Abstract
Enteric infections, enteropathy and undernutrition in early childhood are preventable risk factors for child deaths, impaired neurodevelopment, and later life metabolic diseases. However, the mechanisms linking these exposures and outcomes remain to be elucidated, as do biomarkers for identifying children at risk. By examining the urinary metabolic phenotypes of nourished and undernourished children participating in a case-control study in Semi-Arid Brazil, we identified key differences with potential relevance to mechanisms, biomarkers and outcomes. Undernutrition was found to perturb several biochemical pathways, including choline and tryptophan metabolism, while also increasing the proteolytic activity of the gut microbiome. Furthermore, a metabolic adaptation was observed in the undernourished children to reduce energy expenditure, reflected by increased N-methylnicotinamide and reduced β-aminoisobutyric acid excretion. Interestingly, accelerated catch-up growth was observed in those undernourished children displaying a more robust metabolic adaptation several months earlier. Hence, urinary N-methylnicotinamide and β-aminoisobutyric acid represent promising biomarkers for predicting short-term growth outcomes in undernourished children and for identifying children destined for further growth shortfalls. These findings have important implications for understanding contributors to long-term sequelae of early undernutrition, including cognitive, growth, and metabolic functions.
Childhood undernutrition is a major global health problem with both acute and long-term consequences. One in five children in the developing world is undernourished and malnutrition is an underlying cause of more than half (53%) of all deaths worldwide in children aged younger than five years1,2. In addition to its role in childhood deaths, undernutrition in early life leads to poor ponderal and linear growth, which reflects the combined effects of both acute (wasting) and chronic undernutrition (stunting). Nutritional interventions have reduced mortality in undernourished children but the long-term consequences for children who survive remains a major challenge. Two decades of accumulating evidence link low birth weight to an increased risk for both cardiovascular disease (CVD) and type 2 diabetes. More recent studies indicate that early childhood stunting is associated with cognitive impairment and increased risk of obesity and CVD in later life3. However, the mechanisms underlying these long-term outcomes have not yet been defined.
One variable that may contribute to these outcomes is the collection of microbes residing in the gastrointestinal tract. These gut microbiota have been implicated in gut inflammation, obesity, CVD risk and through the gut-brain axis have been shown to influence brain development4,5,6. By providing the host with a range of extra-genomic metabolic functionality these intestinal microbes extend the biochemical flexibility of the host and hold a significant role in human nutrition. Increasing evidence is emerging to show that early-life malnutrition retards the maturation of the gut microbiota and host acquisition of these metabolic capabilities7,8,9. It is plausible that modulations in gut microbial community assembly could contribute the outcomes associated with early-life nutrition.
Metabolic phenotyping involves the measurement of global sets of metabolites within a biological system and their variation in response to stimuli. Metabolic profiles are the product of genetic and environmental (diet, lifestyle, gut microbial activity) contributions and studying these profiles enables the overall metabolic status of this multi-factorial metabolic system to be assessed. These signatures contain information reflecting healthy physiological processes and pathological events and can be studied to identify metabolic pathways underlying disease risk and for discovering molecular diagnostic and predictive biomarkers. Large-scale metabolic phenotyping (molecular epidemiology) for biomarker discovery has been previously applied to identify unique metabolic signatures of nutrition and aging, and also disease states including autism, prenatal disorders, type 2 diabetes, stress, depressive disorders, hypertension, and CVD10,11,12,13,14,15,16,17,18,19.
In the present work, a metabolic phenotyping approach was applied to characterize the biochemical modulation induced by undernutrition to understand the biomolecular mechanisms underlying undernutrition associated growth and potential cognitive impairment. We have applied a 1H nuclear magnetic resonance (NMR) spectroscopy-based metabolic profiling approach to study the urinary metabolites of children enrolled in a case-control study in Northeast Brazil. This study was supported as a part of the ‘Etiology, Risk Factors and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development’ (MAL-ED) study. Urinary metabolic profiles were studied to elucidate potential biomarkers of undernutrition and to predict subsequent “catch-up” growth in these children. We report here a potential mechanistic role for altered choline and tryptophan metabolism in the effects associated with undernutrition and a potential role for increased nicotinamide N-methyltransferase (NNMT) in catch-up growth with N-methylnicotinamide serving as a biomarker for this adaptation.
