Abstract
APOBEC3A and APOBEC3B, cytidine deaminases of the APOBEC family, are among the main factors causing mutations in human cancers. APOBEC deaminates cytosines in single-stranded DNA (ssDNA). A fraction of the APOBEC-induced mutations occur as clusters (“kataegis”) in single-stranded DNA produced during repair of double-stranded breaks (DSBs). However, the properties of the remaining 87% of nonclustered APOBEC-induced mutations, the source and the genomic distribution of the ssDNA where they occur, are largely unknown. By analyzing genomic and exomic cancer databases, we show that >33% of dispersed APOBEC-induced mutations occur on the lagging strand during DNA replication, thus unraveling the major source of ssDNA targeted by APOBEC in cancer. Although methylated cytosine is generally more mutation-prone than nonmethylated cytosine, we report that methylation reduces the rate of APOBEC-induced mutations by a factor of roughly two. Finally, we show that in cancers with extensive APOBEC-induced mutagenesis, there is almost no increase in mutation rates in late replicating regions (contrary to other cancers). Because late-replicating regions are depleted in exons, this results in a 1.3-fold higher fraction of mutations residing within exons in such cancers. This study provides novel insight into the APOBEC-induced mutagenesis and describes the peculiarity of the mutational processes in cancers with the signature of APOBEC-induced mutations.
Carcinogenesis is associated with elevated mutation rates due to abnormal metabolic activities in the cell, disruption of repair systems, or environmental factors such as UV light, radiation, and chemical damage (Roberts and Gordenin 2014a,b). However, some normal protein enzymatic activities can also be a source of DNA damage and mutations. Recently, it was shown that some homologs of APOBEC (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like), cytidine deaminases that function as viral protecting agents as well as in RNA editing, may be a major factor causing mutations in human cancers (Nik-Zainal et al. 2012; Burns et al. 2013b; Roberts et al. 2013). Deamination of cytidine residues by APOBEC occurs in single-stranded DNA (ssDNA) (Nowarski et al. 2008; Roberts et al. 2012; Smith et al. 2012). Two members of the APOBEC family, APOBEC3A and APOBEC3B, contribute substantially to mutations in cancers (Burns et al. 2013a,b; Roberts et al. 2013; Chan et al. 2015) by deaminating cytosines in the TpC context (henceforth, the mutated nucleotide is underlined) (Nik-Zainal et al. 2012; Burns et al. 2013a,b; Roberts et al. 2013; Taylor et al. 2013; Roberts and Gordenin 2014b; Chan et al. 2015). The APOBEC cytidine deaminase converts cytosines to uracils, which usually results in C → T or C → G mutations, and much less frequently, in C → A mutations (Taylor et al. 2013). The fact that the APOBEC shows the highest specificity for the TpCpW (where W denotes A or T) context was shown in cancer genomic studies and in experimental systems (Burns et al. 2013a,b; Roberts et al. 2013; Taylor et al. 2013).
APOBEC-induced mutations are unevenly distributed along the genome. For example, under experimental conditions in yeasts, 26% of them are located in clusters spanning 6–15 kb (Taylor et al. 2013, 2014). This phenomenon, called kataegis, was described for many cancer types and is believed to be the result of APOBEC-induced mutagenesis (Nik-Zainal et al. 2012; Burns et al. 2013a,b; Roberts et al. 2013). Clustered mutations are frequently strand coordinated, i.e., are comprised of mutations in the TpC context that occur in one of the two strands (Nik-Zainal et al. 2012; Roberts et al. 2012, 2013). Although the majority of clusters carry mutations in one strand, 13% of the clusters exhibit strand switches, e.g., when the 5′ part of the cluster carries TpC coordinated mutations on the forward strand, and the 3′ part, on the reverse strand (corresponding to GpA mutations on the forward strand) (Roberts et al. 2012; Taylor et al. 2013). It was shown in cancers and in yeast experimental models that both coordinated and switching clusters are associated with DNA double-stranded breaks (DSBs) (Nik-Zainal et al. 2012; Roberts et al. 2013; Taylor et al. 2013) as a result of the activity of exonucleases causing long stretches of ssDNA near the DSB, which become a target for APOBEC enzymes (Roberts et al. 2012; Taylor et al. 2013). Alternatively, it was suggested that APOBEC enzymes can induce DSBs (Landry et al. 2011; Burns et al. 2013a). Another putative cause of kataegis is the expansion of ssDNA at the 5′ upstream region of a mismatch during base excision repair (BER) (Taylor et al. 2013; Chen et al. 2014). However, the majority of APOBEC mutations are dispersed (Nik-Zainal et al. 2012; Roberts et al. 2012, 2013; Taylor et al. 2013), and the source of ssDNA that may be a substrate for them in cancer still lacks explicit mechanistic explanation.
During replication, DNA exists for some time in a single-stranded state. Although such ssDNA should be protected by the replication protein A (RPA), it may be a substrate for APOBEC-induced deamination, especially under replication stress (Roberts et al. 2012; Roberts and Gordenin 2014a). The lagging strand is single-stranded for a longer period of time than the leading strand due to discontinuous synthesis (Okazaki et al. 1968) and is also enriched in mutations (Reijns et al. 2015). Despite that firing of individual replication origins is stochastic (Rhind et al. 2010), genomic regions vary in mean time of the replication during the S phase (Ryba et al. 2010; Pope et al. 2014) and in their propensity to be replicated unidirectionally, and the preferential fork direction is conserved among human tissues (Baker et al. 2012).
