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:96–97. [Google Scholar]
- 40.Wu BP, Zhang YL, Zhang ZS, Zhou DY, Gao CF, Guo W, Zhang LL. Study on microsatellite instability in colorectal cancerwith familial predisposition. Jiefangjun Yixue Zazhi. 1999;24:354–357. [Google Scholar]