Results
Metabolic signature of undernutrition in the urinary metabolome
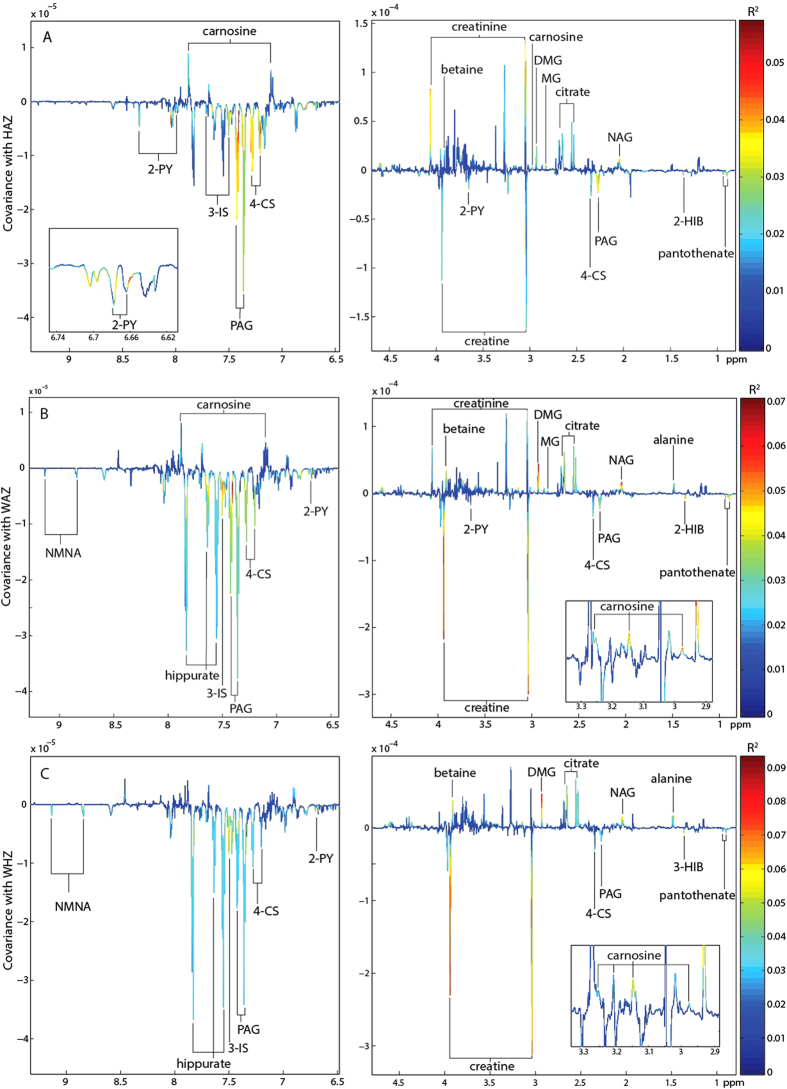
Orthogonal projections to latent structures (OPLS) models with one predictive component and no orthogonal components were calculated to identify urinary metabolites associated with undernutrition in children from Northeastern Brazil (n = 326; 6.2–25.9 months old; 161 male and 165 female). Descriptive information on the children included in these models can be found in the Supplementary Information (Tables S1 and S2). Unit-variance-scaled NMR spectra acquired from urine samples served as the descriptor (X) variables and metrics of nutritional status including height-for-age z scores (HAZ), weight-for-age z scores (WAZ), and weight-for-height z scores (WHZ) were used individually as the response (Y) variable. Valid OPLS models were returned for all metrics analyzed identifying metabolic variation associated with nutritional status (Table 1). Those metabolites associated with stunting (inversely associated with HAZ; where HAZ < −2 indicates moderate stunting) are shown in the coefficients plot extracted from the respective OPLS model (Fig. 1A). Here, stunting was positively associated with creatine, glycerophosphocholine (GPC), N-methyl-2-pyridone-5-carboxamide (2-PY), and pantothenate excretion. In addition, a positive correlation was observed with metabolites arising from gut bacterial-host co-metabolism. This included 2-hydroxyisobutyrate (2-HIB), phenylacetylglutamine (PAG), 4-cresyl sulfate (4-CS), and 3-indoxyl sulfate (3-IS). In contrast, stunting was negatively associated with the excretion of N-acetyl glycoproteins (NAG), citrate, methylguanidine (MG), dimethylglycine (DMG), carnosine, creatinine, betaine, and unknown resonances observed at δ 3.95 and 4.60. Similar observations were found for the models considering wasting (WHZ) and underweight (WAZ). In addition, WAZ and WHZ were also found to positively correlate with N-methyl-nicotinic acid (NMNA) and hippurate excretion and negatively correlate with alanine excretion (Fig. 1B,C). The correlation coefficients indicating the association of these metabolites with the various metrics of undernutrition are summarized in Fig. 2. Spectral peak integrals have been calculated for the metabolites identified to change with relation to these measures of nutritional status and are presented in the Supplementary Information Table S3.
Table 1. Summary of the orthogonal projection to latent structures (OPLS) models returned for the various measures of nutritional status.
| Model | n | R2X | R2Y | Q2Y | P |
|---|---|---|---|---|---|
| height-for-age z score | 326 | 0.045 | 0.14 | 0.035 | 0.001 |
| weight-for-age z score | 326 | 0.039 | 0.18 | 0.039 | 0.001 |
| weight-for-height z score | 326 | 0.035 | 0.20 | 0.016 | 0.002 |
| ΔHAZ | 252 | 0.031 | 0.19 | −0.021 | — |
| ΔHAZ (HAZ0>−1) | 79 | 0.027 | 0.37 | −0.41 | — |
| ΔHAZ (HAZ0<−1) | 173 | 0.048 | 0.20 | 0.028 | 0.0040 |
| ΔHAZ (HAZ0<−2) | 108 | 0.044 | 0.28 | 0.030 | 0.0190 |
ΔHAZ, change in height-for-age z-score over two to five months. HAZ0, height-for-age z-score at baseline.
Figure 1. Orthogonal projection to latent structures (OPLS) model identifying metabolic associations with measures of undernutrition.
Coefficients plot extracted from the OPLS model constructed from the urinary 1H NMR profiles and corresponding (A) height-for-age z (HAZ)-scores (B) weight-for-age z (WAZ)-scores and (C) weight-for-height z (WHZ)-scores. 2HIB, 2-hydroxyisobutyric acid; 2-PY, N-methyl-2-pyridone-5-carboxamide; 3-IS, 3-indoxyl sulfate; 4-CS, 4-cresyl sulfate; DMA, dimethylamine; DMG, dimethylglycine; MG, methylguanidine; NAG, N acetylglycoprotein; NMNA, N-methylnicotinic acid; PAG, phenylacetylglutamine.