Here, we hypothesize that the APOBEC-induced mutagenesis is associated with the lagging strand. By the analysis of large genomic and exomic cancer data sets, we investigate the source of ssDNA targeted by APOBEC in cancer as well as the other APOBEC mutational properties.
Results
APOBEC mutational signatures in whole genome and whole exome cancer data sets
To study the dispersed APOBEC-induced mutations, we used 433 whole genome–sequenced (WGS) cancers from Alexandrov et al. (2013a) and 3000 whole exome–sequenced (WES) cancers from The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/). We considered only those samples that included at least 100 single-nucleotide mutations (Supplemental Tables S1, S2). Similarly to others (Roberts et al. 2013; Chan et al. 2015), we stratified tumors by the prevalence of the dispersed APOBEC mutational signature, calculated as the ratio (rapo) of the frequencies of nonclustered C → K mutations in the TpCpW and in the VpCpW contexts (where K denotes T or G; W denotes A or T; and V denotes A, C, or G). We excluded the APOBEC mutations in the TpCpS context (where S denotes C or G) from the main analysis and treated them separately. This enabled us to avoid a bias due to spontaneous deamination of 5-methylcytosines in the CpG context or to UV-light-induced mutations in the TpCpC context (Alexandrov et al. 2013a; Lawrence et al. 2013; Roberts and Gordenin 2014b).
In order to assess the efficacy of the chosen metric of enrichment of APOBEC mutations (rapo), we first compared the prevalence of this APOBEC signature with the number of kataegistic clusters, the well-documented APOBEC signature (Nik-Zainal et al. 2012; Roberts et al. 2013; Taylor et al. 2013). We observed a strong correlation both in WGS (ρ = 0.69, P < 2.2 × 10−22) (Fig. 1A) and WES (ρ = 0.37, P < 2.2 × 10−22) (Fig. 1B) data sets. Next, we asked whether the enrichment of the APOBEC signature (rapo) was dependent on the level of APOBEC3B expression. In line with other studies (Roberts et al. 2013), we observed a moderate, but significant positive correlation (ρ = 0.26, P = 3.6 × 10−10) (Fig. 1C). Moreover, in line with previous studies (Burns et al. 2013b; Roberts et al. 2013), we found that the rapo APOBEC signature is particularly prevalent in cancer types with a high expression of APOBEC3B (Fig. 1D).
Figure 1.
Properties of cancers associated with the APOBEC mutational signature (rapo). (A,B) Correlation between rapo and the number of kataegistic mutations +0.5 per tumor in WGS (A) and WES (B) data sets. (C) Correlation of rapo and the level of expression of APOBEC3B per tumor in the WES data set. (D) Box plots representing rapo per tumor type; WES data set: (ACC) adrenocortical carcinoma; (BLCA) bladder cancer; (BRCA) breast cancer; (CESC) cervical squamous cell carcinoma; (ESCA) esophageal carcinoma; (HNSC) head and neck squamous cell carcinoma; (LUAD) lung adenocarcinoma; (LUSC) lung squamous cell carcinoma; (READ) rectum adenocarcinoma; (SKCM) cutaneous melanoma. (E) Distributions of rapo in WGS and WES data sets. Tumors with rapo < 1 were categorized as APOpoor, and tumors with rapo > 5, as APOrich. (F) Enrichment of dispersed and clustered APOBEC signature mutations in the proximity of rearrangement breakpoints (used as a proxy for DSBs) in the WGS data set. The observed mutation rates in the proximity (up to 100 kb) of rearrangement breakpoints were compared with the average mutation rates per tumor in APOrich tumors.
We selected a subset of tumors with a strong APOBEC signature (APOrich), in which >80% of the C → K mutations in the TpCpW context are associated with APOBEC (rapo > 5). We also selected a subset of tumors with a low prevalence of APOBEC mutations (APOpoor), in which the frequency of C → K mutations in the TpCpW context is lower than in the VpCpW context (rapo < 1) as a control data set (Fig. 1E). This yielded 23 APOrich and 167 APOpoor tumors in the WGS data set and 587 APOrich and 904 APOpoor tumors in the WES data set. APOrich tumors from the WGS data set harbored 168,326 APOBEC mutations, including 146,600 dispersed and 21,726 clustered mutations. To confirm that this definition of APOrich cancers is robust, we asked how well it matches the set of cancers with APOBEC signatures deciphered by computational methods (http://cancer.sanger.ac.uk/cosmic/signatures) (Alexandrov et al. 2013a,b, 2015). In all 23 APOrich cancers in the WGS data set, and in 563 out of 585 cancers in the WES data set, the 2 + 13 (APOBEC) signatures comprise at least one-third of all mutations and were more than five times more prevalent than a combination of the 7 + 10 + 11 + 19 + 23 + 30 (confounding non-APOBEC) signatures. In contrast, none of the 167 WGS APOpoor cancers and only one of the 683 WES APOpoor cancers met this criterion, indicating that our definitions of APOrich and APOpoor cancers are robust.
