Abstract
The application of genomics technologies to medicine and biomedical research is increasing in popularity, made possible by new high-throughput genotyping and sequencing technologies and improved data analysis capabilities. Some of the greatest genetic diversity among humans, animals, plants, and microbiota occurs in Africa, yet genomic research outputs from the continent are limited. The Human Heredity and Health in Africa (H3Africa) initiative was established to drive the development of genomic research for human health in Africa, and through recognition of the critical role of bioinformatics in this process, spurred the establishment of H3ABioNet, a pan-African bioinformatics network for H3Africa. The limitations in bioinformatics capacity on the continent have been a major contributory factor to the lack of notable outputs in high-throughput biology research. Although pockets of high-quality bioinformatics teams have existed previously, the majority of research institutions lack experienced faculty who can train and supervise bioinformatics students. H3ABioNet aims to address this dire need, specifically in the area of human genetics and genomics, but knock-on effects are ensuring this extends to other areas of bioinformatics. Here, we describe the emergence of genomics research and the development of bioinformatics in Africa through H3ABioNet.
Functional genomics approaches aiming to improve human health are revolutionizing medicine. Next generation sequencing (NGS) approaches have been used to identify genetic polymorphisms that underlie susceptibility to a rapidly expanding list of diseases, and sequencing and interpretation of genomes as part of public health studies are becoming increasingly commonplace. Africa is the cradle of mankind and is host to the greatest human genetic diversity in the world (Schlebusch et al. 2012), as well as unique flora, fauna, and microbiota. A better understanding of the extensive genetic and microbiota diversity in African populations presents a compelling opportunity for the delivery of more accurate diagnoses, new drugs, precision medicine, and a deeper understanding of susceptibility and resistance to infections and metabolic disease. In stark contrast to this opportunity, Africa's genomic analysis capacity has yet to reach a viable critical mass.
The development and application of effective genomic medicine is heavily dependent upon the ability to aggregate and analyze large data sets and to interpret and disseminate knowledge across multiple biomedical disciplines. In Africa, there are few centers of expertise where large numbers of clinicians, genome scientists, and bioinformaticians are sited to jointly perform competitive genomic medical research. Over the last 10 years, African bioinformatics groups have been collaborating to develop the capacity to perform globally competitive research on public and local data sets, in spite of the geographical distances separating these groups (Ramsay et al. 2011). These efforts recently received a major funding boost that has catalyzed the nascent African genomics research community. The H3Africa Bioinformatics Network (H3ABioNet) has been established with a grant from the National Institutes of Health (NIH) Common Fund, as part of its contributions to the Human Heredity and Health in Africa (H3Africa; http://www.h3africa.org) initiative (The H3Africa Consortium 2014). In this article, we describe the development of bioinformatics in Africa through H3ABioNet, which is tightly coupled to the emergence of genomics research on diseases relevant to African populations.
Genomics research in Africa
Until recently, African populations were poorly represented in human genome studies (Bustamante et al. 2011; Schlebusch et al. 2012), despite the enormous genetic diversity found on the continent (Sirugo et al. 2008; Rotimi and Jorde 2010). A small number of genome-wide association studies (GWAS) have been undertaken in African populations without significant European admixture or in African populations outside the United States, for example, to identify genetic determinants of malaria susceptibility in children in West Africa (Jallow et al. 2009) as well as tuberculosis susceptibility (El Baghdadi et al. 2006; Thye et al. 2010; Chimusa et al. 2014). Recently, this type of research led to the discovery of two novel resistance loci for severe malaria (Timmann et al. 2012). Genome-wide single nucleotide polymorphism (SNP) analysis has also been infrequently used to study population diversity in Africa (Henn et al. 2011; Pagani et al. 2012). The HapMap project has subsequently brought African genetic diversity into the public domain and has provided genome-wide SNP data for Yoruba (Nigeria), Luhya, and Maasai (Kenya) populations (The International HapMap Consortium 2007). The first genome of a Nigerian individual was only sequenced in 2008 (Bentley et al. 2008), seven years after the first human genome sequence was published, and five Southern African genomes were subsequently published in 2010 (Schuster et al. 2010). The 1000 Genomes Project has taken up the challenge of bringing more complete low-coverage African genomes into the public domain by including additional populations from Malawi, The Gambia, and Ghana (The 1000 Genomes Project Consortium 2010). More recently a larger data set of African genomic data has been published (Gurdasani et al. 2015).