Figure 2. Summary of the metabolites associated with the OPLS models given by the correlation coefficient (R) with the response variable.
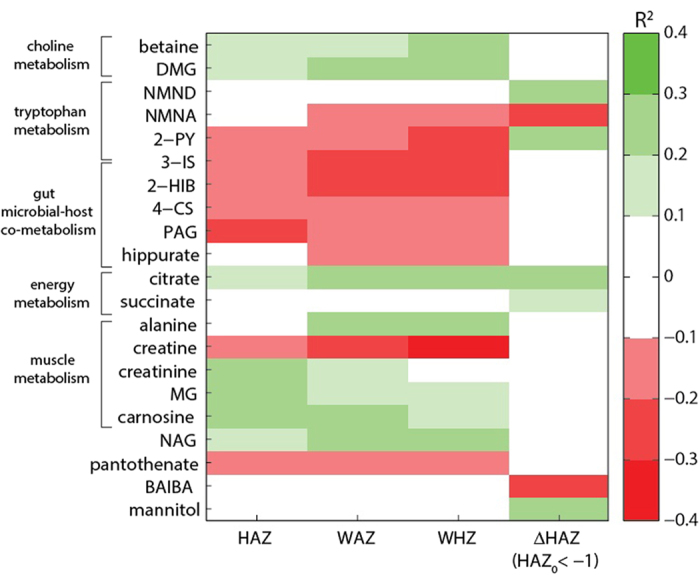
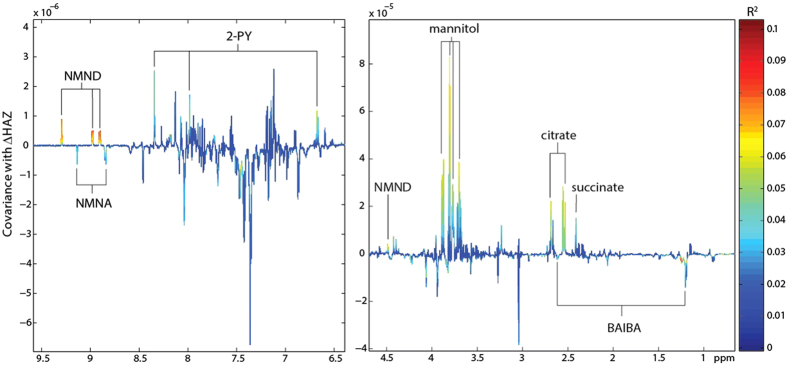
HAZ, height-for-age z-score; WAZ, weight-for-age z-score; WHZ, weight-for-height z-score; ΔHAZ, change in height-for-age z-score over two to five months. 2HIB, 2-hydroxyisobutyric acid; 2-PY, N-methyl-2-pyridone-5-carboxamide; 3-IS, 3-indoxyl-sulfate; 4-CS, 4-cresyl sulfate; BAIBA, β-aminoisobutyric acid; DMA, dimethylamine; DMG, dimethylglycine; MG, methylguanidine; NAG, N acetylglycoprotein; NMNA, N-methylnicotinic acid; NMND, N-methylnicotinamide; PAG, phenylacetylglutamine.
Urinary N-methylnicotinamide (NMND) predicts catch-up growth (ΔHAZ)
Measures of the change in HAZ (ΔHAZ) from baseline to 2–5 month follow-up were available for 252 children whose urinary metabolic phenotypes were defined at baseline. Missing data analyses were performed and no significant differences were observed in gender (p = 0.52), age (p = 0.26) or HAZ status (p = 0.49) at baseline for children with or without follow-up anthropometry. OPLS models were constructed on baseline urinary metabolic phenotypes to elucidate metabolic information associated with subsequent growth 2–5 months later (ΔHAZ). No significant metabolic associations were observed with ΔHAZ when all children were modeled. However, stratifying the sample set to examine metabolic correlates with ΔHAZ in stunted children or those at risk of stunting (HAZ < −1 (more than one standard deviation below the population median); n = 173) returned a significant OPLS model (ΔHAZ range: −0.92–1.39; Fig. 3; metabolic associations (correlation coefficients) are summarized in Fig. 2). This model identified a positive relationship between the excretion of N-methylnicotinamide (NMND), 2-PY, succinate and citrate at baseline and growth (ΔHAZ) measured 2 to 5 months later. The baseline excretion of β-aminoisobutyric acid and N-methylnicotinic acid were also negatively correlated with ΔHAZ in these children. In addition, the excretion of mannitol, provided as part of the lactulose-mannitol test to assess small bowel surface area, was also positively associated with growth. Further refinement of the sample set to look at metabolic associations with ΔHAZ in stunted children (baseline HAZ < −2; n = 108; ΔHAZ range: −0.92–1.39) also returned a significant model (see Supplementary Fig. S1 online). The metabolic alterations observed with this model were consistent with those identified in the earlier model (baseline HAZ < −1). No association was identified between baseline metabolic signatures and ΔHAZ in the non-stunted (HAZ > −1; n = 82) children.
Figure 3. OPLS coefficients plot identifying metabolic predictors of growth in stunted children or those at risk of stunting.