Dispersed APOBEC-induced mutations are less associated with double-stranded breaks than clustered mutations
First, we asked whether dispersed APOBEC-induced mutations occur during the repair of double-stranded breaks (DSBs), as previously identified for clustered APOBEC-induced mutations (Nik-Zainal et al. 2012; Roberts et al. 2012, 2013; Taylor et al. 2013). To investigate this, we calculated the enrichment of APOBEC-induced clustered and dispersed mutations near DNA rearrangement breakpoints, used as a proxy for the locations of DSBs (Fig. 1F). We categorized APOBEC clusters by strand colocalization of mutations into strand-coordinated clusters, clusters with one strand switch, and short or noncoordinated clusters (details in Methods). In line with the previous observations (Nik-Zainal et al. 2012; Roberts et al. 2012, 2013; Taylor et al. 2013), mutations in strand-coordinated clusters and clusters with strand switches were highly enriched within 10 kb from breakpoints (∼80-fold enrichment). In contrast, the enrichment of dispersed APOBEC mutations near DSBs was 11.4 times weaker (sevenfold enrichment) (Fig. 1F). Mutations in noncoordinated clusters were also less enriched near DSBs (20-fold enrichment) than mutations in other types of clusters.
APOBEC mutations are at least two times more frequent on the lagging DNA strand
We next estimated the preferential direction of the replication fork from the replication timing (RT) data (Koren et al. 2012). The RT is highly conserved between human tissues and cell types (Ryba et al. 2010; Pope et al. 2014); therefore, we utilized the RT data from one cell type (lymphoblastoid cell line) to different cancer types. To validate this approach, we used data on replication timing for five different cell types from the ENCODE Project Consortium (2012) (https://www.encodeproject.org/search/?type=Experiment&assay_term_name=Repli-seq&limit=all) and estimated the correlations between RT for these cell types. In all comparisons, correlation coefficients exceeded 0.7 (Supplemental Table 3), while using the cell type with the lowest correlation, HeLa, as the source of information on RT still produced the same results (see below).
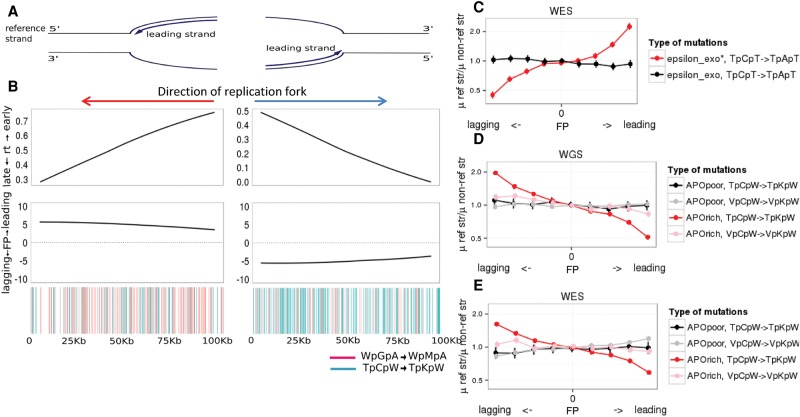
We reconstructed replication fork polarity (FP) as the derivative of the RT (Chen et al. 2011; Baker et al. 2012) and predicted for each genomic region whether the reference strand is replicated more frequently as leading (FP > 0) or lagging (FP < 0) (Fig. 2A,B). The FP values reflect the ratio between the frequencies of passages of the replication fork in forward and reverse directions (Chen et al. 2011; Baker et al. 2012).
Figure 2.
APOBEC mutational signature as a function of fork polarity in cancer genomes. (A) Diagram explaining the correspondence between the reference strand being replicated as leading or lagging and the direction of the replication fork. (B) Characteristics of the 50 genomic regions of lengths of 100 kb each with the highest (left) and lowest (right) FP on WGS data set for APOrich tumors. Horizontal axis: coordinates within the considered genomic regions. (Upper) average of the replication times (RT) across these 50 regions; (middle) fork polarity values reconstructed as derivatives of RT represented in the upper panel; FP > 0 corresponds to the leading strand, and FP < 0 corresponds to the lagging strand; (bottom) color-coded APOBEC signature mutations on reference strand within these regions. Each vertical line corresponds to a mutation. (C,D,E) The ratio of the mutation rates of the considered mutation type to its reverse compliment on the reference strand as a function of the propensity of the replication fork to replicate the reference strand as lagging or leading. Horizontal axis: genome split by FP values into nine bins from low FP (bin 1) to high FP (bin 9). Vertical bars represent 95% confidence intervals. (C) Pol ε_exo* produces mutations on the leading strand; APOBEC causes mutations on the lagging strand in WGS (D) and WES (E) data sets.