Genomic studies in African populations promise to be useful in the prediction and development of specific therapies and vaccines. Low levels of funding and a lack of infrastructure and data analysis capacity have, however, hampered the expansion of genomic studies in Africa and have resulted in a one-way flow of samples, data, and skills out of African countries. Only recently have African governments begun to regulate this outflow (de Vries and Pepper 2012). To date, few African scientists have been involved in genomics studies on African individuals, and there has concomitantly been poor capacity development for genomics within the continent (Wonkam et al. 2011). In this regard, bioinformatics is one of the key areas to be developed toward competitive genomics research.
The H3Africa initiative
Responding to the challenge of making human genomics research in Africa a truly African endeavor, the H3Africa initiative was developed through substantial funding from both the NIH Common Fund and the Wellcome Trust. H3Africa is intended to encourage a contemporary research approach by African investigators to study the genomic and environmental determinants of common diseases for improving the health of African populations (The H3Africa Consortium 2014). Projects funded to date cover a wide range of communicable and noncommunicable diseases as well as the study of microbiomes. Specific goals of H3Africa include establishing collaborative networks of African researchers pursuing genomics-based, disease-oriented projects and creating or expanding infrastructure for genomic medicine research. One of the key areas identified was the need to develop the infrastructure that would enable scientists in Africa to handle large data sets and increase the capacity to analyze them. This infrastructure includes the bioinformatics network, H3ABioNet, which forms the backbone of the H3Africa initiative.
Results and Discussion
Establishment of H3ABioNet
Prior to H3ABioNet, bioinformatics groups on the continent were active, but expertise was relatively sparse and mostly localized in South Africa. Founded in 2004, the African Society for Bioinformatics and Computational Biology (ASBCB) was the first continent-wide network of researchers in the field of bioinformatics; it is now a regional network affiliated with the International Society of Computational Biology (ISCB). A formal organization to develop African bioinformatics based on a system of collaborating nodes, ABioNet (African Bioinformatics Network), was proposed at a WHO-supported meeting in Abuja in 2008. Without follow-up funding, however, the development of ABioNet was stifled until recently, when the announcement of the H3Africa initiative provided the springboard for the establishment of H3ABioNet. The network, which is run from a central node at the University of Cape Town, consists of more than 30 nodes across 15 African countries (see http://www.h3abionet.org/home/consortium for complete list) (Fig. 1) with one partner in the United States (USA) and one in the United Kingdom (UK). The institutions range in their current capacity from full nodes with a track record in bioinformatics research, training, and support; through associate nodes with some bioinformatics activities; to development nodes with little or no bioinformatics capacity. Altogether, the network funds more than 40 staff and students and includes more than 80 additional members who contribute to H3ABioNet activities. The nodes collectively provide excellent expertise in different areas of bioinformatics including functional genomics, human population genetics, GWAS and NGS analysis, microbiome analysis, SNP linked protein structure analysis, and biomedical and clinical data storage and management.
Figure 1.
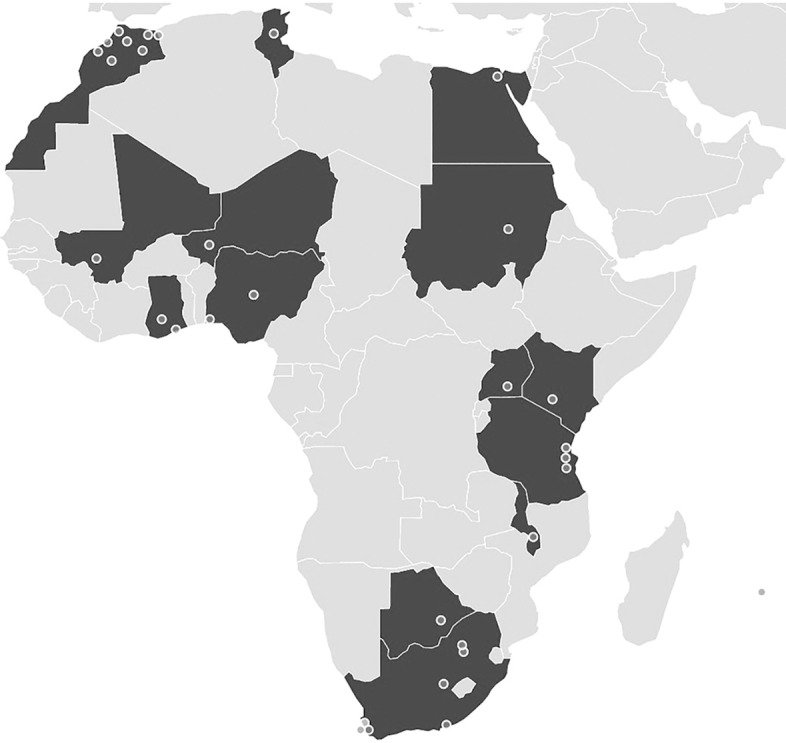
Map of Africa showing the distribution of nodes in the H3ABioNet network. The dot on the right is for Mauritius, and two additional sites not shown include one in the USA and one in the UK.
