Abstract
Cis-regulatory elements (CREs, e.g., promoters and enhancers) regulate gene expression, and variants within CREs can modulate disease risk. Next-generation sequencing has enabled the rapid generation of genomic data that predict the locations of CREs, but a bottleneck lies in functionally interpreting these data. To address this issue, massively parallel reporter assays (MPRAs) have emerged, in which barcoded reporter libraries are introduced into cells, and the resulting barcoded transcripts are quantified by next-generation sequencing. Thus far, MPRAs have been largely restricted to assaying short CREs in a limited repertoire of cultured cell types. Here, we present two advances that extend the biological relevance and applicability of MPRAs. First, we adapt exome capture technology to instead capture candidate CREs, thereby tiling across the targeted regions and markedly increasing the length of CREs that can be readily assayed. Second, we package the library into adeno-associated virus (AAV), thereby allowing delivery to target organs in vivo. As a proof of concept, we introduce a capture library of about 46,000 constructs, corresponding to roughly 3500 DNase I hypersensitive (DHS) sites, into the mouse retina by ex vivo plasmid electroporation and into the mouse cerebral cortex by in vivo AAV injection. We demonstrate tissue-specific cis-regulatory activity of DHSs and provide examples of high-resolution truncation mutation analysis for multiplex parsing of CREs. Our approach should enable massively parallel functional analysis of a wide range of CREs in any organ or species that can be infected by AAV, such as nonhuman primates and human stem cell–derived organoids.
Cis-regulatory elements (CREs, e.g., promoters and enhancers) are DNA regions that regulate gene expression, and variants within CREs can contribute to phenotypic diversity, including disease susceptibility (Wray 2007; Albert and Kruglyak 2015). In the past several years, vast amounts of genomic data have been generated that predict the locations of hundreds of thousands of CREs in cell lines and primary tissues (The ENCODE Project Consortium 2012; Shen et al. 2012; Romanoski et al. 2015). As an avenue for the experimental validation of these predictions, massively parallel reporter assays (MPRAs, e.g., CRE-seq) have been developed, in which barcoded plasmid reporters are introduced into cells. Next-generation sequencing of the resulting barcoded transcripts provides a quantitative measure of CRE activity (Kwasnieski et al. 2012; Melnikov et al. 2012; Patwardhan et al. 2012; Arnold et al. 2013; White et al. 2013; Levo and Segal 2014; Shlyueva et al. 2014). Thus far, MPRAs have been largely restricted to assaying short CRE fragments (<150 bp) synthesized as oligonucleotide libraries on microarrays (Patwardhan et al. 2009; Baker 2011; White et al. 2013) and delivered into select mammalian cells accessible by transfection or electroporation. However, CREs are often hundreds of base pairs in length, and CRE activity depends crucially on the assayed cell type and its particular complement of transcription factors (TFs) (Davidson 2001). Therefore, we sought to expand the biological relevance and applicability of MPRAs by increasing the length of assayed CREs and by widening the repertoire of assayable cell types.
The retina and cerebral cortex are two parts of the central nervous system (CNS) with a shared forebrain origin, whose gene regulatory networks are topics of intense research interest (Swaroop et al. 2010; Wright et al. 2010; Bae et al. 2015; Nord et al. 2015). The genome-wide locations of putative CREs have been mapped in both tissues, using methods such as ChIP-seq and DNase-seq (Visel et al. 2009; Corbo et al. 2010; The ENCODE Project Consortium 2012; Wilken et al. 2015). Compared to the cortex, the retina is more experimentally amenable to cis-regulatory analysis, in part because its cellular composition is more completely understood (Livesey and Cepko 2001; London et al. 2013). Electroporation can be used to efficiently deliver plasmid DNA into rod photoreceptors, which constitute the majority (∼80%) of the cells in the retina (Jeon et al. 1998). We previously conducted CRE-seq by electroporating thousands of short CREs into the neonatal mouse retina ex vivo (Kwasnieski et al. 2012; White et al. 2013). Although hundreds of putative developmental forebrain enhancers have been assayed with one-at-a-time transgenic mouse reporter assays (Nord et al. 2013; Visel et al. 2013), never before has massively parallel cis-regulatory analysis been conducted in the mammalian CNS in vivo.
Here, we sought to overcome current technological hurdles by developing a “capture-and-clone” approach for synthesizing CRE-seq libraries with a selectable range of fragment sizes for targeted cis-regulome analysis. As a built-in feature, our approach allows for truncation mutation analyses, which can identify regions within CREs that are critical for activity. We furthermore demonstrate the feasibility of conducting in vivo CRE-seq in the adult cerebral cortex by AAV-mediated delivery. Our approach provides a framework for the massively parallel functional analysis of CREs in a broad repertoire of organs and species in vivo.
Results
Identification and characterization of candidate CRE regions
The genomic locations of CREs can be predicted by the patterns of phylogenetic conservation, the occurrence of transcription factor binding sites, and the presence of various chromatin features (Levo and Segal 2014; Shlyueva et al. 2014). DNase I hypersensitive (DHS) sites, which demarcate regions of open chromatin, are one of the most informative predictive features of active CREs (Arvey et al. 2012; Natarajan et al. 2012; Kwasnieski et al. 2014). Moreover, DNase-seq data for a variety of primary mouse tissues are available as part of the Mouse ENCODE Project (Yue et al. 2014). To facilitate the direct comparison of a given CRE-seq library in retina and cerebral cortex, we generated a list of tissue-specific candidate CREs based on mouse DNase-seq data, corresponding to 1000 DHS regions from adult retina and 1000 DHS regions from adult whole brain. Additionally, we included DHSs from two adult mouse non-neural tissues (1000 DHSs from heart and 1000 DHSs from liver) as controls (Supplemental Table S1). Together, this yielded 4000 target DHS regions.
We first examined the genome-wide distributions of the 4000 target DHS regions using GREAT and HOMER, two computational tools for annotating coding and noncoding regions (Heinz et al. 2010; McLean et al. 2010). The majority (75%) of the DHS regions were distal elements located >10 kb away from the nearest transcriptional start site (TSS) (Supplemental Fig. S1A). Almost all of the DHS regions fell within introns (46%) or intergenic regions (45%) (Supplemental Fig. S1B), similar to the genome-wide distribution of DHS regions in other cell types (Shu et al. 2011). A small number of DHSs (156/4000 or 4%) were “promoter-proximal,” i.e., falling within −1 kb to +100 bp relative to the nearest TSS (Supplemental Fig. S1A). Among these, 77/156 (49%) were retinal DHSs, consistent with the previous observation that photoreceptor CREs often cluster around TSSs (Corbo et al. 2010).
Tissue-specific CREs are often enriched for the binding of TFs important for cell identity and function (Davidson 2001). Accordingly, we used HOMER (Heinz et al. 2010) to quantify enrichment of TF motifs in the target regions (Supplemental Table S2). For each set of tissue-specific target DHSs, we found strong enrichment of putative binding sites for TFs known to be important in that tissue. For example, among the top statistically significant enrichments for the retina, brain, heart, and liver DHSs were putative motifs for CRX (Chen et al. 1997; Freund et al. 1997), ASCL1 (Kim et al. 2008), MEF2C (Edmondson et al. 1994), and ONECUT1 (also known as HNF6) (Clotman et al. 2005), respectively.
Since tissue-specific CREs are often associated with genes specifically expressed in the corresponding tissue (Natarajan et al. 2012; Heinz et al. 2015), we also examined the genes associated with the target DHSs based on the nearest TSS (Supplemental Table S1). Gene Ontology (GO) analysis (Carbon et al. 2009) revealed an enrichment for tissue-specific functions that corresponded to the tissue of DHS origin. For instance, among the top significant hits for the retina, brain, heart, and liver target DHSs were “sensory perception of light stimulus,” “nervous system development,” “cardiovascular system development,” and “organic substance metabolic process,” respectively (Supplemental Table S3). Thus, the 4000 target DHS regions were likely enriched for tissue-specific CREs.
‘Capture-and-clone’ allows synthesis of targeted cis-regulome libraries
To overcome the length restrictions imposed by oligonucleotide array synthesis of CRE fragments (Cleary et al. 2004), we took advantage of DNA capture, a technique routinely used for exome sequencing. For exome capture, biotinylated RNA baits are designed to selectively hybridize with DNA fragments containing sequences of interest, i.e., exonic regions (Gnirke et al. 2009). Here, we adapted this technology to target our CREs of interest (a subset of the putative “cis-regulome”) instead of the exome. This approach offers important advantages. First, the input DNA pool can derive from any genomic DNA source. Hence, the cis-regulome of any single individual or groups of individuals can be assessed. Second, the input DNA pool can be size-selected for a range of fragment lengths, enabling inclusion of long CREs.
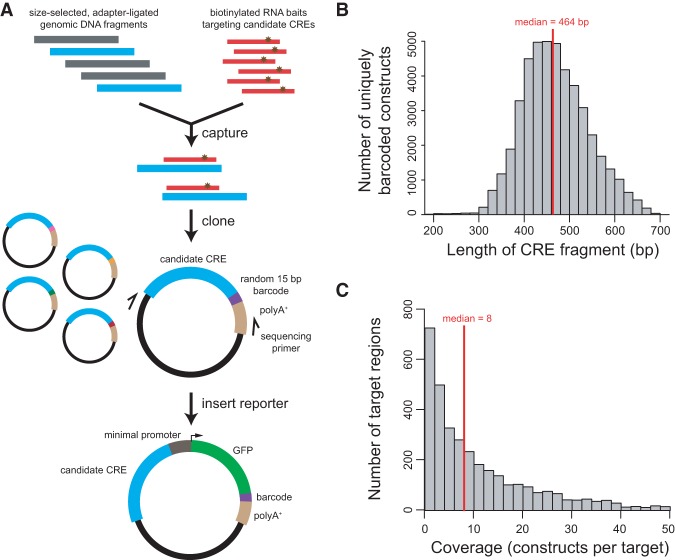
Using mouse (C57BL/6J) genomic DNA that was sheared by sonication and then size-selected to be ∼400–500 bp (excluding adapter sequence), we captured with RNA baits tiling the central 300 bp (which is the median size of DHSs) (Natarajan et al. 2012) of the 4000 target DHS regions. We amplified the captured fragments with primers containing restriction sites for cloning into a barcoded vector library (Fig. 1A). Since the cloning was nondirectional, both orientations were roughly equally represented, as expected (49% and 51% of fragments mapped to the plus and minus strands of the mm9 reference genome, respectively). Paired-end sequencing revealed a distribution of CRE fragment sizes with a median length of 464 bp (SD = 72 bp) (Fig. 1B). Using two successive rounds of capture, we achieved a very high “on-target” rate: 98.5% of the captured fragments overlapped a target region. The median overlap for on-target fragments was 282 bp out of the 300-bp target, i.e., 94% of the target region length (Supplemental Fig. S2). Overall, 3483 of the 4000 (87%) targeted regions were represented, with a median coverage of eight barcodes per represented region, for a total of 45,670 uniquely barcoded constructs (Fig. 1C).
Figure 1.
“Capture-and-clone” allows synthesis of CRE-seq libraries with long CREs. (A) Schematic of the capture-and-clone approach. Size-selected, adapter-ligated genomic DNA was hybridized to biotinylated RNA baits that tiled across candidate CRE regions of interest. Captured fragments were cloned into a barcoded vector library with unique 15-mer barcodes. Paired-end sequencing revealed the CRE-barcode correspondence. A minimal promoter-GFP reporter cassette was subsequently cloned into the library. (B) Histogram showing the distribution of the lengths of captured fragments that were cloned into the barcoded vector library, based on paired-end sequencing. The median length was 464 bp. (C) Histogram showing the distribution of target coverage, i.e., the number of captured fragments that overlapped a 300-bp target region. Of the 4000 targeted regions, 3483 regions were represented by at least one construct. The median coverage among represented regions was 8. Not shown in graph: 517 nonrepresented regions and 114 target regions with a coverage of more than 50.