In order to validate this approach for cancer data sets, we first tested the FP metric using polymerase epsilon (pol ε). This DNA polymerase specifically duplicates the leading strand during DNA replication (Shinbrot et al. 2014). Somatic mutations in the proofreading exonuclease domain of pol ε cause an extremely high rate of TpCpT → TpApT mutations that occur during replication specifically on the leading strand (Shinbrot et al. 2014). We took advantage of the set of tumor genomes (Shinbrot et al. 2014) with such somatic mutations in pol ε and investigated the distribution of the TpCpT → TpApT mutations as a function of FP. If our approach was able to correctly predict the preferential fork direction, we expected to observe the enrichment of TpCpT → TpApT mutations on the leading strand in tumors with the somatic mutations in pol ε. Indeed, we have found a 2.25-fold (P < 2.2 × 10−16) enrichment of the TpCpT → TpApT mutations on the leading strand in the 10% of genomic regions with the highest FP. No such enrichment was observed in cancers with unaffected pol ε (Fig. 2C).
Having confirmed that our FP statistic accurately reflects the propensity of the reference strand to be replicated as leading or lagging in cancer genomes, we set out to study its relationship with the APOBEC signature. In APOrich tumors from the WGS data set, the fraction of dispersed APOBEC mutations that occurred on the reference strand grows monotonically with FP. In the 10% of the genome with the highest FP, APOBEC mutations were approximately twofold less abundant on the reference strand than on the nonreference stand (P < 2.2 × 10−16). Similarly, in the 10% of the genome with the lowest (negative) FP, APOBEC mutations were approximately twofold more abundant on the reference strand than on the nonreference stand (P < 2.2 × 10−16, χ2 test) (Fig. 2D). This indicates that 33% of APOBEC signature mutations genome-wide occur on the lagging strand during DNA replication. This estimate is conservative, as even in the regions with the highest and the lowest FP values, the replication fork is not strictly unidirectional (Baker et al. 2012). A similar profile was observed in the WES data set (e.g., a 1.69-fold depletion for the top 10% FP, and 1.62-fold enrichment for the bottom 10% FP) (Fig. 2E) and in tumors with increased APOBEC expression (Supplemental Fig. 1A). In order to control for the potential effect of the transcribed strand in genes, we replicated the analyses only on intergenic regions of the WGS data set and obtained similar results (Supplemental Fig. 1B). Moreover, the results were also very similar when HeLa cells were used to estimate FP (Supplemental Fig. 1C), confirming that tissue specificity of replication fork direction does not affect our results.
We next investigated how the bias of APOBEC signature mutations in the TpCpW context toward the lagging strand depends on the prevalence of rapo signature in the tumors. As expected, the strand bias of this mutation type in the 10% of the genome with the lowest FP monotonically increases with rapo (Supplemental Fig. 1D,F), confirming involvement of APOBEC in the observed patterns. The group of APOBEC signature mutations in the TpCpW context in APOrich tumors was the only one to reveal a strong strand bias (Fig. 2D,E). A marginal association with FP was also observed for mutations in the VpCpW context in APOrich (1.18-fold, P = 0.0039) but not in APOpoor cancers (0.97-fold, P = 0.24) (Fig. 2D), implying that some of these mutations may also be induced by APOBEC.
APOBEC-dependent and APOBEC-independent mutation rates in APOrich tumors are moderately dependent on replication timing
Point mutation rates in cancer are increased in the genomic loci that are replicated during the late S phase compared to earlier-replicating regions (Lawrence et al. 2013; Supek and Lehner 2015). We therefore asked whether APOBEC mutations are also enriched in regions of late RT. APOpoor tumors exhibited a 3.11-fold increase of mutation rates in late RT regions (Fig. 3A), consistent with previous observations (Lawrence et al. 2013). In contrast, in APOrich tumors, we observed only a very moderate (on average, 1.09-fold) increase of the rates of APOBEC-induced mutations in late RT. In some of these tumors, the APOBEC mutations rate was, in fact, decreased in late RT (Supplemental Table 1).
Figure 3.
Mutations in early versus late replicating regions. Horizontal axis: genome split into 10 bins of equal size from early to late RT. (A) Mutation rates as a function of replication time (RT), relative to the mutation rate in the early RT bin. (B) Percentage of mutations in exons that fall into this bin (among all mutations in the genome) as a function of RT. Three percent of mutations in APOrich tumors and only 2.3% of mutations in APOpoor tumors occur in exons. Vertical bars represent 95% confidence interval.
In order to investigate if this difference is specific to APOBEC mutations, we performed a similar comparison for non-APOBEC mutations: T → V and VpCpW → VpKpW. Unexpectedly, we observed the same trend for these types of mutations as for APOBEC mutations: a moderate (1.32- and 1.34-fold, respectively) increase of mutation rates with RT in APOrich tumors versus a strong (3.71- and 2.27-fold, respectively) increase in APOpoor tumors (Fig. 3A; Supplemental Fig. 3). Some of the mutations in cancers with deficiencies in the mismatch repair (MMR) are nearly independent of RT (Supek and Lehner 2015). However, the rates of TpCpN → TpApN mutations depend on RT even in MMR-deficient cancers (Supek and Lehner 2015), and these mutations are also moderately dependent on RT in APOrich cancers (Supplemental Fig. 2). This suggests that the lack of dependence on RT of mutations in APOrich tumors is not related to MMR deficiency and represents a novel unexplained phenomenon. The fact that both APOBEC-induced and other mutations are nearly independent of RT in APOrich tumors suggests that this lack of dependence is not specific to APOBEC-related mechanisms, but rather is a consequence of another perturbation of the mutational process in APOrich tumors.