The mandate of H3ABioNet (http://www.h3abionet.org/) is to develop and roll out a coordinated bioinformatics research infrastructure that is tightly coupled to a sophisticated pan-African bioinformatics training program. In modern genome sequence analysis, analytical tools are moving to where the high dimensional data are stored, so the expertise for genome data manipulation and analysis in Africa are being developed at the source of these data. The network seeks to exploit the development and implementation of best practices in genome bioinformatics in local centers while keeping an eye on the rapidly evolving field through collaboration with centers of expertise in the United States (Harvard University and University of Illinois) and elsewhere.
H3ABioNet faces a number of high priority challenges that need to be overcome to enable genomics research and competitiveness on the continent. These include, but are not limited to, poor internet connectivity for communication, data access, transfer and remote computing; lack of significant computing infrastructure for data storage and processing; lack of bioinformatics skills in clinical genetics and genomics teams performing genomics research; and disparate pockets of bioinformatics expertise across the continent. Some of the major objectives of H3ABioNet are thus to develop human resources through the training of bioinformaticians and researchers in computational techniques and to develop a robust continent-wide research infrastructure that provides access to bioinformatics tools, computing resources, and technical and data management expertise. The network activities are being achieved through dedicated working groups and task forces comprising representatives from multiple countries. Full nodes, including those situated abroad, are helping to build capacity in the less resourced nodes, thus, ensuring the transfer and dissemination of knowledge and skills within Africa. Also, some nodes have already or plan to set up their own bioinformatics centers dedicated to training and research in bioinformatics. An example is the Moroccan Center of Bioinformatics, which was launched in Tangier in September 2015 with the support of H3ABioNet.
Building computing infrastructure
Analysis and management of large data sets requires computing resources and internet bandwidth, both of which can be limiting in Africa. An infrastructure working group developed a set of specifications required for different types of genomic data analyses, and nodes purchased the equipment relevant to their needs. A total of 15 servers, 512 cores, 2384 GB RAM, and 120 TB storage have been placed across the continent in the last two years. Systems administrators at these nodes were trained on how to install and manage their equipment, as well as in high-performance and cloud computing. A systems administrator task force was set up to provide ongoing support to ensure that the equipment is used effectively. This task force is also available, through requests to our help desk, to address the information technology needs of development and associate nodes as well as H3Africa project sites with little expertise.
Internet connections in Africa are often slow and unreliable. However, there is a clear need for research groups across the continent to be able to share data sets larger than those that can be attached to a standard e-mail. The Globus Online service, based on the GridFTP data transfer protocol, provides a transparent, reliable, secure, and fault-tolerant method for transferring large data sets over the internet, regardless of the bandwidth and availability of the path between sender and recipient (Foster 2011). H3ABioNet has installed and tested Globus Online endpoints at a gradually increasing number of nodes, with the goal to reach all of them by early 2016. In parallel, it has developed tools based on the iPerf toolbox (http://iperf.sourceforge.net/) to monitor the performance of network connections and is installing these at the same time.