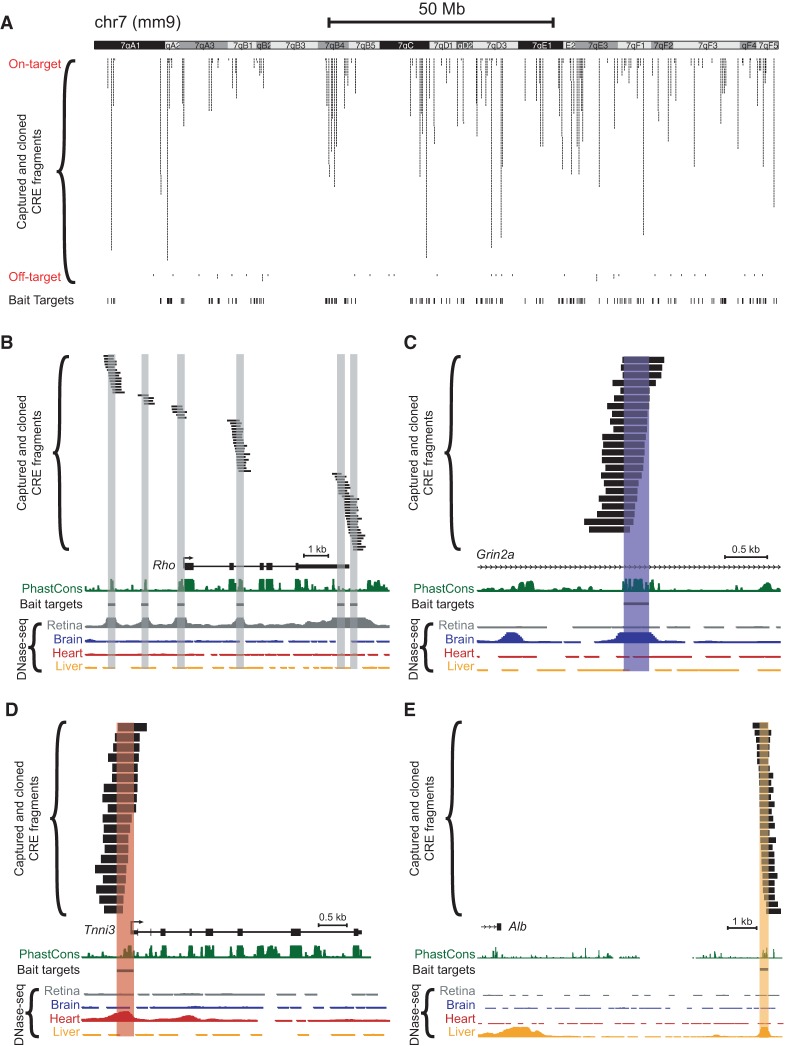
The distribution of captured fragments across a representative chromosome is shown in Figure 2A. Notably, many loci exhibited a multiplicity of captured fragments corresponding to a single target region, resulting in a tiling of the DHS peak, as exemplified in Figure 2B–E. Hence, the ability to conduct CRE truncation mutation analysis at a given locus is a key built-in feature of our capture-and-clone approach.
Figure 2.
Tiling of captured fragments across target regions. Capture baits were designed based on adult (8-wk-old C57BL/6J) DNase-seq data from Mouse ENCODE (Yue et al. 2014). Paired-end sequencing revealed the locations of individual barcoded, captured-and-cloned fragments. The UCSC Genome Browser (mm9) (Karolchik et al. 2014) screenshots depict: (A) Captured fragments for an entire representative chromosome (Chr 7). Off-target fragments, i.e., those that did not overlap a 300-bp target bait region, are also shown. Examples of captured fragments around a retina-specific locus (B), in an intron of a brain-specific locus (C), in the 5′ UTR/promoter region of a heart-specific locus (D), and downstream from a liver-specific locus (E): (Rho) rhodopsin; (Grin2a) glutamate receptor, ionotropic, NMDA2a (epsilon 1); (Tnni3) troponin I, cardiac 3; (Alb) albumin. Note that some DNase-seq peaks visible in the screenshots were not included as targets for capture. PhastCons depict 30-way vertebrate phylogenetic conservation (Siepel et al. 2005).
AAV packaging and delivery preserves CRE-seq library composition
We next considered how to expand the repertoire of cell types accessible by CRE-seq. Whereas efficient plasmid delivery is limited to mitotic cells amenable to chemical transfection or electroporation (Mortimer et al. 1999; Karra and Dahm 2010), the ideal CRE-seq delivery vehicle would permit access to a variety of tissues, including post-mitotic tissues, and in a range of species. We reasoned that adeno-associated virus (AAV), a nonpathogenic virus commonly used for gene therapy studies, would be suitable for this purpose. AAV causes long-lasting infection in rodents and primates, and its tissue tropism ranges by serotype from promiscuous to cell-type selective (Mingozzi and High 2011). Moreover, unlike DNA delivered by lentivirus, the AAV-delivered DNA remains almost exclusively episomal, thereby permitting cis-regulatory analysis without the insertion site effects associated with integration into the host genome (McCarty et al. 2004).
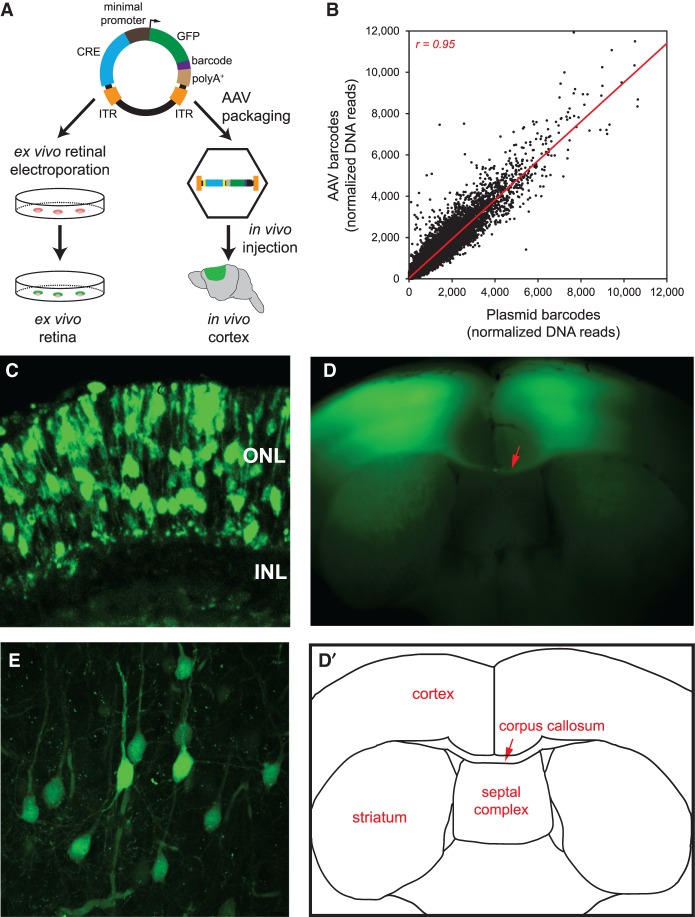
After cloning in a TATA box-containing minimal promoter-green fluorescent protein (GFP) cassette (Fig. 1A), we transferred the library into a vector with inverted terminal repeats (ITRs), which are necessary for AAV packaging (Yan et al. 2005). This yielded the final plasmid library (Fig. 3A). To deliver the library into the retina, we conducted ex vivo electroporation of the plasmid library into the neonatal mouse retina, as in our past CRE-seq studies (Kwasnieski et al. 2012; White et al. 2013). We generated three biological replicates, each consisting of multiple electroporated retinas.
Figure 3.
Delivery of capture CRE-seq library into mouse retina ex vivo and cerebral cortex in vivo. (A) Schematic of the CRE-seq library delivery approach. The plasmid library can be directly electroporated into the retina ex vivo. Alternatively, the library can be packaged into AAV and delivered via stereotactic injection into the cerebral cortex in vivo. (B) Scatterplot comparing the relative abundance of approximately 45,000 individual barcoded constructs in the plasmid library delivered into the retina, and in the AAV-packaged library delivered into cortex, as measured by barcode DNA reads summed across the three biological replicates for each tissue and then normalized to the total number of barcode DNA reads. Each data point represents a unique barcoded construct. DNA reads were well-correlated (Pearson r = 0.95), indicating fidelity of barcode representation after AAV packaging and delivery. Off-target constructs and constructs with zero reads in all samples were excluded. Four points falling outside the depicted plot range (included in the calculation of Pearson r) are not shown. (Red line) linear regression. (C) Confocal image of a retina that was electroporated with the plasmid library and cryosectioned after 8 d in culture: (ONL) outer nuclear layer; (INL) inner nuclear layer. (D) Flat-mount image of a coronal slice from a brain injected with the AAV-packaged library bilaterally into the primary motor cortex and harvested ∼4 wk later. (D′) Schematic corresponding to the flat-mount image. Note the bilateral GFP-positive regions in the cortex as well as bundles of GFP-positive axons in the corpus callosum (red arrow). (E) Confocal image of a cortical region infected with the AAV-packaged library.
To deliver the library into the cerebral cortex, we packaged the plasmid library into AAV9(2YF) and conducted in vivo stereotactic injections to infect adult primary motor cortex. AAV9 is a serotype that exhibits broad tissue tropism, and its tyrosine-mutated derivative AAV9(2YF) transduces neurons of the CNS with high efficiency and minimal host-mediated degradation of viral particles (Zhong et al. 2008; Zincarelli et al. 2008; Dalkara et al. 2012; Aschauer et al. 2013). We generated three biological replicates, each consisting of cerebral cortex tissue from a single injected mouse.
As evidence that AAV packaging and stereotactic injection did not adversely affect the composition of the library, we observed a strong correlation (Pearson r = 0.95) between the relative abundance of individual barcoded constructs in the retina after delivery of the plasmid CRE-seq library and in the cerebral cortex after infection with the AAV-packaged CRE-seq library (Fig. 3B). Furthermore, 76% (34,824/45,670) of the on-target barcodes were “well-represented” (i.e., had at least 10 raw DNA reads) in all six biological replicates (three replicates each for retina and cerebral cortex). These 34,824 barcodes covered 97% (3375/3483) of the targeted DHS regions that were represented in the initial post-capture library. These results indicated good preservation of barcode abundance and diversity throughout the procedure, from the initial post-capture cloning to the delivery of the library.
We then examined the tissues histologically for evidence of library expression, as visualized by fluorescence microscopy. Upon examination of the electroporated retinas, we observed GFP-positive cells in the outer nuclear layer (ONL) of the retina, where the rod photoreceptor cell bodies reside (Fig. 3C). Moreover, the GFP-positive cells coexpressed the rod-specific Rho-CBR3-DsRed reporter (Supplemental Fig. S3A; Corbo et al. 2010). These findings indicated that the GFP-positive cells were rod photoreceptors, which are the predominant cell type assayed by neonatal retinal electroporation.
Upon histological examination of the AAV-injected brains, we observed bilateral GFP-positive regions throughout all layers of the cerebral cortex (Fig. 3D), corresponding to GFP-expressing cells seen under higher magnification (Fig. 3E). Many of the GFP-positive cells were morphologically consistent with pyramidal neurons, with an apically oriented primary dendrite and an axon. Furthermore, GFP expression colocalized with RBFOX3 (also known as NeuN) (Mullen et al. 1992), a widely expressed marker of mature neurons (Supplemental Fig. S3B). Interestingly, there were bundles of GFP-positive axons crossing the midline in the corpus callosum (red arrow in Fig. 3D), indicating that interhemispheric projection neurons were among the cells that expressed the CRE-seq library.
AAV-mediated CRE-seq demonstrates tissue-specific CRE activity of DHSs in vivo
Given the histological evidence for expression of the library in both tissues, we next quantified the cis-regulatory activity of individual constructs by next-generation sequencing. As quality control measures, we verified that the samples overall clustered by the assayed tissue type (retina versus cerebral cortex). We also observed that the RNA read counts for individual barcodes were correlated among the three biological replicates for each tissue, although greater variability was observed among the cerebral cortex samples than the retinal samples (Supplemental Fig. S4; Supplemental Table S4).
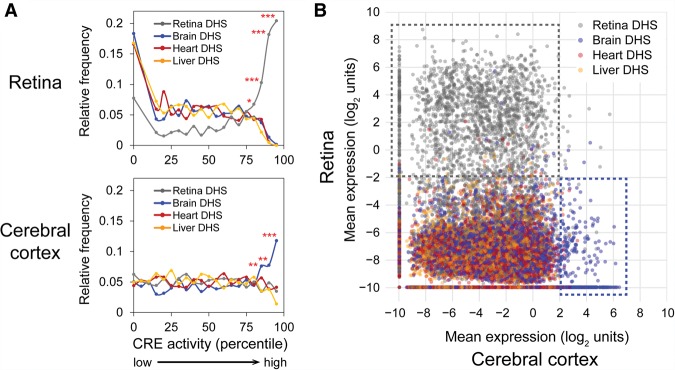
Since tissue-specific DHSs are believed to mediate tissue-specific cis-regulatory activity (Natarajan et al. 2012; Heinz et al. 2015), we first asked whether this was the case. For this analysis, we assigned the “overall” cis-regulatory activity of a given DHS by averaging across corresponding barcoded constructs (as well as across biological replicates). Here, we included the roughly 3000 DHSs with at least two barcoded constructs. When we examined the relationship between the DHS type (i.e., the tissue origin of the DHS) and CRE activity as assayed in the retina, we observed strong enrichment of retinal DHSs among highly expressed DHSs, especially among the top ∼20% most highly expressed DHSs in the retina (Fig. 4A). Since averaging across barcoded constructs may not necessarily be the best metric of cis-regulatory activity for a given DHS, we also examined the expression of individual barcoded constructs. This again revealed the strong preference of the retina for expressing retinal DHSs (Fig. 4B).