Protein-coding genes are preferentially located in early RT regions (Supplemental Fig. 3), which are less exposed to mutagenesis than late RT regions (Farkash-Amar et al. 2008). As a result, in APOrich tumors, where mutation rates are similar between early and late RT, the fraction of mutations that occurred in coding exons is 1.27-fold higher than in APOpoor tumors (Fig. 3B). Therefore, the increased probability of occurrence of functional mutations in cancer driver genes in APOrich cancers (Roberts et al. 2013; Henderson and Fenton 2015) is associated not only with the higher rate of mutations, but also with a higher fraction of genic mutations among them.
TpCpS → TpKpS and TpCpN → TpApN mutations are likely associated with APOBEC activity
C → K mutations in the TpCpW context are considered to be the canonical APOBEC mutational signature. Nevertheless, we also investigated the propensity of APOBEC for mutating cytosines in other contexts (Fig. 4A–D; Supplemental Table 4; Burns et al. 2013a; Taylor et al. 2013). Assuming that APOBEC context preferences in cancers are independent of the extent of APOBEC mutational activity, we searched for additional signatures. For that, we defined measures analogous to rapo and strength of the lagging strand mutational bias for noncanonical mutational contexts (see Methods).
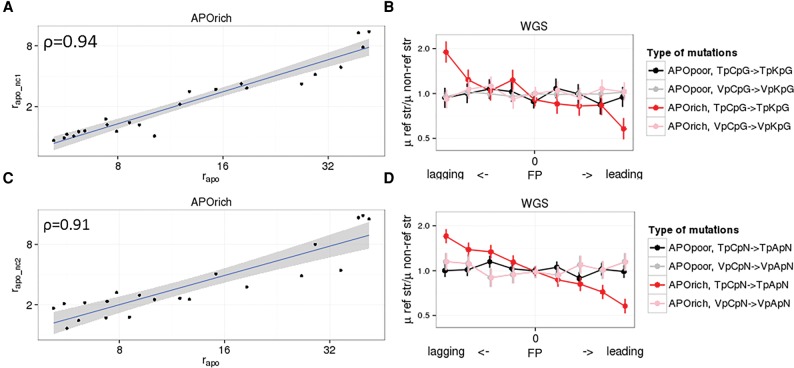
Figure 4.
Noncanonical mutation signatures of APOBEC3. Properties of TpCpS → TpKpS (noncanonical mutation type 1, nc1) (A,B) and TpCpN → TpApN (noncanonical mutation type 2, nc2) (C,D) induced by APOBEC3. (A,C) Correlations of enrichments of noncanonical (rapo_nc1 and rapo_nc2) and canonical rapo APOBEC3-induced mutations across tumors. (B,D) The ratio of the mutation rates of the considered mutation type to its reverse compliment on the reference strand as a function of the propensity of the replication fork to replicate the reference strand as lagging or leading. Horizontal axis in C and D: genome split by FP values into nine bins from low (bin 1) to high (bin 9). Vertical bars represent 95% confidence interval. Analysis performed using WGS data set.
Specifically, we considered the remaining mutations in the TpC contexts: C → K mutations in the TpCpS context, and C → A mutations in the TpCpN context (Supplemental Table 3). For the analysis of C → A mutations, we excluded lung cancers due to the known high prevalence of APOBEC-independent C → A mutations in them (Alexandrov et al. 2013a; Lawrence et al. 2013; Roberts and Gordenin 2014b). We found very strong correlations for APOrich samples (WGS data set) between rapo and enrichments of mutations estimated analogically to rapo for TpCpS → TpKpS (rapo_nc1) (ρ = 0.94, P = 3.2 × 10−6) (Fig. 4A) and TpCpN → TpApN mutations (rapo_nc2) (ρ = 0.91, P = 3.9 × 10−6) (Fig. 4C). Moreover, for these mutations, we also observed a strong lagging strand bias of 1.8- and 1.7-fold, respectively (Fig. 4B,D). These analyses confirm a high prevalence of APOBEC-induced TpCpS → TpKpS and TpCpN → TpApN mutations. The mean values of rapo_nc1 and rapo_nc2 across all APOrich cancers were 3.12 and 4.28, implying that 68% and 77% of respective mutations are associated with APOBEC compared to 95% of TpCpW → TpKpW mutations. The rates of TpCpS → TpKpS mutations caused by APOBEC were 5.34 lower than the rates of TpCpW → TpKpW mutations, resulting in 12.38 times fewer such mutations per genome. The rates of TpCpN→TpApN mutations caused by APOBEC were 16.27 lower than the rates of TpCpW → TpKpW mutations, resulting in 8.73 times fewer such mutations per genome. For comparison, in yeast experiments, APOBEC caused C → A mutations at a rate 1/40 that of C → K mutations (Taylor et al. 2013). Finally, based on the analysis of lagging strand bias, we observed that at least 9% of VpCpW → VpKpW mutations may also be caused by APOBEC (Supplemental Table 4).