Tool development and user support
H3ABioNet is working on addressing specific training and development needs of the H3Africa Consortium for all aspects of projects, from patient databases to laboratory information management system (LIMS) interoperability to genomic data processing, analysis and interpretation. As an example, the National Institute of Hygiene node is setting up a LIMS application in the National Institute of Hygiene of Morocco. This project, cofinanced by the European Union is taking advantage of the training received by the members of the node through H3ABioNet. Some consortium-wide needs being addressed include mapping of biospecimen, phenotype and experimental data, to ontologies and development of a query interface for these data to provide a harmonized catalog of the consortium products that will be available in biorepositories and public databases. It is also developing a tool for tracking patient recruitment from different sites across projects and an H3Africa data archive to facilitate submission to the European Genome-phenome Archive (EGA), ensuring compliance with the consortium data sharing, access, and release policy.
H3ABioNet has facilitated transfer of large data sets from collaborating sites abroad using Globus Online and provided support for trivial and complex data analysis queries posed to the help desk. In order for H3ABioNet nodes to effectively contribute to the analysis of high-throughput data generated by H3Africa projects, it is essential that they first demonstrate that they have mastered the requisite analytical and computational methods for this analysis. An ad hoc task force was created to develop standard operating procedures (SOPs) for analysis of data that will be produced by H3Africa projects, i.e., genotypes derived from SNP arrays, and high-throughput data from whole-exome or whole-genome sequencing. These SOPs are available from the website (http://www.h3abionet.org/tools-and-resources/sops) and include suggested software and computing resources required for the pipeline. The task force has also produced practice and test data sets for the nodes to hone their skills. The nodes are being evaluated on their performance for specific workflows using an evaluation protocol and independent external experts who judge the quality of the analysis. This assessment will go a long way toward establishing the expertise of African groups in the analysis of human genomic data and instilling confidence in our clinical genetics colleagues that this analysis can be performed to a high standard on the continent. For general bioinformatics support, a user-friendly online help desk is available (http://www.h3abionet.org/support), backed by a team with wide ranging expertise.
Human capacity development
A key aim of H3ABioNet is to enable African scientists to analyze their own data. This is being achieved through a well-coordinated training and support program. The training program for bioinformatics users (usually biologists and clinicians) includes short specialized courses and internships in the nodes (Tastan Bishop et al. 2015). This approach is also being used to train bioinformaticians, with a stronger focus on the technical aspects of the topics as well as on computational biology, statistics, and computer science. Where internet is limited, we make use of eBioKits (http://www.ebiokit.eu/), which are stand-alone devices for bioinformatics data analysis, and we also try to ensure the participants have access to comparable tools and resources when they return home. Some nodes with a better internet connectivity work with Galaxy (https://galaxyproject.org/), which is a web-based analysis platform, where researchers can perform, reproduce, and share data and complete workflows. H3ABioNet has developed training-related resources to aid with the development and organization of new courses and is actively following up with trainees to determine the impact of courses through a trainer–trainee database. In order to reach as wide an audience as possible, we have used the Vidyo system (www.vidyo.com) to live stream courses to distant classrooms and record lectures to make them available to others in the network. To enhance and sustain scientific cooperation between trainees within H3ABioNet and H3Africa, we launched a bioinformatics webinar series involving H3ABioNet staff that is open to the whole H3Africa Consortium. H3ABioNet members also work with the Global Organisation for Bioinformatics Learning, Education and Training (GOBLET) (Attwood et al. 2015) to ensure training activities are aligned with international education and training best practices.