Figure 4.
Tissue-specific cis-regulatory activity of DHSs. (A) Frequency distribution of DHSs ranked by cis-regulatory activity (bin size: 5 percentiles) as measured in the retina (top) or cerebral cortex (bottom). In the retina, ∼15% DHSs had undetectable activity and hence were binned together. Averages were taken across biological replicates and barcodes for a given target DHS. Only DHSs with at least two barcoded constructs were included in this analysis (about 3000 DHSs). Frequencies were normalized to the total number of DHSs in each category. To test for enrichment, a χ2 test was performed (one-tailed): (*) P<0.05; (**) P < 0.01; (***) P < 10−4. (B) Scatterplot showing the expression of individual barcoded constructs as assayed in the cerebral cortex (x-axis) versus retina (y-axis). Each dot represents an individual construct. For each construct, the average measurement across the three biological replicates for each tissue was taken. The approximately 35,000 barcodes that were well-represented (at least 10 DNA reads) in all six samples were included in the analysis. Gray, blue, red, and orange dots denote constructs with CRE fragments that overlap retina, brain, heart, and liver DHSs, respectively. The dotted gray box encompasses constructs that are strongly active in the retina, and the dotted blue box encompasses constructs that are strongly active in the cortex.
Similarly, in the cerebral cortex, there was an enrichment of brain DHSs among highly expressed DHSs, especially among the top ∼15% most highly expressed DHSs in the cortex (Fig. 4A). However, this enrichment was less pronounced than for retina: Among the top 15% most highly expressed DHSs in the retina, 79% were retinal DHSs, whereas among the top 15% most highly expressed DHSs in the cerebral cortex, 42% were brain DHSs (P < 0.0001, Fisher's exact test). As seen from the individual barcoded constructs (Fig. 4B), there was a clear preference for brain DHSs among the most active constructs, but there was overall more promiscuous (less selective) activity of constructs in the cortex. The activity profile of nonbrain DHSs in the cortex was right-shifted (increased) and overlapped to a greater extent with the activity profile of brain DHSs in the cortex, compared to the activity profile of nonretinal versus retinal DHSs in the retina. Overall, these findings indicated that there was tissue-specific cis-regulatory activity of DHSs in the retina and the cortex, with the retina exhibiting a stronger preference for retinal DHSs than the cortex exhibited for brain DHSs.
Parameters that predict cis-regulatory activity
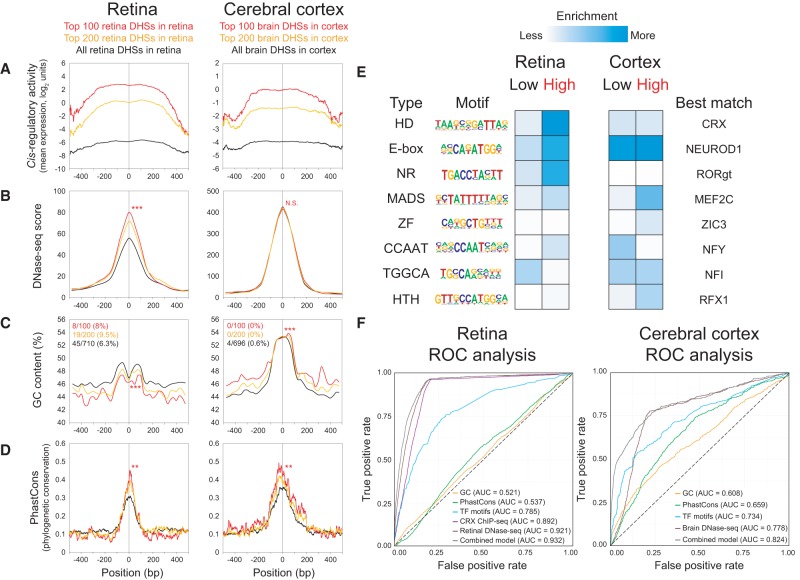
We next asked whether certain parameters previously found to be associated with cis-regulatory activity were predictive of high activity in our assay. For each parameter examined in Figure 5, A–D, we considered the top 100 and top 200 most highly expressed DHSs for the tissue-appropriate DHS type (i.e., for the retina, we restricted our analysis to retinal DHSs; and for the cerebral cortex, we restricted our analysis to brain DHSs). Corresponding data for the liver and heart DHSs are provided in Supplemental Figure S5. We first surveyed expression as a function of position relative to the center of the DHS target region, within a 1-kb window (Fig. 5A). Although DNase-seq signals had a relatively narrow peak (∼300-bp width) (Fig. 5B), cis-regulatory activity in both the retina and cortex had a much broader peak, plateauing in the central ∼500 bp. The breadth of the cis-regulatory activity peaks likely reflects the longer length of the captured fragments (median length of 464 bp) and the large extent of overlap with the central 300 bp of the DHS regions (median overlap of 94%). Notably, we did not find a substantial relationship between the length of individual CRE fragments and CRE activity (Supplemental Fig. S6) or between distance from the nearest TSS and CRE activity (Supplemental Fig. S7).
Figure 5.
Parameters that predict CRE activity. (A–D) Retinal DHSs as assayed in the retina (left) and brain DHSs as assayed in the cerebral cortex (right). Each panel shows a 1-kb centered window. Only DHSs with at least two barcodes were included in this analysis, i.e., 710 retinal DHSs in retina (black lines, left) and 696 brain DHSs in cortex (black lines, right). The top 100 (red lines, left) and top 200 (orange lines, left) retinal DHSs expressed in the retina and the top 100 (red lines, right) and top 200 (orange lines, right) brain DHSs expressed in the cortex are shown. To compare the top 100 DHSs versus the rest of the DHSs in each group, a two-tailed Student's t-test was calculated for the means within the 1-kb window, except for PhastCons scores, which was calculated within the central 100 bp: (**) P < 0.01; (***) P < 0.001; (N.S.) not significant. (A) Cis-regulatory activity, as measured by mean expression in log2 units. For each assayed DHS, at each base position across the 1-kb window, the expression values of the individual barcoded constructs whose CREs overlapped the position were averaged across biological replicates. (B) DNase-seq score (Yue et al. 2014). (C) GC content, calculated in 50-bp windows, sliding 25 bp at a time. The fractions denote the proportion of DHSs that were promoter-proximal (i.e., located within −1 kb to +100 bp relative to the nearest TSS) based on GREAT annotations (McLean et al. 2010). (D) Phylogenetic conservation as measured by 30-way vertebrate PhastCons (Siepel et al. 2005). (E) Enrichment for TF motifs among low- versus high-expressing DHSs in each tissue, without restriction on the type of DHS (see Methods). Only significant motifs are shown (P < 0.05 in at least one category). For motifs enriched in both tissues, the logo from the tissue with the more significant enrichment is shown: (HD) homeodomain; (NR) nuclear receptor; (ZF) zinc finger; (HTH) helix-turn-helix. (F) Receiver operator characteristic (ROC) curves show the performance of logistic regression models for GC content, PhastCons, TF motifs, retina or brain DNase-seq, or a combined model. A model based on CRX ChIP-seq (Corbo et al. 2010) was included for the retina only. The area under the curve (AUC) for each model is indicated. For cross-validation results, see Supplemental Table S6.
Interestingly, higher DNase-seq scores were significantly associated with higher cis-regulatory activity in the retina but not in the cortex (Fig. 5B). A possible explanation is that the retinal DNase-seq data primarily reflect the chromatin state of rods, since they constitute the vast majority of cells in the retina (Jeon et al. 1998), and the most strongly expressed DHSs are rod CREs. By comparison, the brain DNase-seq data reflect the chromatin state of a heterogeneous cell population, and the most strongly expressed DHSs in the cortex may be cell-type–specific CREs highly active in only a subset of cells.
Next, we investigated GC content, which has been reported to be elevated within CREs. This elevation in GC content is thought to favor nucleosome occupancy in tissues where the CRE is not active, thereby repressing cis-regulatory activity in those tissues (Tillo and Hughes 2009; Tillo et al. 2010; Fenouil et al. 2012; Wang et al. 2012; Hughes and Rando 2014). We previously published an enhancer study, in which short (84 bp) synthetic CREs were cloned upstream of a photoreceptor-specific proximal promoter. This study revealed a positive correlation between GC content and enhancer activity in the retina (White et al. 2013). Thus, we were surprised to find that here, the most active retinal DHSs had significantly lower GC content (Fig. 5C). However, a recent CRE-seq study using a minimal promoter also found lower GC content in highly active enhancers (Kwasnieski et al. 2014). Therefore, GC content appears to have distinct roles when the CRE acts as an autonomous element with a minimal promoter or as an enhancer with an active proximal promoter. Brain DHSs had a different pattern, with markedly elevated GC content centrally, and further increased GC content was seen among the most active brain DHSs in the cortex (Fig. 5C). The different effects of GC content in the two tissues may reflect AT-rich versus GC-rich motifs of tissue-specific TFs, and/or the distinct preferences of tissue-specific TFs for AT-rich versus GC-rich “environments” surrounding the TF motif (Dror et al. 2015).
An ongoing debate in the field of genomics is the degree to which phylogenetic conservation at the DNA sequence level is an accurate predictor of functional CREs, given that there is rapid turnover of individual TF binding sites in the course of evolution (Dermitzakis and Clark 2002; Vierstra et al. 2014). We observed significantly higher vertebrate conservation (as measured by PhastCons scores) (Siepel et al. 2005) for the most strongly expressed retinal and brain DHSs in the retina and cortex, respectively. This elevated phylogenetic conservation occurred primarily within the central ∼100 bp of DHSs (Fig. 5D). This distribution of phylogenetic conservation is consistent with the previous observation that highly local (<100 bp) sequences confer substantial CRE activity (White et al. 2013).
We then considered TF motif content, which has been found to be predictive of cis-regulatory activity (Kwasnieski et al. 2014; Blatti et al. 2015). Here, we examined the enrichment of TF motifs among the DHSs with the highest or lowest activity in the retina and cortex, regardless of the type of DHS (Fig. 5E; Supplemental Table S5). In the retina, highly active DHSs were enriched for homeobox, E-box, nuclear receptor (NR), MADS-box, and CCAAT motifs, while in the cerebral cortex, highly active DHSs were enriched for MADS-box, zinc finger (ZF), and helix-turn-helix (HTH) motifs.
To assess the predictive power of these features (DNase-seq scores, GC content, PhastCons scores, and TF motifs), we created logistic regression models and visualized their performance with receiver operating characteristic (ROC) curves, with fivefold cross-validation to control for overfitting (Supplemental Table S6). All constructs assayed in each tissue were classified as “high” (top ∼5% of approximately 36,000 constructs in retina, or top ∼1% of approximately 39,000 constructs in cerebral cortex) versus “not high.” In the retina, DNase-seq was the single most predictive feature (AUC = 0.921), reflecting the strong tendency for highly active constructs to be retinal DHSs. Retinal CRX ChIP-seq peaks (Corbo et al. 2010) performed nearly as well (AUC = 0.892), likely reflecting the fact that CRX ChIP-seq peaks are essentially a subset of retinal DHSs (Wilken et al. 2015). Interestingly, a model based on 15 TF motifs also performed reasonably well (AUC = 0.785). By comparison, in a prior CRE-seq study conducted in cell lines, a model using 50 TF motifs attained an AUC of 0.80 (Kwasnieski et al. 2014). The predictive values of GC content (AUC = 0.521) and PhastCons (AUC = 0.537) were weak. In the cerebral cortex, DNase-seq was likewise the single most predictive feature (AUC = 0.778). A model based on 13 TF motifs performed reasonably well (AUC = 0.734), whereas GC content (AUC = 0.608) and PhastCons (AUC = 0.659) had modest predictive power in the cortex. Notably, in both tissues, the combined model performed only slightly better than DNase-seq alone. Overall, these results reflect the degree of preference of the retina and cerebral cortex for expressing retinal DHSs and brain DHSs, respectively, while underscoring the importance of TF motifs in specifying CRE activity. Furthermore, these results underscore the power of open chromatin mapping techniques such as DNase-seq for identifying functional CREs.