APOBEC preferentially targets nonmethylated cytosines
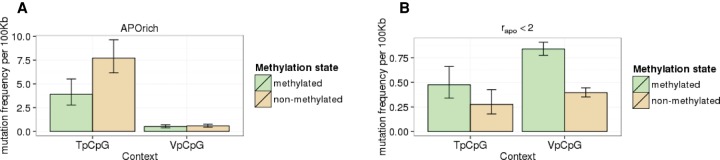
In order to better understand the action of APOBEC in the TpCpS noncanonical context, and specifically, the controversial role of cytosine methylation (Nabel et al. 2012; Caval et al. 2014), we investigated the preferences of APOBEC for 5-methylcytosines (5mC) versus nonmethylated cytosines by using the data on methylation level from Meissner et al. (2008). In APOrich tumors, we observed an approximately twofold lower mutation rate at 5mCs, compared to nonmethylated cytosines, at the TpCpG context (P = 1.4 × 10−4), but not in the VpCpG context (P = 0.64) (Fig. 5A). This is in stark contrast to tumors of the same cancer type with rapo < 2, where a 1.5- to twofold increase of mutation rates in methylated cytosines, both in TpCpG (P = 0.069) and VpCpG contexts (P < 2.2 × 10−16), was observed (Fig. 5B). These data suggest a strong preference of APOBEC for nonmethylated cytosines in the TpCpG context in cancer. The observation that the fraction of mutations in nonmethylated cytosines in the VpCpG context is higher in APOrich tumors than in APOpoor tumors is in line with the residual APOBEC-induced mutagenesis in this context (Burns et al. 2013a; Taylor et al. 2013) and with the weak lagging strand bias specific to APOrich tumors (Fig. 2D, pink curve).
Figure 5.
Mutation rates of 5-methylcytosines versus nonmethylated cytosines at CpG sites. (A) APOrich tumors. (B) Tumors with rapo < 2. Methylation (Meissner et al. 2008) and mutation (WGS) data sets are tissue matched. Vertical bars represent 95% confidence intervals.
Discussion
APOBEC-dependent mutagenesis is a frequent phenomenon in cancer. Of TCGA samples, 18% have a strong APOBEC signature; and in some tumor types, such as bladder cancer, the fraction of APOrich tumors may reach 37% (Nordentoft et al. 2014). In some samples, up to 70% of all mutations are associated with the APOBEC mutational signature (Supplemental Table 1), confirming that APOBEC mutagenesis may be an important factor for cancer progression and tissue transformation (de Bruin et al. 2014; Henderson et al. 2014; McGranahan et al. 2015). Recently, it was shown that the expression level of APOBEC3B has prognostic potential, and APOBEC was suggested as a target for oncotherapy (Cescon et al. 2015).
DSB repair is the most widely discussed source of ssDNA targeted by APOBEC in cancer (Taylor et al. 2013; Nordentoft et al. 2014; Henderson and Fenton 2015). Our study uncovers the main source of ssDNA substrate of APOBEC, which is related not to DSBs but to DNA replication. There is some evidence that APOBEC may specifically mutate the lagging strand under replication stress in yeast (Roberts et al. 2012). Knowledge about the origin of ssDNA in cancer cells can expand our understanding of mutational processes in cancer and can be medically relevant. Moreover, knowledge about the propensity of regions of DNA strands to be replicated as leading or lagging can be widely utilized for the studies of cancer genomes. For example, here we have used it to show a strong bias of mutations produced by pol ε with damaged proofreading exonuclease domain toward the leading strand. We suggest that our method would be useful for detection of other strand-specific mutational signatures and mechanisms.
In theory, selection could confound inference of mutational patterns. Despite the presence of positive selection in a subset of cancer-related genes (Woo and Li 2012), the vast majority of exonic mutations in cancer are not subject to selection (McFarland et al. 2014), and therefore are unlikely to affect our results. Moreover, in the WGS data set, our results were completely reproduced even when all genes were excluded (Supplemental Fig. 1B). Our conclusions likely concern many different cancer types, as the APOrich WES data set comprises many types of cancers (Supplemental Table 2).
The rates of germline mutations are known to be increased in late RT (Stamatoyannopoulos et al. 2009). A recent study of de novo mutations has uncovered that this pattern is weaker for mutations that originated in older fathers, resulting in a higher fraction of mutations in exonic regions (Francioli et al. 2015). In line with these observations, we show that in APOrich cancers, the fraction of mutations that fall into exons is increased (Fig. 3B). This difference is even stronger after exclusion of CpG mutations (1.27-fold for all mutations versus 1.34-fold for non-CpG mutations, P = 0.016) (Supplemental Fig. 4). The association between the rate of both TpCpW → TpKpW and non-APOBEC signature mutations and RT is stronger in APOpoor than in APOrich cancers (Fig. 3A; Supplemental Fig. 2), suggesting that the prevalence of APOBEC-induced mutations is a marker of perturbations of the mutational processes. These perturbations may be associated with the frequent fork uncoupling or impairment of certain replication mechanisms in APOrich tumors. Enrichment of mutations in early RT should also increase the probability of acquisition of driver mutations and therefore, is directly related to cancer progression (Lawrence et al. 2013).
In summary, our data suggest that the dispersed mutations with the APOBEC signature are not explained by DSBs. In contrast, a high fraction of them arise in the ssDNA produced during replication, particularly, in the lagging strand. We also show that the mutation rates in APOrich tumors are almost independent of RT, suggesting disruption of a range of DNA protection mechanisms in these cancer samples. This difference leads to a higher fraction of genic mutations in APOrich tumors (which comprise 18% of all TCGA cancers), likely affecting the functional consequences of APOBEC-related mutagenesis.