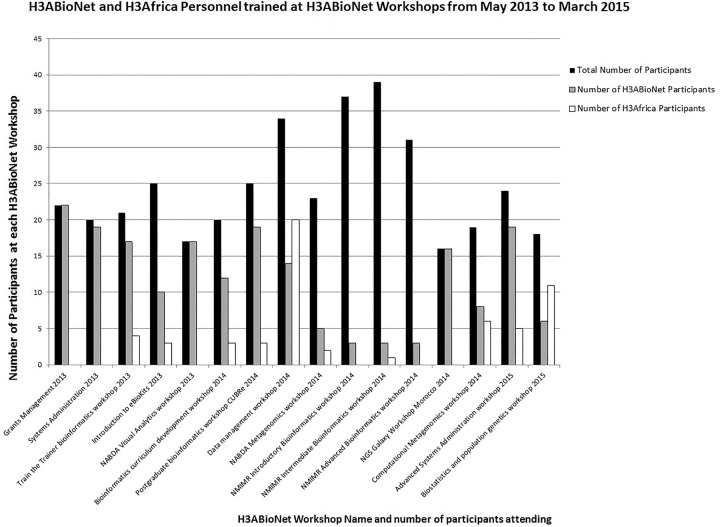
To date, H3ABioNet has run courses ranging from the technical (systems administration, cloud computing, data security), to quantitative (biostatistics, programming), to applied (genome-wide association studies [GWAS], NGS, metagenomics, etc.) areas of computational biology and genomics, training more than 450 African students and researchers (summarized in Fig. 2). Through these and a dedicated train-the-trainer program, courses are increasingly being taught by local trainers. Training faculty is also essential to build the next generation of bioinformatics educators and supervisors, which are currently in short supply on the continent. In parallel with focused training, the network is currently developing curricula for degree programs at African institutions and has established an African Bioinformatics Education Committee to achieve this. In this way, we are addressing the very source of the problem of insufficient trained human capacity in bioinformatics. The first Masters program in bioinformatics in Mali started in early 2015, using the curriculum provided by this committee. Although it reaches a smaller audience, the H3ABioNet internship program has also been successful at enabling skills transfer and analysis of existing data sets. For example, sickle cell disease GWAS data sets generated from prior projects that were not yet analyzed due to lack of skills were analyzed during an internship at an H3ABioNet node. The H3ABioNet training program is closely aligned with that of the larger H3Africa Consortium to ensure coordination of training activities and that the bioinformatics training needs of the consortium are met.
Figure 2.
Summary of H3ABioNet courses run and number of people trained.
Research
Although research activities were not prioritized by the NIH for the bioinformatics network, some research has been necessary, such as development of new algorithms, visualization tools, and workflows relevant to genomic analysis of African populations. Specific examples include the development of a job management system (JMS), a structural analysis pipeline for nonsynonymous coding variants, an improved recombination tool for human sequences, more robust admixture analysis tools for multiway admixed populations, and new databases integrating genotype and phenotype data. The JMS (Brown et al. 2015) has been developed for research groups of the H3Africa Consortium for two distinct purposes: managing and monitoring their servers and using and developing tools and workflows. Currently, a number of H3ABioNet members are working to put pipelines and workflows together with the aim of making them public via JMS. One recently developed pipeline, for ligand docking and molecular dynamics calculations, has already been applied to identify potential hits for malaria (Musyoka et al. 2015).
In addition to the development of new algorithms or pipelines, H3ABioNet is using research projects as a capacity building exercise, promoting inter-node collaborations in which groups with complementary areas of expertise can collectively work on a data set and facilitate cross-pollination of skills. A number of collaborative research projects have been established between participating nodes, which we hope will lead to more efficient addressing of health issues in a resource-poor environment, increased research output from the continent, and promotion of skills retention to ensure long-term sustainability of the network and of genomic medicine research in Africa. One example is a collaboration between CERMES in Niger and the H3ABioNet node at the University of Illinois, working on genetic polymorphism data of Plasmodium falciparum populations infecting patients in different parts of Niger. Their analysis has revealed extensive polymorphisms in the sizes of the parasite's msp1 and msp2 genes, indicating the presence of different strains in different parts of the country as well as the fact that most patients had been multiply infected (O Ouwe Missi Oukem, pers. comm.). The H3ABioNet Node at the National Biotechnology Development Agency (NABDA), Abuja, Nigeria, is conducting research on reference genome sequences of the Human Microbiome Project (Isokpehi et al. 2014). The primary goal is to understand metabolic pathways encoded in microbial genomes that could influence human health. Additionally, in collaboration with the Institute for Genomic Biology at the University of Illinois, metagenomics data have been generated on leg ulcer specimens from individuals with sickle cell disease in Ibadan, Nigeria. The H3ABioNet NABDA node is coordinating the sickle cell leg ulcer study, in which dedicated researchers are analyzing the data sets for biological significance and developing models and workflows for integrating and evaluating data sets (including human genome variants, microbial community diversity, and socioeconomic determinants) to better understand genetic and environmental predispositions to leg ulcers in sickle cell disease.