Baseline urinary metabolic profiles of children with a baseline HAZ <−1 correlated with ΔHAZ measured two to five months later. 2-PY, N-methyl-2-pyridone-5-carboxamide; BAIBA, β-aminoisobutyric acid; NMNA, N-methylnicotinic acid; NMND, N-methylnicotinamide.
Discussion
Combining field studies of undernutrition with a 1H NMR spectroscopy-based metabolic profiling approach we obtained a deeper understanding of the metabolic pathways altered in the setting of early childhood growth stunting and, in turn, identified potential biochemical mechanisms underlying undernutrition associated growth and cognitive impairment. In this study similar metabolic alterations were found across all measures of stunting (HAZ), underweight (WAZ) and wasting (WHZ) with the exception of NMNA and alanine, which were uniquely associated with measures of underweight and wasting, and creatinine which was unique to stunting and underweight. This is consistent with the coexistence of these anthropometric failures in the undernourished children from this cohort.
Undernourished children were found to excrete lower amounts of betaine and dimethylglycine (DMG) compared to the nourished controls, indicating perturbations in choline metabolism (Fig. 4). Dietary choline is rapidly absorbed in the small intestine and catabolized to betaine and then DMG. Reduced excretion of these metabolites in the undernourished children indicates a lower availability of choline (via reduced intake or absorption) compared to nourished controls. Choline can be obtained from many dietary sources and the human infant typically consumes a choline-rich diet, with breast milk containing ~1.5 mmol/L choline and choline esters20. In this study no significant association was identified between breast-feeding and stunting (data not shown), although those children exposed to mixed breast-feeding practices did excrete more betaine and DMG compared to those not receiving breast milk (see Supplementary Fig. S2 online). However, when considering all study children simultaneously, stunting was found to negatively correlate with betaine and DMG excretion, which are surrogate markers of choline bioavailability, irrespective of breast milk exposure.
Figure 4. Metabolic pathways modulated by undernutrition.
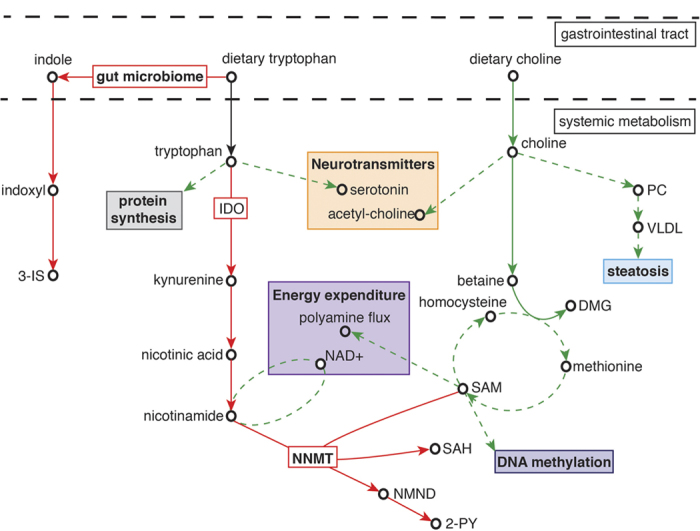
Red solid arrows indicate pathways increased with undernutrition (negatively associated with HAZ, WAZ, WHZ) and green solid arrows indicate pathways decreased with undernutrition (positively associated with HAZ, WAZ, WHZ). Dashed green arrows indicate pathways and biological functions hypothesized to decrease as a result of these metabolic alterations. 3-IS, 3-indoxyl-sulfate; DMG, dimethylglycine; IDO, indoleamine 2,3-dioxygenase; NAD, nicotinamide adenine dinucleotide; NMND, N-methylnicotinamide; NNMT, nicotinamide N-methyltransferase; PC, phosphatidylcholine; SAH, S-adenosyl-L-homocysteine; SAMe, S-adenosyl-L-methionine; VLDL; very-low-density lipoproteins.
Choline and betaine are major sources of methyl groups used to generate the body’s primary methylating agent, S-adenosylmethionine (SAMe). Betaine is converted to DMG as it methylates homocysteine to methionine, which is subsequently used to generate SAMe. Lower excretion of betaine and DMG by undernourished cases is consistent with reduced flux through the betaine pathway to produce DMG and homocysteine. This has potential downstream consequences for SAMe abundance and others have demonstrated a strong positive correlation between plasma choline and plasma SAMe21.
SAMe serves as the sole methyl donor in a multitude of cellular methylation reactions including epigenetic processes such as DNA and histone methylation. Diet related modulation of DNA methylation has been previously shown in animal and human studies (reviewed in McKay et al. 201122). For example, dietary choline deficiency results in DNA hypomethylation of mouse brains23. During early-life epigenetic regulation is dynamic and the epigenome has a developmental plasticity that is susceptible to dietary modulation. Alterations to these developing epigenetic marks have potential to modify gene expression and cellular function. Evidence is emerging that critical windows may exist to allow diet-associated variations in epigenetic programming to exert strong long-term impacts on health24,25. This was demonstrated in a rodent study where sustained methyl-group deficiency resulted in a reduced growth rate and elevated hepatic fat content26, outcomes attenuated by supplemental feeding of choline and methionine27. Exposure to a nutritionally limited environment in early-life is hypothesized to result in the fine-tuning of the epigenome to prime individuals towards metabolic “thrift”. Such early-life programming could compromise individuals in later life when this thrifty phenotype experiences an energy-rich environment. Consistently, deficiencies in both choline and betaine have been suggested to produce epigenetic changes in genes linked to atherosclerosis28 and brain development23,29.