Tiling of captured fragments allows for truncation mutation analysis
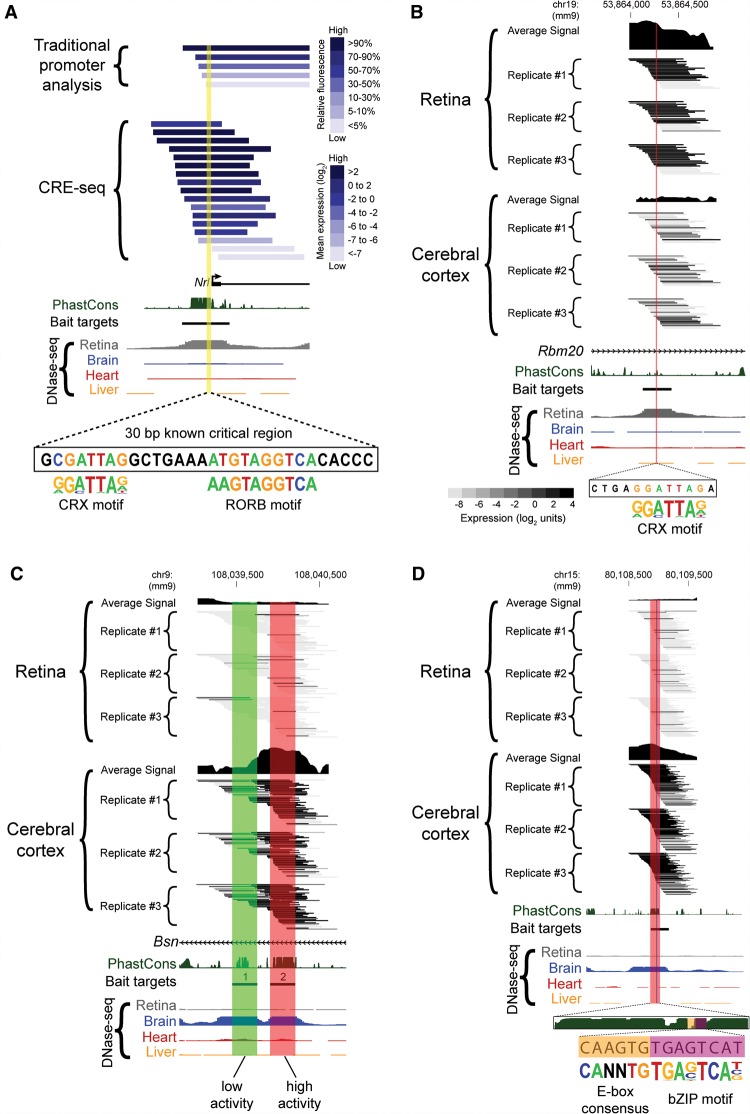
The potential for conducting truncation mutation analysis is an attractive and potentially powerful feature of the capture approach. We therefore sought to determine whether the results were comparable to those of a previously published “traditional” one-at-a-time promoter analysis. NRL is a master regulator of rod photoreceptor development, required both for rod fate determination and maintenance (Mears et al. 2001; Swaroop et al. 2010). Past studies of the Nrl promoter region identified a 30-bp “critical region” that is absolutely required for promoter activity. This critical region contains TF binding sites for CRX and RORB, both of which are required for Nrl expression (Kautzmann et al. 2011; Montana et al. 2011a). Since the Nrl promoter contained a retinal DHS that was targeted in our library, we compared the results of CRE-seq and a traditional promoter analysis that used fluorescence as a readout of cis-regulatory activity (Montana et al. 2011a). Since promoters act directionally (Andersson et al. 2014; Duttke et al. 2015), we compared CRE-seq constructs that were oriented in the same direction as the traditional promoter constructs. We found good agreement between the two assays overall (Fig. 6A), despite differences in construct design (e.g., the CRE-seq constructs contained a minimal promoter, and the 3′ ends of fragments varied). Importantly, both identified the same critical region within a block of phylogenetic conservation (Montana et al. 2011a). Thus, CRE-seq truncation analysis recapitulated the results of a traditional truncation mutation analysis.
Figure 6.
Truncation mutation analysis by CRE-seq. (A) Example of a truncation mutation analysis at the Nrl promoter via a traditional one-at-a-time reporter assay (Montana et al. 2011b) versus capture-and-clone CRE-seq. For the traditional reporter constructs, the 3′ end extends beyond the window depicted in the figure. For the CRE-seq data, only barcoded constructs in the same orientation as the Nrl promoter are shown. The yellow highlighted region corresponds to a known critical region with CRX and RORB motifs (André et al. 1998; Montana et al. 2011b). The minus strand of DNA is displayed. In A and B, the CRX motif (from HOMER) (Heinz et al. 2010) is based on CRX ChIP-seq data (Corbo et al. 2010). The reverse orientation of the CRX motif is displayed. (B–D) Additional examples of CRE-seq truncation mutation analysis: (B) Retinal DHS with retina-specific expression. The critical region identified by CRE-seq (red) contains a putative CRX motif. (C) Two adjacent brain DHSs in the same intron of Bsn exhibit low (DHS #1, green) versus high (DHS #2, red) activity in the cortex. (D) Truncation mutation analysis of a brain DHS. A gradual decrease in activity was observed within the ∼150-bp critical region (red), corresponding to a phylogenetically conserved peak. Within this critical region, a smaller region (vertical blue stripe) was identified that contained an E-box consensus motif (“CANNTG”) and a motif for a bZIP protein, based on AP-1 ChIP-seq data (Heinz et al. 2010). All browser images are from the UCSC Genome Browser (mm9) (Karolchik et al. 2014). DNase-seq data are from Mouse ENCODE (Yue et al. 2014). PhastCons depict 30-way vertebrate phylogenetic conservation (Siepel et al. 2005). The heat map scale shown in B is the same as that used in C and D.
Besides the Nrl promoter, we found additional instances of novel truncation mutation analyses afforded by the capture approach. As seen in Figure 6B, a retinal DHS in the intron of Rbm20 showed strong activity in the retina and weak activity in the cortex. Intriguingly, our assay revealed a 12-bp critical region containing a predicted binding motif for CRX. This motif, “CTAATCCT” (on the negative strand), is a near-perfect match to the consensus motif, “CTAATCCC” (Lee et al. 2010).
Figure 6C depicts another truncation mutation analysis, this time for two brain DHSs (labeled “1” and “2”) located <0.5 kb apart within an intron of Bsn (bassoon). Bassoon is a presynaptic protein that is important for neurotransmitter release from glutamatergic (excitatory) neurons (Altrock et al. 2003). Both of these brain DHSs contained phylogenetically conserved regions, as observed by PhastCons (Siepel et al. 2005). Interestingly, although both had low cis-regulatory activity in the retina, DHS #1 had low activity in the cerebral cortex, whereas DHS #2 had high activity in the cortex. Furthermore, given the extensive tiling of the region, the boundaries of activity could be determined at both the 5′ and 3′ ends of DHS #2.
Next, we present a brain DHS region with high cis-regulatory activity in the cerebral cortex (Fig. 6D). A critical region of ∼150 bp in length was identified that overlapped a block of phylogenetic conservation. Incremental loss of bases in this region resulted in progressive decreases in cis-regulatory activity. Within this critical region, two TF motifs were identified: a consensus E-box motif (recognized by bHLH TFs) (Massari and Murre 2000), immediately next to a motif recognized by basic region leucine zipper (bZIP) proteins of the AP-1 family (Heinz et al. 2010). Like neural bHLH proteins, AP-1 family proteins are known to have important roles in regulating gene expression in the cerebral cortex (Raivich and Behrens 2006; Mongrain et al. 2011).
Additional examples of truncation mutation analysis are presented in Supplemental Figure S8. Overall, we identified 46 retinal DHSs and 13 brain DHSs with examples of truncation mutation analysis, thus representing 4.6% and 1.3% of the 1000 retinal DHSs and 1000 brain DHSs initially targeted in the library, respectively. We observed that for the loci with truncation mutation analyses, at least eight barcoded constructs tiled across the DHS. For DHSs with at least eight assayed barcodes, the fraction of loci with truncation mutation analyses was about threefold higher: 46/363 (12.7%) of retinal DHSs and 13/345 (3.8%) of brain DHSs.
Truncation mutation analyses rely on assaying long CRE fragments that tile across CRE regions. Previously, we conducted a CRE-seq enhancer study (White et al. 2013) in which short (84 bp) CREs (synthesized by oligonucleotide array) were assayed upstream of a rod photoreceptor-specific proximal promoter. These short CREs corresponded to retinal CRX ChIP-seq peaks, which are essentially a subset of retinal DHSs (Wilken et al. 2015). Thus, we wondered whether, for a given CRE, our capture-and-clone approach identified active cis-regulatory sequences beyond the central region tested by the short CRE. Overall, there were 176 CRE regions in the White et al. library that overlapped with assayed regions in the current library, all of which corresponded to retinal DHSs. Most (141/176 or 80%) regions were more active as short enhancers than as long autonomous elements (Supplemental Fig. S9A). This is not surprising, as it is known that some photoreceptor CREs exhibit strong activity as enhancers but minimal activity as autonomous elements (Corbo et al. 2010). Interestingly, in a minority (13/176 or 7%) of cases, the long autonomous elements exhibited substantially more activity, likely because they encompassed functional regions (e.g., critical regions and/or phylogenetically conserved regions) that were not found within the short CREs, as illustrated in Supplemental Figure S9B,C. Although the comparison of these two studies is limited by the differences in assay platforms and the small number of shared CREs, these results indicate that the capture-and-clone approach can provide additional cis-regulatory information beyond that of short CREs.
Together, these examples illustrate that CRE-seq multiplex truncation mutation analysis can identify both known and novel critical regions. In some cases, the spatial resolution is high enough to pinpoint candidate TF motifs required for activity. Thus, our assay has the ability not only to measure the overall activity of a candidate CRE, but also to demarcate the spatial boundaries of cis-regulatory activity.
Traditional reporter assays confirm that critical bases identified by CRE-seq truncation mutation analysis are required for activity
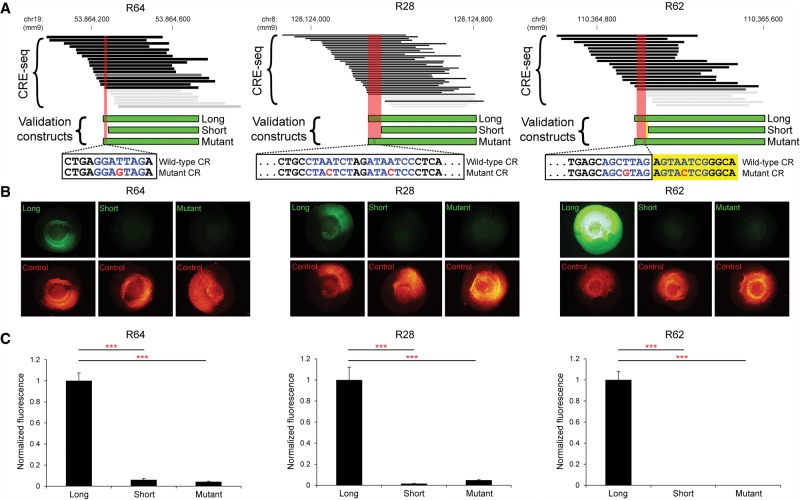
To validate the ability of CRE-seq truncation mutation analysis to identify critical regions de novo, we utilized traditional reporter assays. We previously developed a quantitative fluorescence reporter assay in retinal explants that accurately measures CRE activity (Montana et al. 2011b; Kwasnieski et al. 2012). Thus, we selected three retinal DHS loci (including R64, which is the locus depicted in Fig. 6B) with critical regions identified by CRE-seq truncation mutation analysis to test with the traditional approach (Fig. 7A). These critical regions contained bioinformatically predicted CRX sites, thus allowing us to test whether these CRX sites were required for cis-regulatory activity.
Figure 7.
Validation of individual loci by fluorescence reporter assays. (A) Critical regions (red areas) identified by CRE-seq truncation mutation analysis at three retinal DHSs (R64, R28, and R62) were validated by testing of individual constructs with fluorescence reporter assays. Depicted CRE-seq data are based on expression scores averaged across retinal replicates. Note that R64 is the same locus as in Figure 6B. For each locus, a “long” construct containing the critical region (CR), a “short” construct without the critical region, and a “mutant” construct with point mutations (red font) in predicted CRX sites (blue font) were synthesized. Sequences are shown for the plus strand of DNA in all cases. For R62, one CRX site fell within the critical region, and a second CRX site was immediately adjacent (yellow area). Individual test constructs were directionally cloned upstream of the minimal promoter-GFP cassette in a non-AAV vector. The test constructs were coelectroporated into explant retinas with Rho-CBR3-DsRed (Corbo et al. 2010) as a loading control. (B) Representative whole-mount images of electroporated retinas are shown (exposure times are the same for all images). (C) Quantification of the GFP levels normalized to DsRed levels. Error bars represent SEM (n = 10–12 retinas per test construct). (***) P-value <10−6 (two-tailed Student's t-test).