Another research project was initiated by students attending an H3ABioNet course at Covenant University, Ota, Nigeria. They started a project to study the genetic components underlying uterine fibroid disease among different African populations. They established contacts with relevant research groups around the continent and have submitted a proposal for funding of the project. In a final example of significance to the H3Africa Consortium, members of H3ABioNet are working with international teams to design a new genotyping array that is more relevant to African populations by using more than 3000 full genome sequences from ethnic groups across a broad range of African countries. The array is due to be ready for use by early 2016.
Conclusions
As mentioned previously, the high priority problem H3ABioNet seeks to address is the lack of infrastructure and skills for management and analysis of genomic data. The H3Africa Consortium is preparing to deliver genomic data for more than 50,000 samples, and these data should be analyzed in Africa. The key strategy of H3ABioNet for building bioinformatics capacity for genomics in Africa is pooling of resources and exploiting the skills present to develop the weaker nodes and provide a high-quality training and support platform. This coordinated approach and support provided through mailing lists, working groups, and task forces is a theme that runs through all the H3ABioNet activities to ensure that they are correctly implemented. The network will also contribute to H3Africa and other genomics projects through its legacy of developing and providing access to computing infrastructure. It is responding to the needs of the projects by developing new and supporting existing algorithms, workflows, data management systems, and data integration platforms. A new project on the horizon, which will include contributions from multiple nodes, is the planned development of a resource for mining African variants. It is clear that the H3Africa researchers favor a local resource for their data in addition to the required submission to public repositories.
The establishment of H3ABioNet comes at a time when there has been considerable investment in information communication and technology (ICT) by many African countries. These countries have ensured that the national internet infrastructure is accessible at a subsidized rate to universities and research institutions. Furthermore, the rapid growth of mobile telephony as well as the use of information management systems is changing the way health and biomedical research is being conducted in Africa. As a consequence, strategic investment in bioinformatics in Africa will enable African scientists to become equitable partners in genomic research with scientists in countries outside of Africa. According to the World Bank, sub-Saharan countries comprise one of the fastest growing regions with a projected GDP growth of 4.6–5.1 for 2017. Governments are increasingly investing in higher education and research facilities to tackle their own health and agricultural problems. The African Union (AU) is emphasizing strengthening universities and research institutions with the prevention and control of diseases as one of the priorities. The H3ABioNet initiative is timely and in line with the vision of the AU to enhance the technical competencies of young researchers in the field of bioinformatics for applications in both health and nutrition. Although government support for fields such as bioinformatics is increasing with the increased visibility through projects such as H3ABioNet, most often they do not have funds available to commit. However, with the management and analysis of big data in the biomedical sciences becoming more crucial to research, nodes need to lobby with their governments to encourage investment in developing essential skills in data science for the future. Having said that, involvement in H3ABioNet has had a financial impact on some nodes already, where it has led to cofunding or other institutional support, in a clear demonstration of the positive knock-on effect of the network. H3ABioNet is also building strong informal scientific networks across Africa, which is a very important factor in research capacity development.
Long-term sustainability is a key objective of the network but is not realistically achievable within the first five years of its existence. However, several things have been put in place to contribute to sustainability. The project has increased computing facilities and provided for eBioKits in many nodes, which will remain in place beyond the end of the five-year project. In Egypt, H3ABioNet funds facilitated establishment of an eBioKit-based computer laboratory connected to the internet. Many introductory and advanced bioinformatics workshops are conducted in this laboratory for medical and computer science graduates. These courses are reaching different universities and institutions in Egypt and the Middle East, and course fees are covering the trainers' salaries, stationery, and other logistics. The H3ABioNet train-the-trainer program is contributing to the development of local trainers, and new postgraduate degrees have been developed that are supported by university funds. New staff positions have been created by H3ABioNet, a handful of which have subsequently been taken over by institutions. In addition, joint collaborative project funding proposals are being developed using bilateral agreements between some of the participating countries, and multi-institution research project proposals have been submitted to funding agencies in response to specific calls.
In summary, the funding received from the NIH for the H3ABioNet network provides a unique opportunity to develop durable and sustainable bioinformatics skills among African researchers and to build the necessary computational infrastructure. The enormous genetic and microbiota diversity in Africa, the cradle of humanity, will provide a rich terrain to study the relationship between this diversity and susceptibility to diseases and possibly to suggest new approaches to disease management for the benefit of the continent and the world at large. Through its efforts, H3ABioNet plans to boost biomedical genomics capacity in Africa to accelerate the rate at which African scientists discover the genetic background underlying important diseases.