One advantage of metabolic profiling is the ability to capture the influence of the gut microbiota on the metabolic status of the host. Increased excretion of gut bacterial-host co-metabolites derived from amino acid metabolism in the undernourished cases implies a functional modulation in the gut microbiome towards protein breakdown. This includes the excretion of phenylacetylglutamine (PAG), 4-cresyl sulfate (4-CS), and 3-indoxyl-sulfate (3-IS) derived from phenylalanine, tyrosine and tryptophan, respectively. Here, phenylalanine is converted to phenylacetate by the colonic bacteria and is subsequently conjugated with glutamine in the liver and the gut mucosa to form PAG. 3-IS and 4-CS are the products of the hepatic sulfation of indole and 4-cresol, which are end-products of the bacterial metabolism of tryptophan and tyrosine in the colon, respectively. Greater proteolytic activity of the undernourished microbiome is further supported by the increased excretion of 2-hydroxyisobutyrate, a bacterial degradation product of dietary proteins, in the undernourished children compared to the better nourished, control children30. Given that dietary intake is the main source of substrate for the gut microbiota it is unsurprising that undernutrition would imprint on microbiomic activity. Previous studies have shown gut microbial variation associated with kwashiorkor and with less severe forms of malnutrition8,9. A recent study in Indian children found that carbohydrate-active enzyme (CAZymes) families in the gut microbiome were positively associated with the nutritional status of the children7, consistent with our findings identifying a proteolytic enterotype associated with undernutrition. This may have consequences for the host given that metabolites generated from carbohydrate fermentation are generally considered beneficial for the host while those arising from protein fermentation tend to be more toxic31.
Greater urinary excretion of 3-IS and N-methyl-2-pyridone-5-carboxamide (2-PY) by the undernourished children suggests a dysregulation of tryptophan metabolism associated with undernutrition. There are three major metabolic routes for tryptophan degradation: the kynurenine, serotonin, and the indolic pathway. The kynurenine pathway is the major (>95%) catabolic route for tryptophan, resulting in the production of nicotinic acid, its amine form nicotinamide and several neuroactive intermediates (kynurenic acid, quinolinic acid, picolinic acid), whereas 1–2% of dietary tryptophan is used for the synthesis of the neurotransmitter serotonin. Approximately 4–6% of tryptophan undergoes bacterial degradation in the gut to produce indole, which is subsequently metabolized in the host to indoxyl and then to 3-IS or indoxyl-β-D glucuronide. In this study, undernutrition was positively associated with greater excretion of 3-IS, N-methylnicotinic acid, and 2-PY, a metabolite of nicotinamide, showing a greater metabolism of tryptophan through the bacterial-mediated indolic pathway and the endogenous kynurenine pathway with undernutrition (Fig. 4).
Tryptophan is an essential amino acid and its use in a wide variety of physiological functions result in a relatively low abundance in the body. Since tryptophan cannot be synthesized a large amount of tryptophan is used in protein synthesis and is essential for growth. In pigs, a moderate deficiency in tryptophan has been shown to significantly reduce growth rate32. Enhanced metabolism of tryptophan in the undernourished children through these two major pathways could reduce the bioavailability of tryptophan with potential implications for growth and serotonin biosynthesis.
Activation of the kynurenine pathway may also be driven by immune responses. Two enzymes are capable of metabolizing tryptophan to kynurenine. Tryptophan 2,3-dioxygenase (TDO) is primarily expressed in the liver and is considered a protective mechanism to prevent toxic amounts of tryptophan accumulating in plasma and tissues. Indoleamine 2,3-dioxygenase (IDO) is an enzyme present in several extra-hepatic tissues and its expression is increased in response to infection and inflammation33,34. One driver of systemic inflammation is increased intestinal permeability or a “leaky gut” frequently observed with undernutrition and persistent infection. This IDO-mediated activation of the kynurenine pathway depletes tryptophan from the local environment and deprives T-cells of tryptophan needed for activation and proliferation upon encountering potentially deleterious microbial antigens. In addition, downstream metabolites of the kynurenine pathway actively suppress the proliferation of activated T cells35. Therefore, inflammation-induced IDO activation down-regulates the initial immune response, leading to “secondary” immunosuppression. This is consistent with the lower excretion of N-acetylglycoprotein and carnosine in undernourished compared to control children. N-acetylglycoproteins are frequently associated with inflammation-induced acute phase proteins and several studies have demonstrated that carnosine has strong and specific antioxidant properties and the ability to promote wound healing36. In this context, carnosine scavenges reactive oxygen species generated by cells during an acute inflammatory response and enhances the process of wound healing by stimulating effusion at the initial stage of inflammation. Hence, chronic inflammation and infections in undernourished children may exert prolonged pressure on systemic tryptophan levels. Coupled with the enhanced consumption of tryptophan by the gut microbiome of these individuals this may have detrimental consequences for the regulation of growth, behavior, cognition and immune responses.