For each locus, we created a “long” construct, a “short” construct missing the critical region, and a “mutant” construct identical to the “long” construct except that a single point mutation was introduced in the predicted CRX site (Fig. 7A). The point mutation was an adenine-to-cytosine substitution at the fourth position of the CRX motif (thymine-to-guanine in the reverse orientation), which is predicted to inactivate the CRX site (Supplemental Table S7; Lee et al. 2010; White et al. 2013). The constructs were directionally cloned upstream of the minimal promoter-GFP cassette in a non-AAV vector without barcodes in the 3′ UTR, thus controlling for any effects of orientation, AAV vector sequence, or barcode sequence.
Each construct was individually electroporated into multiple retinas and quantified relative to a loading control, Rho-CBR3-DsRed (Fig. 7B). We observed that in each case, the long construct showed high activity, whereas the short construct showed extremely low activity. Notably, the mutant construct exhibited a low level of activity comparable to the activity of the short construct (Fig. 7C). Thus, for all three loci, we not only verified that the critical regions are required for activity, but also that these specific CRX sites are required. These experiments demonstrate that our approach identifies bona fide TF binding sites required for activity.
Discussion
Here, we described an innovative “capture-and-clone” approach for synthesizing CRE-seq libraries. We furthermore demonstrated the feasibility of using AAV-mediated CRE-seq to conduct massively parallel cis-regulatory analysis in the cerebral cortex in vivo. By comparing retina and cerebral cortex, we showed tissue-specific cis-regulatory activity of DHSs. By taking advantage of the truncation mutation analysis afforded by the tiling of captured fragments across targeted loci, we illustrated high-resolution, multiplex functional parsing of CREs.
Previously, high-throughput functional assays of CRE activity had been technologically limited with regard to the length of CREs that could be readily assayed (Levo and Segal 2014; Shlyueva et al. 2014). Our capture-and-clone approach provides a strategy for assaying candidate CREs with lengths of a desired range. Moreover, the capture approach can be used in conjunction with any existing MPRA-like approach, including those that already rely on DNA fragmentation (Dickel et al. 2014; Murtha et al. 2014). For example, STARR-seq (Arnold et al. 2013) has been used to assess long DNA fragments obtained by whole-genome shotgun cloning of the Drosophila genome. However, the mouse and human genomes are approximately 25 times larger than the fly genome. Moreover, only ∼5%–10% of the mammalian genome is thought to be functionally constrained (Graur et al. 2013; Kellis et al. 2014; Rands et al. 2014). Therefore, whole-genome shotgun cloning of mammalian genomes for cis-regulatory analysis is impractical. Instead, capture-and-clone permits targeted cis-regulome analysis.
We note that another group has recently coupled capture technology to STARR-seq (i.e., CapSTARR-seq) (Vanhille et al. 2015). Our approach differs from CapSTARR-seq in two key ways (Supplemental Table S8). First, we achieved higher on-target rates of capture (98.5% versus 14%) due to a rigorous capture protocol to avoid nonspecific pull-down of off-target DNA (Gnirke et al. 2009; Lee et al. 2009). Second, we conducted paired-end sequencing of the input library, whereas CapSTARR-seq mapped only one end of the fragments. Thus, we were able to harness the potential of capture-and-clone for truncation mutation analysis.
Capture-and-clone allows the testing of longer CREs, which presumably harbor more cis-regulatory information. However, there was essentially no correlation between fragment length and CRE activity. What accounts for this observation? One consideration is that the size range of assayed CRE fragments was relatively narrow. Another explanation, based on the truncation mutation analyses, is that some long fragments exhibited low activity due to the omission of critical regions. A third possibility is that some long CRE fragments included repressive sequences that decreased activity (Reynolds et al. 2013).
The capture-and-clone approach is particularly well suited for screening thousands of candidate CREs and identifying the most active CREs in a particular tissue of interest, thereby narrowing the list of CREs that may be relevant to a particular phenotype. For instance, genome-wide association studies (GWAS) and whole-genome sequencing studies have generated lists of thousands of disease-associated noncoding variants (Ward and Kellis 2012; Albert and Kruglyak 2015). To prioritize these lists and thereby accelerate the identification of causal variants, the locations of the candidate variants can be intersected with the locations of putative CREs. The cis-regulomes of unaffected and affected individuals can then be screened by capture-and-clone CRE-seq to identify CREs that exhibit the greatest differential activity between the unaffected and affected groups. Capture-and-clone is thus complementary to CRE-by-synthesis, which is better suited to precisely measuring the effects of specific variants (Levo and Segal 2014). Capture-and-clone can be used to assess a broad range of regions in any organism whose DNA and reference genome are available, although certain types of sequences are not amenable to targeted capture, namely repetitive regions (due to nonspecific pull-down) and sequences with very high (>65%) or low (<25%) GC content (Mertes et al. 2011).
Prior to our study, the implementation of MPRAs in mammalian cells had been almost exclusively restricted to immortalized cell lines and cultured tissues (Shlyueva et al. 2014). The only mammalian tissue that had been assayed in vivo was the mouse liver, due to its ability to take up limited amounts of plasmid DNA via a hydrodynamic tail vein assay (Herweijer and Wolff 2007; Patwardhan et al. 2012). Here, we take a step forward by using AAV to conduct CRE-seq in vivo in the mammalian CNS.
One potential drawback of AAV is that packing constraints limit the size of the insert to <4.7 kb (Wu et al. 2010). Lentiviruses have greater carrying capacity (Kumar et al. 2001), but their integration into the host genome poses the risk of integration site cis-regulatory effects (Clark et al. 1994). In contrast, AAV-mediated CRE-seq measures the cis-regulatory potential of elements independent of chromosomal context, thereby interrogating the function of the DNA sequences themselves. Interestingly, there is evidence that despite being episomal, the AAV vector is organized into nucleosomes (Penaud-Budloo et al. 2008). Another limitation of AAV is that the onset of expression is relatively slow, with maximal expression requiring up to several weeks (Day et al. 2014). This delay is due to the required conversion of the genome from single-stranded into double-stranded DNA. Recently, self-complementary AAV (scAAV) serotypes have been developed that exhibit more rapid transgene expression (McCarty 2008). As novel AAV serotypes for gene therapy continue to emerge (Wu et al. 2006; Daya and Berns 2008), AAV-mediated CRE-seq will become increasingly powerful.
Why are some tissue-specific DHSs active and others inactive, even when assayed in the appropriate tissue? One reason is that DHSs demarcate not only active enhancers but also other types of regulatory elements (e.g., silencers and insulators) (Gross and Garrard 1988; Thurman et al. 2012). Here, we used a TATA-box containing minimal promoter to assay the autonomous cis-regulatory activity of the tested elements, rather than a tissue-specific proximal promoter to assay for enhancer/silencer activity (Butler and Kadonaga 2002). Only a minority (∼10%–20%) of mammalian promoters contain TATA boxes (Sandelin et al. 2007). Future use of tissue-specific proximal promoters may allow for more sensitive assays, especially as enhancer–promoter compatibility and TATA-box versus DPE-containing promoters become better understood (Sandelin et al. 2007; van Arensbergen et al. 2014; Zabidi et al. 2015). Additionally, since some enhancers become active only in response to particular stimuli (Ostuni et al. 2013; Shlyueva et al. 2014), environmental perturbations may be necessary to unmask their cis-regulatory potential. Furthermore, the cis-regulatory landscape of a given tissue is dynamic across development, as illustrated by DNase-seq in the developing mouse retina and brain (Wilken et al. 2015). Future CRE-seq experiments at multiple developmental stages will help elucidate the temporal dynamics of CREs. Nonetheless, even with the TATA-box–containing minimal promoter assayed in steady-state conditions, we demonstrated tissue-specific CRE activity.
Assaying autonomous activity and assaying enhancer activity are complementary approaches, as they appear to reflect different biological activities and properties of a given CRE. In the current study, we observed that GC content was associated with decreased autonomous CRE activity in the retina. Given the differences in the assays, this finding does not contradict our earlier retinal CRE-seq study (White et al. 2013), in which we observed a positive association between GC content and enhancer activity. In fact, the current result is consistent with a recent CRE-seq study in which GC content was associated with decreased autonomous activity of predicted enhancers in cell culture (Kwasnieski et al. 2014).
In our study, the retina exhibited a stronger preference for retinal DHSs than the cerebral cortex exhibited for brain DHSs. Several explanations are possible. First, the cellular complexity of the brain is likely a major factor (Wurmbach et al. 2002). A recent DNase-seq study in the mouse brain observed that DHSs could be found around genes expressed in only a small percentage of neurons, such as cortical laminar-specific genes (Wilken et al. 2015). Thus, a given “brain DHS” may actually be a cell-type–specific DHS that is active in a small population of cells. When averaged over the entire population of assayed cells, the cell-type–specific activity of the DHS may be obscured. For tissues with highly heterogeneous cell populations such as the cerebral cortex, it should be possible to target specific subpopulations by combining AAV-mediated CRE-seq with fluorescence-activated cell sorting (FACS) of defined cell types (Okaty et al. 2011; Gisselbrecht et al. 2013; Dickel et al. 2014). Second, the minimal promoter used in this study contains a possible weak CRX site, whose affinity is predicted to be ∼10% that of the CRX consensus motif (Chen and Zack 1996; Lee et al. 2010). Lastly, although DNA barcode representation was similar in the retina and cerebral cortex, the difference in delivery methods for the two tissues may have been a contributing factor.
In summary, we have developed a powerful and efficient strategy for constructing CRE-seq libraries that extends the size range of the CREs that can readily be assayed, using targeted cis-regulome capture. At the same time, we have demonstrated the feasibility of conducting CRE-seq in vivo in a mammalian tissue using AAV. As new assays for rapidly identifying the locations of putative cell-type–specific CREs are developed, e.g., ATAC-seq (Buenrostro et al. 2013), our study sets the stage for the high-throughput functional screening of thousands of candidate CREs in a range of cell types and in a variety of model systems, including nonhuman primates and human induced pluripotent stem cell (iPSC)-derived organoids (Lancaster et al. 2013).
Methods
Animals
Mice were maintained on a 12-h light/dark cycle at ∼20°C–22°C with free access to food and water. Neonatal mice were euthanized by decapitation, and adult animals were euthanized with CO2 anesthesia followed by cervical dislocation, unless otherwise stated. All experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council 2011), and were approved by the Washington University in St. Louis Institutional Animal Care and Use Committee.
Reference genome
The mouse reference genome used throughout was mm9.
Identification of target tissue-specific DHS peaks
We downloaded DHS data in narrowPeak format from the Mouse ENCODE Project (Yue et al. 2014) for the following tissues (GEO sample accessions are listed): whole brain age E14.5 (GSM1014197, replicate 1), whole brain age E18.5 (GSM1014184, replicate 1), whole brain age 8 wk (GSM1014151, replicate 1), retina age P1 (GSM1014188), retina age P7 (GSM1014198), retina age 8 wk (GSM1014175), liver age E14.5 (GSM1014183, replicate 1), liver age 8 wk (GSM1014195, replicate 1), lung age 8 wk (GSM1014194, replicate 1), kidney age 8 wk (GSM1014193, replicate 1), thymus age 8 wk (GSM1014185, replicate 1), and heart age 8 wk (GSM1014166, replicate 1). We parsed these data using custom Perl scripts, tallying the number of reads per 150-bp block across the mouse genome to give a DHS “score.” We then examined the top roughly 4000 tissue-specific peaks each for brain age 8 wk, retina age 8 wk, heart age 8 wk, and liver age 8 wk. For a peak to be identified as “tissue-specific,” it was required to have a DHS score of greater than 25 in the 8 wk tissue of interest and less than 25 in samples derived from other tissues (but the peak score for samples deriving from different developmental stages of the same tissue type were not required to be less than 25). For instance, if the score for a retina age 8 wk peak was greater than 25 and the score for the corresponding retina age P7 peak was greater than 25, but all nonretinal peaks were less than 25, then that peak was called “retina-specific.” After removing any tissue-specific peaks that overlapped repetitive genomic sequences (∼10% of peaks), we selected the 1000 peaks with the highest tissue-specific peak scores from each of adult brain, retina, heart, and liver for inclusion as capture targets.