Methods
Data sets and APOBEC signature
Somatic point mutations from 433 whole cancer genomes were obtained (Supplemental Table 1; Alexandrov et al. 2013a). Three thousand whole cancer exomes with more than 100 mutations per sample were obtained from TCGA (https://tcga-data.nci.nih.gov/tcga/) (Supplemental Table 2). All regions annotated as repeats by RepeatMasker were excluded.
Similar to other studies (Burns et al. 2013a,b; Roberts et al. 2013), we considered TpCpW → TpKpW mutations as the APOBEC signature. As a measure of the prevalence of APOBEC signature mutations (rapo), we used the ratio of rates of C → K mutations in the TpCpW context to C → K mutations in the VpCpW context
which is similar to the metric used in Roberts et al. (2013). For the comparative analyses, we selected the APOBEC-rich tumors with rapo > 5, and APOBEC-poor tumors with rapo < 1 (Supplemental Tables 1, 2). The fraction of APOBEC-induced mutations was calculated as (rapo−1)/rapo. Similarly, the prevalence of noncanonical signature mutations was defined as
and
In order to validate that the rapo signature is specific to APOBEC mutations, we utilized the Non-Negative Matrix Factorization (NMF) method (Alexandrov et al. 2013b). This analysis was applied to all 23 WGS APOrich and to 167 WGS APOpoor cancers, to 585 of the 587 WES APOrich cancers, and to 683 WES APOpoor cancers; the remaining two WES APOrich cancers and 221 WES APOpoor cancers did not have any signatures detected by NMF.
Clusters of APOBEC-induced mutations
Kataegistic clusters were defined as at least three APOBEC signature mutations with distance between adjacent mutations not exceeding 20 kb; mutations not falling into any cluster were considered dispersed. A cluster was categorized as strand coordinated if it carried at least four mutations, and all mutations occurred in the same orientation (TpCpW or WpGpA); switched if it carried at least two mutations in one orientation and two adjacent mutations in the other orientation; and noncoordinated otherwise.
Double-strand breaks
To investigate the relationship between double-strand breaks (DSB) and clusters of different types as well as dispersed mutations, we used data on rearrangements (Nik-Zainal et al. 2012) and considered rearrangement breakpoints as a proxy for DSBs. We used a relaxed criterion for APOBEC signature enrichment in this analysis (rapo > 2), yielding 13 of the 21 cancers represented in Nik-Zainal et al. (2012); using rapo > 5 as a criterion would yield only three cancers.
Fractions of different types of APOBEC mutations were calculated for regions with lengths between 20 and 200 kb (in 2 kb increments) centered at the rearrangement breakpoints. The enrichments of APOBEC clusters and dispersed mutations near DSBs were calculated as their frequencies in the windows near DSBs divided by the genome average frequencies in the corresponding category.
Estimates of polarity of replication fork
Replication timing (RT) was obtained from Koren et al. (2012). Fork polarity (FP) for a genomic coordinate was calculated as replication timing (RT) increment between points 5 kb upstream of and 5 kb downstream from the coordinate (Supplemental data). To ensure that this estimation is robust, we recalculated FP from RT at distances ranging between 0.5 and 15 kb from the current coordinate; all resulting values were strongly correlated (Spearman's ρ > 0.995). Absolute values of RT change reflect the propensity of FP toward unidirectionality (Baker et al. 2012). We divided the genome into nine bins according to the values of FP: eight bins each containing 10% of all nucleotides, and one bin centered at FP = 0, containing 20% of all nucleotides. For each bin, we then measured the rates of selected mutation types on the reference and nonreference strands. To verify conservation of RT between cell types, we downloaded the RT track for five more cell types (https://www.encodeproject.org/search/?type=Experiment&assay_term_name=Repli-seq&limit=all) and measured the correlations of the RT values in 10-kb windows between each of the five tracks and the track from the lymphoblastoid cell line that was used in our analyses (Supplemental Table 4). The fraction of APOBEC-induced mutations was calculated as the ratio of APOBEC-induced mutations on the lagging strand to those on the leading strand in the ninth bin (y-axis) (Fig. 2C,D,E). The ratio of C → K mutations in the TpCpW context and their reverse compliment on the lagging strand is
where x is the fraction of the APOBEC-induced mutations that specifically occurred on the lagging strand. Therefore, sb = 2 corresponds to x = 0.33.
For the analysis of the relationship between the RT and the mutation rates, we categorized the genome into 10 equal bins by RT values. The TpCpN → TpApN mutation were considered as MMR-independent (Supek and Lehner 2015).
Stratification of the cancer genomes based on expression and methylation
APOBEC3B expression normalized by TBP expression for TCGA cancer samples was obtained from Roberts et al. (2013). The subset of cancers with high APOBEC3B expression was defined as the 20% of cancers with the highest APOBEC3B/TBP expression ratios, and the subset with low APOBEC3B expression was defined as the 50% of cancers with the lowest ratio.