The principal branch of the kynurenine pathway generates quinolinic acid and nicotinamide. In humans, nicotinamide is methylated by the SAMe-dependent enzyme, nicotinamide N-methyltransferase (NNMT), to form N-methylnicotinamide (NMND), which is further metabolized to 2-PY. Recently, NNMT has been identified as a novel regulator of energy expenditure where knockdown of NNMT expression increased nicotinamide and SAMe37. Elevations in nicotinamide, a NAD+ precursor, have been shown to increase energy expenditure and by entering the polyamine flux, SAMe has also been shown to have a similar metabolic effect38. By increasing energy expenditure NNMT knockdown protected against diet-induced obesity. Here, undernourished children excreted greater amounts of 2-PY than the control children. This may reflect a metabolic adaptation in these children to increase NNMT activity and reduce nicotinamide and SAMe abundance, thereby lowering energy expenditure. Intriguingly, stunted children or those at risk of stunting who excreted greater amounts of NMND and 2-PY at baseline were found to have better growth 2 to 5 months later. These children also excreted lower amounts of β-aminoisobutyric acid at baseline, another metabolite involved in the regulation of energy expenditure. Through PPARα-mediated increases in white adipocyte thermogenesis and increased hepatic fatty acid β-oxidation, β-aminoisobutyric acid has been shown to increase energy expenditure and protect against diet-induced obesity39,40. Similarly, children with better growth excreted lower amounts of this metabolite. As with the changes observed in NMND and 2-PY excretion, this metabolic alteration could potentially reflect a reduction in energy expenditure. Hence, the better growers may adapt more effectively than those with poorer growth to their nutritionally limited environment by reducing energy expenditure in favor of growth. This metabolic adaption could predispose the individual to higher risk of obesity or insulin resistance in later life. Indeed, up-regulation of NNMT activity and a subsequent increase in plasma NMND has been observed with atherosclerosis and insulin resistance and increases in urinary NMND and 2-PY have been reported with type 2 diabetes15,41,42. Up-regulation of NNMT in cancer cells has also been found to impair SAMe-mediated methylation of DNA and histones43. As such, increased NNMT activity in these children could place additional pressure on their methylation capacity with epigenetic consequences.
Using a metabolic phenotyping approach we have shown disruptions to choline and tryptophan metabolism that could explain later-life outcomes of early-life undernutrition and potential effects on the epigenome due to the reduced methylation capacity of undernourished children. Furthermore, metabolic adaptation may occur in stunted children to restrict energy expenditure to promote growth with potential implications for later life. As markers of this adaptation, urinary NMND and β-aminoisobutyric acid may provide useful non-invasive biomarkers for growth propensity in children at risk of stunting.
Methods
Population and study design
The children participating in this study were mainly from Fortaleza and neighbouring municipalities in Ceará, Brazil. The study was conducted from August 2010 to September 2013 at the Institute for the Promotion of Nutrition and Human Development (IPREDE) in collaboration with the Institute of Biomedicine, Faculty of Medicine, Federal University of Ceará. This case-control study was part of the Malnutrition-Enteric Diseases (MAL-ED) network and the protocol and consent form were approved by the local institutional review board at the Federal Universisty of Ceará, the national IRB Conselho Nacional de Ética em Pesquisa (CONEP), and the University of Virginia in the United States. All work was done in accordance with these relevant guidelines and regulations and informed consent was obtained from the responsible guardians of all subjects.
Children who were being seen in the IPREDE clinic whose screening WAZ was <−2 were invited to participate, consent (parental or guardian) and to return to enrol. Approximate (i.e. within 6 months) age and gender matched ‘control’ children whose WAZ was >−1 were then invited to participate, consent and return to enrol. The inclusion criteria were as follow: (a) 6–24 months of age; (b) cases defined as CDC/WHO weight-for-age z score <−2 and controls >−1; (c) being healthy (i.e., without any specific illness or fever); (d) mother/primary care-giver present and have legal custody of the child. The exclusion criteria were as follows: (a) children with serious health issues or who require prolonged hospitalization; or (b) a parent or care-giver with cognitive deficits or who was <16 year old. Children were enrolled after the parents or guardians of children who met the entry criteria provided informed consent. Cases and controls had their height and weight measured at enrolment and, when possible, at follow up 2–5 months later at the IPREDE clinic. An initial power analysis was used to estimate that 200 each of Cases and Controls should be sufficient to detect expected differences between undernourished and nourished children. The same number of Cases (201) and Controls (201) as well as equal numbers of boys and girls were enrolled. At their first visit, urine was collected and a 4-hour lactulose-mannitol urinary excretion test of gut function was performed. Of the 402 children enrolled, 337 provided adequate urine samples for metabolic analyses.
1H NMR spectroscopy
All urine samples were analyzed by 1H nuclear magnetic resonance (NMR) spectroscopy. For spectroscopic analysis, 400 μL of urine was combined with 200 μL of phosphate buffer (pH 7.4; 100% D2O) containing 1 mM of 3-trimethylsilyl-1-[2,2,3,3-2H4] propionate (TSP) as an external standard and 2 mM sodium azide as a bacteriocide. Samples were mixed by vortex and spun (13000 g) for 10 minutes prior to transferring 550 μL to a 5 mm NMR tube. Quality control samples were prepared by combining aliquots of urine (10 μL) from randomly selected individuals. Specimens were randomized and interspersed with quality control aliquots (using a total of 10 aliquots) in order to assess data quality and variation over the analytical measurement period. Spectroscopic analysis was carried out on a 700 MHz Bruker NMR spectrometer equipped with a cryo-probe. Standard one-dimensional 1H NMR spectra of the urine samples were acquired [recycle delay (RD)−90°−t1−90°−tm−90°-acquire free induction decay]. The water signal was suppressed by irradiation during the RD of 2 s, with a mixing time (tm) of 10 μs. The acquisition time was set to 2.91 s and the 90° pulse length was 15.87 μs. For each sample, 8 dummy scans were followed by 128 scans and collected in 64K data points using a spectral width of 16 ppm. Two-dimensional 1H–1H correlation spectroscopy, 1H–1H total correlation spectroscopy, 1H–13C heteronuclear single quantum coherence and J-resolved NMR spectra were acquired to aid metabolite identification in addition to statistical total correlation spectroscopy44.