Capture bait library design and synthesis
For each of the 4000 target regions, seven 80-bp baits were designed to tile across the 300-bp region (sliding 37 bp at a time), for a total of 1.2 Mb and 28,000 baits. To check for potential off-target bait hybridization, bait candidates were BLASTed against the mm9 genome, which was masked for the regions from which baits were designed. By definition, Tm is the temperature at which 50% of the molecules are hybridized. Bait candidates were accepted only if no BLAST hits (Altschul et al. 1990) with a predicted Tm > 40.0°C were found.
GREAT analysis and gene ontology
GREAT v2.0.2 analysis with mm9 as the reference genome was implemented, using the “single nearest gene” within 1000 kb as the algorithm for associating genomic regions to genes, and using the whole genome as background and excluding the “include curated regulatory domains” option (McLean et al. 2010). The input to the GREAT analysis was the list of 4000 target DHS regions. Gene Ontology (GO) (Ashburner et al. 2000) enrichment analysis for “biological process” in Mus musculus was implemented using PANTHER (Mi et al. 2005) with AmiGO 2 v2.1.4 (Carbon et al. 2009). The input to the GO analysis was the GREAT-generated list of genes associated with target DHSs (“region-to-gene” associations).
Restriction enzymes and PCR reagents
Unless otherwise indicated, restriction enzymes were from New England BioLabs, and Phusion Hot Start Flex 2× Master Mix (New England BioLabs) was used for PCR. Primer sequences are listed in Supplemental Table S9.
Preparation of gDNA for capture
Genomic DNA was purified from liver tissue of C57BL/6J mice and sonicated with Covaris E210 (duty 10%, intensity 4, cycles/burst 200, time 100 sec). The freshly sonicated DNA was end repaired, 3′ adenylated, ligated to commercial adapters, and enriched by PCR, using the TruSeq LT or TruSeq Nano Kit (Illumina) according to the manufacturer's instructions (1 µg or 200 ng input gDNA, and 10 or 8 cycles of PCR, respectively). For final size selection and purification prior to capture, the samples were gel electrophoresed on 2% low melting point agarose and gel extracted with MinElute (Qiagen). To concentrate the samples in preparation for capture, the samples were speed vacuumed in LoBind tubes (Eppendorf).
Cis-regulome capture and preparation for cloning
Capture was conducted in a similar manner as previously described (Gnirke et al. 2009). Two rounds of sequential capture were conducted to achieve high on-target rates (Lee et al. 2009). Briefly, for the first round of capture, a 9-μL library mix was prepared, consisting of ∼300 ng input (TruSeq LT or TruSeq Nano gDNA library), 2.5 μg human Cot-1 DNA, 2.5 μg salmon sperm DNA, and 0.6 μL adapter blocking agent (MYcroarray). This solution was denatured for 5 min at 95°C. Meanwhile, a 36.8-μL hybridization mix was prepared, consisting of 5 μL 20X SSPE (instead of the standard 20 µL), 0.8 μL 0.5 M EDTA, 8 μL 50X Denhardt's, 8 μL 1% SDS, and 15 μL RNase-free water. This solution was prewarmed for 3 min at 65°C. A 6-μL capture bait mix was prepared, consisting of 50 ng (instead of the standard 500 ng) biotinylated baits and 1 μL SUPERase-In (Ambion). This solution was prewarmed for 2 min at 65°C. Finally, 7 μL of the library mix, 13 μL of the hybridization mix, and all 6 μL of the capture bait mix were incubated for ∼24 h at 65°C. The reaction was then applied to Dynabeads MyOne Streptavidin C1 (Invitrogen) with washing and elution as described (Gnirke et al. 2009). Each capture reaction was purified with MinElute (Qiagen), with an elution volume of 30 µL. Each eluate was speed vacuumed in a LoBind tube (Eppendorf) down to a volume of 3–4 μL and used as the library “input” for a single reaction in the second round of capture. The second round of capture was otherwise identical to the first. No PCR was conducted between the first and second rounds of capture. After the second round of capture, PCR was conducted using Ill_NotI_1XL and Ill_NotI_2XL primers (for 1 min at 98°C, 14–16 cycles: for 10 sec at 98°C, for 30 sec at 58°C, for 1 min at 72°C, followed by 5 min at 72°C). The samples were PCR purified with MinElute (Qiagen), digested with NotI-high fidelity (HF) and gel extracted with MinElute (Qiagen). Two independent pools of capture products were generated, with each pool deriving from multiple capture reactions.
CRE-seq library construction
To minimize the likelihood of cleaving captured fragments, the 8-bp cutters NotI, FseI, and AscI were used. To create the barcoded vector library for insertion of NotI-ended captured fragments, the Rho basal-DsRed construct (Hsiau et al. 2007) was modified with linkers on the 3′ end of DsRed to replace a former NotI site with an EagI site and to add NsiI, FseI, and AscI sites, and on the 5′ end of the Rho basal promoter to add XbaI, NotI, and KpnI sites.
To add 15-mer barcodes, two pools of 30 nmol oligos were synthesized with random 15-bp sequences (Integrated DNA Technologies) as BC_F and BC_R. The two pools were annealed and ligated into the AscI and NsiI sites of the vector. After transformation of 5-alpha chemically competent E. coli (New England BioLabs) and overnight growth in liquid culture, a total of approximately 9.5 × 106 colonies were harvested (as estimated from plating a small aliquot) and purified with the PureLink HiPure Plasmid Maxiprep Kit (Invitrogen). The barcoded vector library was then digested with EagI-HF and dephosphorylated with alkaline phosphatase (Roche). The captured fragments were digested with NotI-HF and cloned into the EagI site of the vector library with 5-alpha chemically competent E. coli (New England BioLabs). A total of about 80,000 colonies were scraped from LB/ampicillin agar plates, grown for ∼2 h in liquid LB/ampicillin culture, and purified with the PureLink HiPure Plasmid Maxiprep Kit (Invitrogen).
After paired-end sequencing to determine the CRE-barcode correspondence (described below), the minimal promoter-eGFP cassette was cloned into the FseI and AscI sites. The minimal promoter is the previously described “Rho basal” minimal promoter, which contains a TATA box (“CATAA”) and which by itself does not have detectable activity in electroporated retina (Hsiau et al. 2007). The minimal promoter-eGFP cassette was created by replacing DsRed with eGFP (Zhang et al. 1996) in the Rho basal-DsRed construct (Hsiau et al. 2007). After transformation with 5-alpha chemically competent E. coli (New England BioLabs) and overnight growth in liquid culture, a total of about 2.7 × 106 colonies were harvested (as estimated by plating a small aliquot) and purified with the PureLink HiPure Plasmid Maxiprep Kit (Invitrogen).
The AAV-ITR vector was prepared by digesting the pAAV2.1-RHO-eGFP vector (Allocca et al. 2007) with NheI and XhoI, and replacing the RHO-eGFP cassette with a linker containing an EagI site. To transfer the library into the AAV-ITR vector, the entire CRE-minimal promoter-eGFP-polyA cassette was subjected to PCR using 5′ Tak and NotI_polyA_R1 primers (for 1 min at 98°C, 10 cycles: for 10 sec at 98°C, for 30 sec at 64°C, for 1 min 30 sec at 72°C, followed by 5 min at 72°C). The PCR product was digested with NotI-HF (New England BioLabs) and cloned into the EagI site of the AAV-ITR vector. After transformation of 5-alpha chemically competent E. coli (New England BioLabs) and overnight growth in liquid culture, a total of about 2.5 × 106 colonies (as estimated by plating a small aliquot) were harvested and purified with the PureLink HiPure Plasmid Maxiprep Kit (Invitrogen). ITR integrity was verified by restriction digest. Note that the final NotI digestion removes any captured fragments initially cloned in as NotI multimers, leaving only the 3′-most captured fragment.
Paired-end sequencing for CRE-barcode correspondence
Prior to insertion of the promoter-reporter cassette, the library was prepared for paired-end sequencing as follows. PCR amplification was conducted using primers LibPCR_F and LibPCR_R (for 1 min at 98°C, 8 cycles: for 10 sec at 98°C, for 30 sec at 64°C, for 1 min at 72°C, followed by 5 min at 72°C). The product was digested with NotI-HF and SacII, gel purified with MinElute (Qiagen), and ligated to P1_NotI and PE2_SacII adapters with T4 DNA ligase (New England BioLabs) using an equimolar mix of P1_NotI indexed adapters to facilitate nucleotide balance. The ligation products were PCR amplified to enrich for molecules that had both P1 and PE2 adapters, using primers JKP4F and JKP4R (for 1 min at 98°C, 14 cycles: for 10 sec at 98°C, for 30 sec at 65°C, for 1 min at 72°C, followed by 5 min at 72°C). The final product was gel-extracted on 2% low melting point agarose and verified on an Agilent Bioanalyzer. Two lanes of MiSeq 2 × 250-bp sequencing were run at a loading concentration of 1.6–2 pM and 12%–15% spiked-in Phi-X DNA (Illumina).
Analysis of paired-end sequencing for CRE-barcode correspondence
Barcodes and captured fragment sequences were extracted based on flanking bases. Captured fragment sequences were aligned as paired reads to mm9 using Bowtie 2 v2.1.0 (Langmead and Salzberg 2012) with an allowed maximum insert size of 1000 bp (“-X 1000” setting). SAM files were converted to BAM files using SAMtools v0.1.19 (Li et al. 2009) and then to BED files using BEDTools v2.22.1 (Quinlan and Hall 2010). Only paired reads that mapped concordantly were used. Fragments were examined for overlap with the 4000 target DHS regions (which were each 300 bp). If a fragment overlapped two adjacent target regions, it was assigned to the target region with the most bases of overlap. Barcodes were required to be 14–16 bp in length. Barcodes with multiple CRE fragment associations, and PCR-duplicate CRE fragments associated with multiple barcodes (∼1.6% of fragments), were discarded. A list of “on-target” CRE correspondences for 45,670 barcoded constructs (minimum 10 reads) resulted. To determine the “off-target” rate, the number of barcoded constructs that did not overlap a target DHS was found to be 712. Hence, ∼98.5% of fragments were on-target.
Retinal explant electroporation and culture for CRE-seq
Electroporation and explant culture of mouse retinas were performed as described previously (Montana et al. 2011b). In brief, retinas were dissected from newborn (P0) CD-1 mouse pups and coelectroporated with 0.5 μg/μL AAV-ITR plasmid CRE-seq library and 0.5 μg/μL Rho-CBR3-DsRed, a rod-specific construct for visualizing electroporation efficiency (Corbo et al. 2010). Retinas were grown in explant culture and harvested 8 d later. Five retinas were pooled for each CRE-seq biological replicate.
Viral production
Recombinant AAV9(2YF) was produced and purified as previously described (Grieger et al. 2006). To summarize, HEK293 cells at ∼80% confluency were cotransfected with the AAV-ITR plasmid CRE-seq library, p-Helper plasmid, and AAV9(2YF) rep/cap plasmid (Dalkara et al. 2012). Cells were harvested 72 h after transfection, and the virus was purified by Iodixanol gradient ultracentrifugation, followed by buffer exchange. The viral titer, as determined by dot blot or quantitative PCR, ranged from 5 × 1012 to 1 × 1014 vg/mL (Zolotukhin et al. 2002; Aurnhammer et al. 2012).
Stereotactic cortical injection
Stereotactic cortical injections were performed in a manner similar to that described (Cetin et al. 2006). Briefly, female CD-1 mice (age 4–6 wk) were anesthesized with isoflurane. Each mouse received bilateral injections. For each injection, a small craniotomy was performed, and 1 μL of AAV9(2YF) CRE-seq library was delivered into the primary motor cortex (stereotactic coordinates: dorsal/ventral axis 0.52 mm, anterior/posterior axis 1 mm, medial/lateral axis 1.5 mm). Animals were harvested 4–5 wk after injection. The brain was sliced coronally, and a fluorescent dissecting scope (Leica MZ16 F) was used to visualize GFP-positive regions, which were isolated by microdissection. Each CRE-seq biological replicate consisted of GFP-positive cortical tissue from a single animal.