Cytosine methylation levels for noncancer tissues were obtained from http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeHaibMethylRrbs (Meissner et al. 2008). Because all APOrich WGS tumors were from lung, breast, and liver cancer types, tissue-matched comparisons of the methylation and mutational profiles were performed only for these three tissues. We merged biological replicates of methylation analyses and considered only sites with at least 10 reads. To maximize the statistical power of our tests, we divided TpCpG sites into two equal subsets depending on the fraction of methylated reads, resulting in the methylation threshold of 0.013. VpCpG sites were divided analogously, resulting in the methylation threshold of 0.037. For APOpoor cancers, we used a relaxed threshold of rapo < 2 due to the low number of mutations at sites with a known level of methylation.
To map genic and exonic regions, we used KnownGene annotation (UCSC Genome Browser).
Supplementary Material
Acknowledgments
We thank Dmitry Gordenin for useful discussions; Nadezhda Terekhanova for data preprocessing; Maria Andrianova for help in figure preparation; Federico Santoni for NMF analysis; and Ludmil Alexandrov for providing the NMF mutational signatures. This work was performed in IITP RAS and supported by the Russian Science Support Foundation (grant no. 14-50-00150). S.I.N. was supported by the Swiss Cancer League (LSCC 2939-03-2012) and Dinu Lipatti grants.
Author contributions: V.B.S. and S.I.N. designed the project; V.B.S. performed the main analyses; R.A.S. calculated FP and helped to analyze the data; K.Y.P. helped in data preprocessing; V.B.S., R.A.S., S.E.A., G.A.B., and S.I.N. wrote the manuscript; and S.I.N. retrieved and prepared the data.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.197046.115.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. 2013a. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. 2013b. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, Stratton MR. 2015. Clock-like mutational processes in human somatic cells. Nat Genet 47: 1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A, Audit B, Chen CL, Moindrot B, Leleu A, Guilbaud G, Rappailles A, Vaillant C, Goldar A, Mongelard F, et al. 2012. Replication fork polarity gradients revealed by megabase-sized U-shaped replication timing domains in human cell lines. PLoS Comput Biol 8: e1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. 2013a. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494: 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. 2013b. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caval V, Suspène R, Shapira M, Vartanian JP, Wain-Hobson S. 2014. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3′UTR enhances chromosomal DNA damage. Nat Commun 5: 5129. [DOI] [PubMed] [Google Scholar]
- Cescon DW, Haibe-Kains B, Mak TW. 2015. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci 112: 2841–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, et al. 2015. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 47: 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Duquenne L, Audit B, Guilbaud G, Rappailles A, Baker A, Huvet M, d'Aubenton-Carafa Y, Hyrien O, Arneodo A, et al. 2011. Replication-associated mutational asymmetry in the human genome. Mol Biol Evol 28: 2327–2337. [DOI] [PubMed] [Google Scholar]
- Chen J, Miller BF, Furano AV. 2014. Repair of naturally occurring mismatches can induce mutations in flanking DNA. eLife 3: e02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. 2014. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I. 2008. Global organization of replication time zones of the mouse genome. Genome Res 18: 1562–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I; Genome of the Netherlands Consortium, van Duijn CM, Swertz M, Wijmenga C, et al. 2015. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet 47: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S, Fenton T. 2015. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med 21: 274–284. [DOI] [PubMed] [Google Scholar]
- Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. 2014. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 7: 1833–1841. [DOI] [PubMed] [Google Scholar]
- Koren A, Polak P, Nemesh J, Michaelson JJ, Sebat J, Sunyaev SR, McCarroll SA. 2012. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet 91: 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Narvaiza I, Linfesty DC, Weitzman MD. 2011. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep 12: 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland CD, Mirny LA, Korolev KS. 2014. Tug-of-war between driver and passenger mutations in cancer and other adaptive processes. Proc Natl Acad Sci 111: 15138–15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. 2015. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 7: 283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. 2012. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol 8: 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149: 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordentoft I, Lamy P, Birkenkamp-Demtröder K, Shumansky K, Vang S, Hornshøj H, Juul M, Villesen P, Hedegaard J, Roth A, et al. 2014. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep 7: 1649–1663. [DOI] [PubMed] [Google Scholar]
- Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. 2008. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat Struct Mol Biol 15: 1059–1066. [DOI] [PubMed] [Google Scholar]
- Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. 1968. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci 59: 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Kemp H, Ding J, de Procé SM, Jackson AP, Taylor MS. 2015. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Yang SC, Bechhoefer J. 2010. Reconciling stochastic origin firing with defined replication timing. Chromosome Res 18: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Gordenin DA. 2014a. Clustered and genome-wide transient mutagenesis in human cancers: hypermutation without permanent mutators or loss of fitness. Bioessays. 10.1002/bies.201300140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Gordenin DA. 2014b. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer 14: 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 46: 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinbrot E, Henninger EE, Weinhold N, Covington KR, Göksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, et al. 2014. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res 24: 1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. 2012. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol 23: 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. 2009. Human mutation rate associated with DNA replication timing. Nat Genet 41: 393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Lehner B. 2015. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 521: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. 2013. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2: e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Wu YL, Rada C. 2014. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. eLife 3: e03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YH, Li WH. 2012. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun 3: 1004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.