Spectra were manually phased, corrected for baseline distortions and referenced to the TSP signal at δ 0.00. 1H NMR spectra (δ 0.2–10.0) were digitized into consecutive integrated spectral regions (~20,000) of equal width (0.00055 ppm). The regions between δ 4.7–6.10 containing the residual water resonance and the urea peak were removed from all spectra in order to minimize the effect of baseline effects caused by imperfect water suppression. For each spectrum a recursive segment-wise peak alignment (RSPA) algorithm was applied to minimize chemical shift variation due to residual pH differences within samples45. Each spectrum was then normalized to unit area to account for variation in sample concentration.
Multivariate data analysis
Multivariate modeling was performed in Matlab using in-house scripts. This included principal components analysis using pareto scaling and orthogonal projection to latent structures (OPLS) constructed using unit variance scaling. Principal components analysis was first applied to visualize the global variance of the data sets to reveal intrinsic similarities in the spectral profiles and to identify outliers. Here, 11 samples were identified as outliers and removed from further analyses with 326 metabolic profiles remaining (data not shown). OPLS models were constructed to identify features in the urinary spectra associated with undernutrition. These features were then assigned to metabolites using in-house databases and 2D NMR experiments. Here, 1H NMR spectroscopic profiles were used as the descriptor matrix (X) and continuous anthropometric measures (e.g. height-for-age z-score (HAZ)) were used individually as the response variable (Y). Orthogonal signal correction filters remove the variation in the descriptor matrix that is unrelated to the response variable and thus assist model interpretation and aid identification of metabolites associated with the response variable. Loading coefficient plots were generated by back-scaling transformation to display the covariance between the Y-response matrix and the signal intensity of the metabolites in the NMR data (X). Hence, the direction and magnitude of the signals relate to the covariation of the metabolites with the Y-response in the model. Colors projected onto the coefficient plot indicate the correlation coefficient (r2) between each metabolite and the Y-response variable, with red indicating strong significance and blue indicating weak significance. The predictive performance (Q2Y) of the model was calculated using a seven-fold cross validation approach, model validity was established by permutation testing (1000 permutations) and the results are given as p values. ΔHAZ was calculated using follow-up HAZ measurements collected two to five months after baseline and used as a measure of growth in the children. An OPLS model was built using the metabolic profiles from children with a HAZ <−1 at baseline as the X-descriptor matrix and ΔHAZ as the Y-response variable. This allowed metabolic associations with catch-up growth in these children to be identified. This analysis was repeated using children with a HAZ <−2.
Additional Information
How to cite this article: Mayneris-Perxachs, J. et al. Urinary N-methylnicotinamide and β-aminoisobutyric acid predict catch-up growth in undernourished Brazilian children. Sci. Rep. 6, 19780; doi: 10.1038/srep19780 (2016).
Supplementary Material
Acknowledgments
This work was supported by a grant from the Bill and Melinda Gates Foundation entitled, “Novel metabonomic biomarkers of gut function and health: Models of enteropathy (EE) and field validation”, as well as in part as the case-control component of the “Etiology, Risk Factors and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development” Project (MAL-ED) carried out as a collaborative project supported by the Bill & Melinda Gates Foundation, the Foundation for the NIH, and the National Institutes of Health, Fogarty International Center. S.R.M. was supported by Fogarty International Center/NIH award no. K02 TW008767. The authors thank the staff and participants of the MAL-ED Network for their important contributions.
Footnotes
Author Contributions J.M.P., A.A.M.L., R.L.G., A.H., S.M. and J.R.S. wrote the main manuscript. A.A.M.L., R.L.G., A.M.L., A.F.M., N.L.L., A.M.S. and J.R.S. were involved in the study design and sample collection. J.M.P., A.H. and R.P. undertook the experimental work and J.M.P., A.H., R.P., S.M. and J.R.S. contributed to the data analysis. All authors reviewed the manuscript.