Isolation of RNA and DNA and preparation for sequencing
Tissues were rapidly harvested and rinsed in cold sterile HBSS with calcium and magnesium (Gibco) and stored at −80°C in TRIzol (Invitrogen). Samples were homogenized in TRIzol, and RNA and DNA were isolated according to the manufacturer's instructions. RNA samples were treated with TURBO DNase (Ambion) to remove potential DNA contamination. RNA and DNA were prepared for sequencing essentially as previously described (Kwasnieski et al. 2012). RNA was reverse-transcribed with SuperScript III (Invitrogen) using oligo-dT primers. The resulting first-strand cDNA was treated with RNase H. Both the cDNA and DNA samples were subjected to PCR to amplify the barcode sequence in the 3′ UTR of GFP using the forward primer SSP1F and the reverse primer JKP3R (for 1 min at 98°C, 22 cycles for DNA or 26 cycles for cDNA: for 10 sec at 98°C, for 30 sec at 60°C, for 30 sec at 72°C, followed by 5 min at 72°C). This resulted in PCR products flanked by EagI and EcoRI restriction enzyme sites. The products were purified with PureLink PCR Purification Kit (Invitrogen) and digested with EagI-HF and EcoRI. After digestion, the samples were gel purified with Qiagen Gel Extraction Kit and ligated to P1_EagI and PE2_EcoRI adapters using T4 DNA ligase (New England BioLabs). To enrich for molecules that had both P1 and PE2 adapters, the ligation products were PCR amplified with primers JKP4F and JKP4R (for 1 min at 98°C, 20 cycles: for 30 sec at 98°C, for 30 sec at 65°C, for 30 sec at 72°C, followed by 5 min at 72°C). The final product was gel purified from 2% low melting point agarose and verified on an Agilent Bioanalyzer.
Illumina sequencing for CRE-seq barcode abundance
For each tissue, the three cDNA samples and three corresponding DNA samples were multiplexed and run on a single lane of Illumina HiSeq 2000 (1 × 50 bp) at a loading concentration of 8 pM with 10% spiked-in Phi-X DNA.
CRE-seq data analysis
Samples were demultiplexed, and the barcode was extracted based on flanking sequences. Reads were tabulated to obtain the raw RNA and DNA counts for each barcode. Only barcodes with at least 10 raw DNA reads in all three biological replicates of a tissue were included (36,005 barcodes for retina and 38,826 barcodes for cerebral cortex). For each barcode, the RNA count was normalized to the total RNA counts in the sample, and the DNA count was normalized to the total DNA counts in the sample. The normalized expression was the ratio of the normalized RNA count to the normalized DNA count. A pseudocount of 0.001 was added to the normalized expression, and the log2 was taken. The average of the log2 values across biological replicates was the “mean expression (log2 units).”
Histology
Retinal explants were rinsed twice with PBS and fixed in 4% paraformaldehyde/PBS for 30–60 min at room temperature, equilibrated in 30% sucrose/PBS, and embedded in Tissue-Tek O.C.T. (Sakura). Retinal cryosections (12–14 μm) were prepared and stored at −20°C until imaging. For stereotactically injected brains, animals were deeply anesthesized with ketamine/xylazine and then transcardially perfused with heparin/PBS followed by 4% paraformaldehyde/PBS. Animals were decapitated and the brains were dissected in PBS and post-fixed in 4% paraformaldehyde/PBS for at least a day at 4°C. Vibratome sections (200 μm) were prepared from agarose-embedded brain slices and then optically cleared with glycerol/PBS (Selever et al. 2011). Brain slices were treated with sodium borohydride to minimize autofluorescence (Clancy and Cauller 1998). For anti-RBFOX3 (also known as anti-NeuN) staining of free-floating vibratome sections, the sections were blocked with 4% normal donkey serum (NDS)/0.25% Triton X-100/PBS for at least 1 h at room temperature with gentle agitation, incubated with rabbit anti-RBFOX3 antibody (ABN78; EMD Millipore) (1:50, diluted in 4% NDS/0.1% Triton X-100/PBS) overnight at 4°C with gentle agitation, washed with 0.1% Triton X-100/PBS, incubated with Alexa Fluor 555 donkey anti-rabbit (A-31572; Molecular Probes) (1:800, diluted in 4% NDS/0.1% Triton X-100/PBS) for 1 h at room temperature with gentle agitation, and washed with 0.1% Triton X-100/PBS. All brain slices were stored in PBS at 4°C until imaging. For imaging, tissue was mounted with Vectashield (Vectorlabs) and coverslipped. Confocal imaging was conducted with a laser confocal microscope (Zeiss LSM 700) and ZEN 2009 software (Zeiss). Flat-mount imaging of an untreated brain slice (Fig. 3D) was conducted with an inverted fluorescent microscope (Nikon Eclipse TE300) and MetaMorph software (Molecular Devices). Images were processed with Adobe Photoshop.
Cluster analysis of biological replicates
Hierarchical clustering and principal component analysis (PCA) were used to assess the underlying structure of CRE expression across retina and brain replicates. For hierarchical clustering, the sample distance was defined as one minus the Pearson correlation coefficient (calculated across the normalized expression of the roughly 35,000 barcodes with at least 10 DNA reads in all six samples), and clustering was implemented using average linkage. PCA was performed via singular value decomposition on scaled, centered expression data (i.e., zero-centered values with unit variance).
Analysis of TF motif enrichment in low versus high-expressing DHSs
To compare the motif content of low- and high-expressing constructs (Fig. 5E), a list of brain and retina TF motifs were obtained as follows. DNase-seq reads for adult brain (GSM1014151, replicate 1) and adult retina (GSM1014175) were downloaded and aligned to mm9 with Bowtie 2 v2.2.3 (Langmead and Salzberg 2012). DNase-seq peaks were then called using MACS2 v2.1.0 (Zhang et al. 2008). For de novo motif discovery, peaks were first partitioned by HOMER v4.7 annotations (“promoter,” “intronic,” and “intergenic”) (Heinz et al. 2010). De novo motif discovery was then performed independently for each of these classes of peaks from brain and retina, with the final motif list consisting of all motifs identified at a threshold of P < 1 × 10−50. To compare similar numbers of DHSs in the “high” and “low” categories, individual barcoded constructs were ranked by average expression in each tissue. The highest-expressing constructs that constituted 100 distinct DHS target regions (regardless of DHS tissue origin) were classified as “high” in that tissue, and the lowest-expressing constructs that constituted 100 distinct DHS target regions (regardless of DHS tissue origin) were classified as “low” in that tissue (DNA read count was used to break ties). Finally, overlapping intervals were merged, and the resulting regions were scored for motif enrichment (binomial test, via HOMER) relative to a background of approximately 50,000 random mm9 sequences matched for size and dinucleotide content.
Receiver operating characteristic (ROC) curves
To quantify the extent to which sequence features and epigenomic data could predict expression (Fig. 5F), we implemented multiple logistic regression as a means of classifying whether or not individual constructs were among those with the highest expression, similar to the approach described by Kwasnieski et al. (2014). Briefly, all assayed constructs (approximately 36,000 constructs for retina and about 39,000 constructs for cerebral cortex) were partitioned by expression into “high” and “not high” expression groups. “High” was defined here as mean expression across replicates (log2 units) of >−2 for constructs assayed in the retina (∼95th percentile), and > 2 for constructs assayed in the cerebral cortex (∼99th percentile) (see Fig. 4B). Our model included terms for GC content (averaged across the CRE fragment), phylogenetic conservation (30-way vertebrate PhastCons, averaged across the CRE fragment) (Siepel et al. 2005), brain or retina DNase-seq data {log2[(read depth+1)/CRE size]}, retina CRX ChIP-seq data {log2[(1/2) × (read depth of two WT CRX ChIP-seq replicates + 1)/CRE size]} (Corbo et al. 2010), and individual TF motifs (the number of each motif in each CRE fragment, as identified by HOMER). CRX ChIP-seq data were only included in the retina model, and distinct TFs were considered for retina and cerebral cortex models. TF motifs for each tissue were identified as described above (17 motifs for retina and 13 motifs for cerebral cortex) (Supplemental Table S5). Two retinal motifs (YY1 and ZBTB33) were omitted from the model, as they were observed fewer than 100 times across the roughly 36,000 constructs, and hence 15 motifs were in the retina TF motif model. The performance (AUC) of models was quantified using the ROCR package in R (Sing et al. 2005). Fivefold cross-validation was used to control for overfitting.
Expression scores for browser screenshots
For Figure 6A, the scales for the heat maps are indicated. Elsewhere, heat maps were generated according to the default grayscale on the UCSC Genome Browser (Karolchik et al. 2014), using custom BED tracks that were generated as follows. For each biological replicate, a BED track was created using the useScore=1 attribute for intensity shading of individual barcoded constructs using a “BED score.” The “BED score” was obtained by adding 10 to the log2 expression and multiplying by 75. For each tissue, an “average signal” bedGraph track was created by segmenting the tiled regions and averaging the BED scores across replicates and barcodes. A segment was required to be encompassed by at least two barcoded constructs to be included in the “average signal” track. The windowing function was set to “mean.” A smoothing window function (10 pixels) was applied to the average signal tracks, which were displayed on the following scales: 0–1400 for retina and 300–1200 for cortex.
Synthesis of individual constructs for validation
The R28 constructs were cloned as EcoRV/KpnI fragments. To create the long and short R28 constructs, the R28_L/R28_R and R28_S/R28_R primer pairs were used, respectively. To create the mutant R28 construct, R28_MT was ordered as a double-stranded gene block (Integrated DNA Technologies). The R62 constructs were cloned as EcoRI/XbaI fragments. To create the long and short R62 constructs, the R62_L/R62_R and R62_S/R62_R primer pairs were used, respectively. To create the mutant R62 construct, R62_MT was ordered as a double-stranded gene block (Integrated DNA Technologies). The R64 constructs were cloned as EcoRV/KpnI fragments. To create the long, short, and mutant R64 constructs, the R64_L/R64_R, R64_S/R64_R, and R64_MT/R64_R primer pairs were used, respectively. For the PCR reactions, C57BL/6J gDNA was the template. The CREs were digested and cloned upstream of the minimal promoter-eGFP cassette in the Rho basal-eGFP vector, which was created from Rho basal-DsRed (Hsiau et al. 2007) by replacing DsRed with eGFP at XmaI and NotI sites. Test constructs were confirmed with Sanger sequencing that encompassed the entire CRE.
Validation of individual constructs by fluorescent reporter assays
Electroporation, explant culture, and quantification of fluorescence were performed essentially as previously described (Montana et al. 2011b). In brief, as for CRE-seq, retinas were dissected from newborn (P0) CD-1 mouse pups. Here, they were coelectroporated with 0.5 μg/μL of the test construct and 0.5 μg/μL Rho-CBR3-DsRed (Corbo et al. 2010). Retinas were cultured for 8 d, fixed, and then whole mounted for quantitative imaging of fluorescent intensity (GFP intensity normalized to DsRed intensity), using a monochromatic camera (Hamamatsu ORCA-AG) and MetaMorph software (Molecular Devices). For each retina, five regions were quantified in ImageJ and averaged. SEM was calculated based on normalized fluorescence measurements across retinas (n = 10–12 retinas per test construct). Representative whole-mount images using a color camera (Olympus DP70) were also taken.
Comparison with CapSTARR-seq
The raw sequence data for the CapSTARR-seq (Vanhille et al. 2015) input library (GEO accession number GSM1463994) were downloaded and mapped to mm9 with Bowtie 2 v2.1.0 (Langmead and Salzberg 2012).
Data access
The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE68247. Custom tracks for the UCSC Genome Browser (Karolchik et al. 2014) are provided in Supplemental Table S10.
Supplementary Material
Acknowledgments
We thank Karen Lawrence and Jennifer Enright for contributing to the design of the barcoded vector library and sequencing adapters; Jean-Marie Rouillard of MYcroarray for capture advice; Ronald Perez of the Animal Surgery Core at the Hope Center for Neurological Disorders for stereotactic cortical injections; Mingjie Li of the Viral Vectors Core at the Hope Center for Neurological Disorders for assistance with viral production; and the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for sequencing services. We also thank Michael A. White for helpful discussion and Shuyi Ma for critical reading of the manuscript. This work was supported by the Foundation Fighting Blindness (J.G.F.), Simons Foundation Autism Research Initiative (grant number 275579 to J.C.C.), and the National Institutes of Health (National Human Genome Research Institute grant HG006790 and National Eye Institute grants EY018826 to J.C.C.; EY022975 to J.G.F.; EY024958 to J.C.C. and J.G.F.; and 5T32EY013360 to S.Q.S.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.193789.115.