H3ABioNet Consortium
Marion Adebiyi, Azza E. Ahmed, Rehab I. Ahmed, Maaike Alearts, Mohamed Alibi, Shaun Aron, Shakuntala Baichoo, Hocine Bendou, Gerrit Botha, David Brown, Emile Chimusa, Alan Christoffels, Jennifer Cornick, Jean-Baka Domelevo Entfellner, Chris Fields, Anne Fischer, Junaid Gamieldien, Kais Ghedira, Amel Ghouila, Shannan Ho Sui, Itunuoluwa Isewon, Raphael Isokpehi, Mahjoubeh Jalali Sefid Dashti, Arox Kamng'ona, Radhika S. Khetani, Anmol Kiran, Benard Kulohoma, Benjamin Kumwenda, Dan Lapine, Liudmila Sergeevna Mainzer, Suresh Maslamoney, Mamana Mbiyavanga, Ayton Meintjes, Flora Elias Mlyango, Bruno Mmbando, Somia A. Mohammed, Phelelani Mpangase, Chisomo Msefula, Siana Nkya Mtatiro, Dunfunk Mugutso, Zahra Mungloo-Dilmohammud, Patrick Musicha, Victoria Nembaware, Victor Chukwudi Osamor, Jelili Oyelade, Gloria Rendon, Gustavo A. Salazar, Samson Pandam Salifu, Raphael Sangeda, Oussema Souiai, Peter Van Heusden, Mamadou Wele
Acknowledgments
H3ABioNet is supported by the National Institutes of Health Common Fund (National Human Genome Research Institute) under grant number U41HG006941. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank all the Consortium members for their contributions to H3ABioNet activities. A full list of H3ABioNet Consortium members can be found at http://www.h3abionet.org/home/consortium.
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.196295.115.
Freely available online through the Genome Research Open Access option.
Covenant University Bioinformatics Research (CUBRe) and Department of Computer and Information Sciences, Covenant University, Ota, Ogun State, Nigeria, P.M.B. 1023
Centre for Bioinformatics and Systems Biology, Faculty of Science, University of Khartoum/Future University of Sudan, Khartoum, Sudan 11115
Malawi-Liverpool-Wellcome Trust Clinical Research Programme, Blantyre, Malawi, 3/Institute of Infection and Global Health, University of Liverpool, Liverpool L69 3BX, UK
Institute Pasteur of Tunis, Tunis, Tunisia 1002
Sydney Brenner Institute for Molecular Bioscience, University of the Witwatersrand, Johannesburg, South Africa 2193
University of Mauritius, Reduit, Mauritius 80837
South African National Bioinformatics Institute/Medical Research Council of South Africa Bioinformatics Unit, University of the Western Cape, Cape Town, South Africa 7530
Computational Biology Group, Department of Integrative Biomedical Sciences, Institute of Infectious Disease and Molecular Medicine, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa 7925
Research Unit in Bioinformatics, Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, South Africa 6140
National Center for Supercomputing Applications and Institute for Genomic Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA
International Centre of Insect Physiology and Ecology, Nairobi, Kenya 00100
Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA
National Biotechnology Development Agency, Abuja, Nigeria 10099
Centre for Proteomic and Genomic Research, Cape Town, South Africa 7925
University of Dar es Salaam, Dar es Salaam, Tanzania, 73
Muhimbili University of Health and Allied Sciences, Dar es Salaam, Tanzania 00255
Kumasi Centre for Collaborative Research in Tropical Medicine/Kwame Nkrumah University of Science and Technology, Kumasi, Ghana, PMB
University of Sciences, Techniques and Technology of Bamako, Bamako, Mali BPE 3206
References
- The 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood TK, Bongcam-Rudloff E, Brazas MD, Corpas M, Gaudet P, Lewitter F, Mulder N, Palagi PM, Schneider MV, van Gelder CWG, et al. 2015. GOBLET: the Global Organisation for Bioinformatics Learning, Education and Training. PLOS Comput Biol 11: e1004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Penkler DL, Musyoka TM, Bishop ÖT. 2015. JMS: an open source workflow management system and web-based cluster front-end for high performance computing. PLoS One 10: e0134273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Burchard EG, De la Vega FM. 2011. Genomics for the world. Nature 475: 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimusa ER, Zaitlen N, Daya M, Möller M, van Helden PD, Mulder NJ, Price AL, Hoal EG. 2014. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum Mol Genet 23: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Pepper MJ. 2012. Genomic sovereignty and the African promise: mining the African genome for the benefit of Africa. Med Ethics 38: 474–478. [DOI] [PubMed] [Google Scholar]
- El Baghdadi J, Orlova M, Alter A, Ranque B, Chentoui M, Lazrak F, Archane MI, Casanova JL, Benslimane A, Schurr E, et al. 2006. An autosomal dominant major gene confers predisposition to pulmonary TB in adults. J Exp Med 203: 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster I. 2011. Globus online: accelerating and democratizing science through cloud-based services. IEEE Internet Comput 15: 70–73. [Google Scholar]
- Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, et al. 2015. The African Genome Variation Project shapes medical genetics in Africa. Nature 517: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The H3Africa Consortium. 2014. Research capacity. Enabling African scientists to engage fully in the genomic revolution. Science 344: 1346–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodríguez-Botigué L, Ramachandran S, Hon L, Brisbin A, et al. 2011. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci 108: 5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isokpehi RD, Udensi UK, Simmons SS, Hollman AL, Cain AE, Olofinsae SA, Hassan OA, Kashim ZA, Enejoh OA, Fasesan DE, et al. 2014. Evaluative profiling of arsenic sensing and regulatory systems in the human microbiome project genomes. Microbiol Insights 7: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, Kivinen K, Bojang KA, Conway DJ, Pinder M, et al. 2009. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet 41: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musyoka TM, Kanzi AM, Loob K, Bishop ÖT. 2015. Analysis of non-peptidic compounds as potential malarial inhibitors against plasmodial cysteine proteases via integrated virtual screening workflow. J Biomol Struct Dyn 10.1080/07391102.2015.1108231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Kivisild T, Tarekegn A, Ekong R, Plaster C, Gallego Romero I, Ayub Q, Mehdi SQ, Thomas MG, Luiselli D, et al. 2012. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet 91: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay M, Tiemessen CT, Choudhury A, Soodyall H. 2011. Africa: the next frontier for human disease gene discovery? Hum Mol Genet 20: R214–R220. [DOI] [PubMed] [Google Scholar]
- Rotimi CN, Jorde LB. 2010. Ancestry and disease in the age of genomic medicine. N Engl J Med 363: 1551–1558. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Skoglund P, Sjödin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MG, et al. 2012. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 338: 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. 2010. Complete Khoisan and Bantu genomes from southern Africa. Nature 463: 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirugo G, Hennig BJ, Adeyemo AA, Matimba A, Newport MJ, Ibrahim ME, Ryckman KK, Tacconelli A, Mariani-Costantini R, Novelli G, et al. 2008. Genetic studies of African populations: an overview on disease susceptibility and response to vaccines and therapeutics. Hum Genet 123: 557–598. [DOI] [PubMed] [Google Scholar]
- Tastan Bishop Ö, Adebiyi EF, Alzohairy AM, Everett D, Ghedira K, Ghouila A, Kumuthini J, Mulder NJ, Panji S, Patterton HG, et al. 2015. Bioinformatics education—perspectives and challenges out of Africa. Brief Bioinform 16: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thye T, Vannberg FO, Wong SH, Owusu-Dabo E, Osei I, Gyapong J, Sirugo G, Sisay-Joof F, Enimil A, Chinbuah MA, et al. 2010. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet 42: 739–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmann C, Thye T, Vens M, Evans J, May J, Ehmen C, Sievertsen J, Muntau B, Ruge G, Loag W, et al. 2012. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature 489: 443–446. [DOI] [PubMed] [Google Scholar]
- Wonkam A, Kenfack MA, Muna WF, Ouwe-Missi-Oukem-Boyer O. 2011. Ethics of human genetic studies in sub-Saharan Africa: the case of Cameroon through a bibliometric analysis. Dev World Bioeth 11: 120–127. [DOI] [PubMed] [Google Scholar]