References
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nat Rev Genet 16: 197–212. [DOI] [PubMed] [Google Scholar]
- Allocca M, Mussolino C, Garcia-Hoyos M, Sanges D, Iodice C, Petrillo M, Vandenberghe LH, Wilson JM, Marigo V, Surace EM, et al. 2007. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J Virol 81: 11372–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altrock WD, tom Dieck S, Sokolov M, Meyer AC, Sigler A, Brakebusch C, Fässler R, Richter K, Boeckers TM, Potschka H, et al. 2003. Functional inactivation of a fraction of excitatory synapses in mice deficient for the active zone protein bassoon. Neuron 37: 787–800. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André E, Gawlas K, Becker-André M. 1998. A novel isoform of the orphan nuclear receptor RORβ is specifically expressed in pineal gland and retina. Gene 216: 277–283. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077. [DOI] [PubMed] [Google Scholar]
- Arvey A, Agius P, Noble WS, Leslie C. 2012. Sequence and chromatin determinants of cell-type–specific transcription factor binding. Genome Res 22: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschauer DF, Kreuz S, Rumpel S. 2013. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One 8: e76310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurnhammer C, Haase M, Muether N, Hausl M, Rauschhuber C, Huber I, Nitschko H, Busch U, Sing A, Ehrhardt A, et al. 2012. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum Gene Ther Methods 23: 18–28. [DOI] [PubMed] [Google Scholar]
- Bae BI, Jayaraman D, Walsh CA. 2015. Genetic changes shaping the human brain. Dev Cell 32: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. 2011. Microarrays, megasynthesis. Nat Methods 8: 457–460. [Google Scholar]
- Blatti C, Kazemian M, Wolfe S, Brodsky M, Sinha S. 2015. Integrating motif, DNA accessibility and gene expression data to build regulatory maps in an organism. Nucleic Acids Res 43: 3998–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Kadonaga JT. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev 16: 2583–2592. [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub, Web Presence Working Group. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. 2006. Stereotaxic gene delivery in the rodent brain. Nat Protoc 1: 3166–3173. [DOI] [PubMed] [Google Scholar]
- Chen S, Zack DJ. 1996. Ret 4, a positive acting rhodopsin regulatory element identified using a bovine retina in vitro transcription system. J Biol Chem 271: 28549–28557. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. 1997. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19: 1017–1030. [DOI] [PubMed] [Google Scholar]
- Clancy B, Cauller LJ. 1998. Reduction of background autofluorescence in brain sections following immersion in sodium borohydride. J Neurosci Methods 83: 97–102. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Bissinger P, Bullock DW, Damak S, Wallace R, Whitelaw CB, Yull F. 1994. Chromosomal position effects and the modulation of transgene expression. Reprod Fertil Dev 6: 589–598. [DOI] [PubMed] [Google Scholar]
- Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. 2004. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods 1: 241–248. [DOI] [PubMed] [Google Scholar]
- Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. 2005. Control of liver cell fate decision by a gradient of TGFβ signaling modulated by Onecut transcription factors. Genes Dev 19: 1849–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Lawrence KA, Karlstetter M, Myers CA, Abdelaziz M, Dirkes W, Weigelt K, Seifert M, Benes V, Fritsche LG, et al. 2010. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res 20: 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D, Byrne LC, Lee T, Hoffmann NV, Schaffer DV, Flannery JG. 2012. Enhanced gene delivery to the neonatal retina through systemic administration of tyrosine-mutated AAV9. Gene Ther 19: 176–181. [DOI] [PubMed] [Google Scholar]
- Davidson EH. 2001. Genomic regulatory systems: development and evolution. Academic Press, San Diego, CA. [Google Scholar]
- Day TP, Byrne LC, Schaffer DV, Flannery JG. 2014. Advances in AAV vector development for gene therapy in the retina. Adv Exp Med Biol 801: 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya S, Berns KI. 2008. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 21: 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG. 2002. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 19: 1114–1121. [DOI] [PubMed] [Google Scholar]
- Dickel DE, Zhu Y, Nord AS, Wylie JN, Akiyama JA, Afzal V, Plajzer-Frick I, Kirkpatrick A, Göttgens B, Bruneau BG, et al. 2014. Function-based identification of mammalian enhancers using site-specific integration. Nat Methods 11: 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror I, Golan T, Levy C, Rohs R, Mandel-Gutfreund Y. 2015. A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res 25: 1268–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttke SH, Lacadie SA, Ibrahim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U. 2015. Human promoters are intrinsically directional. Mol Cell 57: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF, Olson EN. 1994. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120: 1251–1263. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouil R, Cauchy P, Koch F, Descostes N, Cabeza JZ, Innocenti C, Ferrier P, Spicuglia S, Gut M, Gut I, et al. 2012. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res 22: 2399–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, et al. 1997. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91: 543–553. [DOI] [PubMed] [Google Scholar]
- Gisselbrecht SS, Barrera LA, Porsch M, Aboukhalil A, Estep PW III, Vedenko A, Palagi A, Kim Y, Zhu X, Busser BW, et al. 2013. Highly parallel assays of tissue-specific enhancers in whole Drosophila embryos. Nat Methods 10: 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. 2013. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol 5: 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Choi VW, Samulski RJ. 2006. Production characterization of adeno-associated viral vectors. Nat Protoc 1: 1412–1428. [DOI] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. 1988. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57: 159–197. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16: 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herweijer H, Wolff JA. 2007. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther 14: 99–107. [DOI] [PubMed] [Google Scholar]
- Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. 2007. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One 2: e643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Rando OJ. 2014. Mechanisms underlying nucleosome positioning in vivo. Annu Rev Biophys 43: 41–63. [DOI] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. 1998. The major cell populations of the mouse retina. J Neurosci 18: 8936–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2014. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42(Database issue): D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karra D, Dahm R. 2010. Transfection techniques for neuronal cells. J Neurosci 30: 6171–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautzmann MA, Kim DS, Felder-Schmittbuhl MP, Swaroop A. 2011. Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene Nrl, revealed by in vivo promoter analysis. J Biol Chem 286: 28247–28255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, et al. 2014. Defining functional DNA elements in the human genome. Proc Natl Acad Sci 111: 6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Battiste J, Nakagawa Y, Johnson JE. 2008. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Mol Cell Neurosci 38: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Keller B, Makalou N, Sutton RE. 2001. Systematic determination of the packaging limit of lentiviral vectors. Hum Gene Ther 12: 1893–1905. [DOI] [PubMed] [Google Scholar]
- Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. 2012. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci 109: 19498–19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. 2014. High-throughput functional testing of ENCODE segmentation predictions. Genome Res 24: 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, O'Connor BD, Merriman B, Funari VA, Homer N, Chen Z, Cohn DH, Nelson SF. 2009. Improving the efficiency of genomic loci capture using oligonucleotide arrays for high throughput resequencing. BMC Genomics 10: 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Myers CA, Williams N, Abdelaziz M, Corbo JC. 2010. Quantitative fine-tuning of photoreceptor cis-regulatory elements through affinity modulation of transcription factor binding sites. Gene Ther 17: 1390–1399. [DOI] [PubMed] [Google Scholar]
- Levo M, Segal E. 2014. In pursuit of design principles of regulatory sequences. Nat Rev Genet 15: 453–468. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. 2001. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci 2: 109–118. [DOI] [PubMed] [Google Scholar]
- London A, Benhar I, Schwartz M. 2013. The retina as a window to the brain-from eye research to CNS disorders. Nat Rev Neurol 9: 44–53. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM. 2008. Self-complementary AAV vectors; advances and applications. Mol Ther 16: 1648–1656. [DOI] [PubMed] [Google Scholar]
- McCarty DM, Young SM Jr, Samulski RJ. 2004. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu Rev Genet 38: 819–845. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. 2001. Nrl is required for rod photoreceptor development. Nat Genet 29: 447–452. [DOI] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG Jr, Kinney JB, et al. 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 30: 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertes F, Elsharawy A, Sauer S, van Helvoort JM, van der Zaag PJ, Franke A, Nilsson M, Lehrach H, Brookes AJ. 2011. Targeted enrichment of genomic DNA regions for next-generation sequencing. Brief Funct Genomic 10: 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. 2005. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33(Database issue): D284–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F, High KA. 2011. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 12: 341–355. [DOI] [PubMed] [Google Scholar]
- Mongrain V, La Spada F, Curie T, Franken P. 2011. Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PLoS One 6: e26622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana CL, Lawrence KA, Williams NL, Tran NM, Peng GH, Chen S, Corbo JC. 2011a. Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. J Biol Chem 286: 36921–36931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana CL, Myers CA, Corbo JC. 2011b. Quantifying the activity of cis-regulatory elements in the mouse retina by explant electroporation. J Vis Exp 10.3791/2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer I, Tam P, MacLachlan I, Graham RW, Saravolac EG, Joshi PB. 1999. Cationic lipid-mediated transfection of cells in culture requires mitotic activity. Gene Ther 6: 403–411. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. 1992. NeuN, a neuronal specific nuclear protein in vertebrates. Development 116: 201–211. [DOI] [PubMed] [Google Scholar]
- Murtha M, Tokcaer-Keskin Z, Tang Z, Strino F, Chen X, Wang Y, Xi X, Basilico C, Brown S, Bonneau R, et al. 2014. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat Methods 11: 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A, Yardimci GG, Sheffield NC, Crawford GE, Ohler U. 2012. Predicting cell-type–specific gene expression from regions of open chromatin. Genome Res 22: 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. 2013. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Pattabiraman K, Visel A, Rubenstein JL. 2015. Genomic perspectives of transcriptional regulation in forebrain development. Neuron 85: 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Sugino K, Nelson SB. 2011. Cell type-specific transcriptomics in the brain. J Neurosci 31: 6939–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171. [DOI] [PubMed] [Google Scholar]
- Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. 2009. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol 27: 1173–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, et al. 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol 30: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaud-Budloo M, Le Guiner C, Nowrouzi A, Toromanoff A, Chérel Y, Chenuaud P, Schmidt M, von Kalle C, Rolling F, Moullier P, et al. 2008. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J Virol 82: 7875–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G, Behrens A. 2006. Role of the AP-1 transcription factor c-Jun in developing, adult and injured brain. Prog Neurobiol 78: 347–363. [DOI] [PubMed] [Google Scholar]
- Rands CM, Meader S, Ponting CP, Lunter G. 2014. 8.2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet 10: e1004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, O'Shaughnessy A, Hendrich B. 2013. Transcriptional repressors: multifaceted regulators of gene expression. Development 140: 505–512. [DOI] [PubMed] [Google Scholar]
- Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G. 2015. Epigenomics: roadmap for regulation. Nature 518: 314–316. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. 2007. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 8: 424–436. [DOI] [PubMed] [Google Scholar]
- Selever J, Kong JQ, Arenkiel BR. 2011. A rapid approach to high-resolution fluorescence imaging in semi-thick brain slices. J Vis Exp 53: 2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15: 272–286. [DOI] [PubMed] [Google Scholar]
- Shu W, Chen H, Bo X, Wang S. 2011. Genome-wide analysis of the relationships between DNaseI HS, histone modifications and gene expression reveals distinct modes of chromatin domains. Nucleic Acids Res 39: 7428–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. 2005. ROCR: visualizing classifier performance in R. Bioinformatics 21: 3940–3941. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. 2010. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci 11: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes TR. 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. 2010. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One 5: e9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arensbergen J, van Steensel B, Bussemaker HJ. 2014. In search of the determinants of enhancer–promoter interaction specificity. Trends Cell Biol 24: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhille L, Griffon A, Maqbool MA, Zacarias-Cabeza J, Dao LT, Fernandez N, Ballester B, Andrau JC, Spicuglia S. 2015. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat Commun 6: 6905. [DOI] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. 2013. A high-resolution enhancer atlas of the developing telencephalon. Cell 152: 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22: 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. 2012. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol 30: 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Myers CA, Corbo JC, Cohen BA. 2013. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci 110: 11952–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilken MS, Brzezinski JA, La Torre A, Siebenthall K, Thurman R, Sabo P, Sandstrom RS, Vierstra J, Canfield TK, Hansen RS, et al. 2015. DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics Chromatin 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216. [DOI] [PubMed] [Google Scholar]
- Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. 2010. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet 11: 273–284. [DOI] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Samulski RJ. 2006. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther 14: 316–327. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yang H, Colosi P. 2010. Effect of genome size on AAV vector packaging. Mol Ther 18: 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmbach E, González-Maeso J, Yuen T, Ebersole BJ, Mastaitis JW, Mobbs CV, Sealfon SC. 2002. Validated genomic approach to study differentially expressed genes in complex tissues. Neurochem Res 27: 1027–1033. [DOI] [PubMed] [Google Scholar]
- Yan Z, Zak R, Zhang Y, Engelhardt JF. 2005. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J Virol 79: 364–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. 2015. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Gurtu V, Kain SR. 1996. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem Biophys Res Commun 227: 707–711. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Li B, Mah CS, Govindasamy L, Agbandje-McKenna M, Cooper M, Herzog RW, Zolotukhin I, Warrington KH Jr, Weigel-Van Aken KA, et al. 2008. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci 105: 7827–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. 2008. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S, Potter M, Zolotukhin I, Sakai Y, Loiler S, Fraites TJ Jr, Chiodo VA, Phillipsberg T, Muzyczka N, Hauswirth WW, et al. 2002. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28: 158–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.