References
- Black R. E. et al. Maternal and child undernutrition: global and regional exposures and health consequences. Lancet 371, 243–260, doi: 10.1016/S0140-6736(07)61690-0 (2008). [DOI] [PubMed] [Google Scholar]
- Caulfield L. E., de Onis M., Blossner M. & Black R. E. Undernutrition as an underlying cause of child deaths associated with diarrhea, pneumonia, malaria, and measles. The American journal of clinical nutrition 80, 193–198 (2004). [DOI] [PubMed] [Google Scholar]
- Guerrant R. L., DeBoer M. D., Moore S. R., Scharf R. J. & Lima A. A. The impoverished gut—a triple burden of diarrhoea, stunting and chronic disease. Nature reviews. Gastroenterology & hepatology 10, 220–229, doi: 10.1038/nrgastro.2012.239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R. et al. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America 108, 3047–3052, doi: 10.1073/pnas.1010529108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031, doi: 10.1038/nature05414 (2006). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63, doi: 10.1038/nature09922 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T. S. et al. Gut microbiomes of Indian children of varying nutritional status. PloS one 9, e95547, doi: 10.1371/journal.pone.0095547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. I. et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554, doi: 10.1126/science.1229000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421, doi: 10.1038/nature13421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzmann S. S. et al. Metabolic profiling strategy for discovery of nutritional biomarkers: proline betaine as a marker of citrus consumption. The American journal of clinical nutrition 92, 436–443, doi: 10.3945/ajcn.2010.29672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann J. R. et al. Microbial-mammalian cometabolites dominate the age-associated urinary metabolic phenotype in Taiwanese and American populations. Journal of proteome research 12, 3166–3180, doi: 10.1021/pr4000152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap I. K. et al. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. Journal of proteome research 9, 2996–3004, doi: 10.1021/pr901188e (2010). [DOI] [PubMed] [Google Scholar]
- Diaz S. O. et al. Second trimester maternal urine for the diagnosis of trisomy 21 and prediction of poor pregnancy outcomes. Journal of proteome research 12, 2946–2957, doi: 10.1021/pr4002355 (2013). [DOI] [PubMed] [Google Scholar]
- Diaz S. O. et al. Metabolic biomarkers of prenatal disorders: an exploratory NMR metabonomics study of second trimester maternal urine and blood plasma. Journal of proteome research 10, 3732–3742, doi: 10.1021/pr200352m (2011). [DOI] [PubMed] [Google Scholar]
- Salek R. M. et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiological genomics 29, 99–108, doi: 10.1152/physiolgenomics.00194.2006 (2007). [DOI] [PubMed] [Google Scholar]
- Tomassini A. et al. 1H NMR-based urinary metabolic profiling reveals changes in nicotinamide pathway intermediates due to postnatal stress model in rat. Journal of proteome research 13, 5848–5859, doi: 10.1021/pr500748r (2014). [DOI] [PubMed] [Google Scholar]
- Zheng P. et al. A novel urinary metabolite signature for diagnosing major depressive disorder. Journal of proteome research 12, 5904–5911, doi: 10.1021/pr400939q (2013). [DOI] [PubMed] [Google Scholar]
- Holmes E. et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453, 396–400, doi: 10.1038/nature06882 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap I. K. et al. Metabolome-wide association study identifies multiple biomarkers that discriminate north and south Chinese populations at differing risks of cardiovascular disease: INTERMAP study. Journal of proteome research 9, 6647–6654, doi: 10.1021/pr100798r (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel S. H. & Blusztajn J. K. Choline and human nutrition. Annual review of nutrition 14, 269–296, doi: 10.1146/annurev.nu.14.070194.001413 (1994). [DOI] [PubMed] [Google Scholar]
- Imbard A. et al. Plasma choline and betaine correlate with serum folate, plasma S-adenosyl-methionine and S-adenosyl-homocysteine in healthy volunteers. Clinical chemistry and laboratory medicine : CCLM/FESCC 51, 683–692, doi: 10.1515/cclm-2012-0302 (2013). [DOI] [PubMed] [Google Scholar]
- McKay J. A. & Mathers J. C. Diet induced epigenetic changes and their implications for health. Acta physiologica 202, 103–118, doi: 10.1111/j.1748-1716.2011.02278.x (2011). [DOI] [PubMed] [Google Scholar]
- Niculescu M. D., Craciunescu C. N. & Zeisel S. H. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 20, 43–49, doi: 10.1096/fj.05-4707com (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdge G. C. & Lillycrop K. A. Nutrition, epigenetics, and developmental plasticity: implications for understanding human disease. Annual review of nutrition 30, 315–339, doi: 10.1146/annurev.nutr.012809.104751 (2010). [DOI] [PubMed] [Google Scholar]
- Junien C. Impact of diets and nutrients/drugs on early epigenetic programming. Journal of inherited metabolic disease 29, 359–365, doi: 10.1007/s10545-006-0299-7 (2006). [DOI] [PubMed] [Google Scholar]
- Kang-Lee Y. A. et al. Metabolic effects of nicotinamide administration in rats. The Journal of nutrition 113, 215–221 (1983). [DOI] [PubMed] [Google Scholar]
- Sun S., McKee R., Fisler J. S. & Swendseid M. E. Muscle creatine content in rats given repeated large doses of nicotinamide: effects of dietary methionine, choline, carnitine, and other supplements. The Journal of nutrition 116, 2409–2414 (1986). [DOI] [PubMed] [Google Scholar]
- Zaina S., Lindholm M. W. & Lund G. Nutrition and aberrant DNA methylation patterns in atherosclerosis: more than just hyperhomocysteinemia? The Journal of nutrition 135, 5–8 (2005). [DOI] [PubMed] [Google Scholar]
- Zeisel S. H. The fetal origins of memory: the role of dietary choline in optimal brain development. The Journal of pediatrics 149, S131–136, doi: 10.1016/j.jpeds.2006.06.065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proceedings of the National Academy of Sciences of the United States of America 105, 2117–2122, doi: 10.1073/pnas.0712038105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke K. A. et al. Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutrition research reviews 28, 42–66, doi: 10.1017/S0954422415000037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Floc’h N., Otten W. & Merlot E. Tryptophan metabolism, from nutrition to potential therapeutic applications. Amino acids 41, 1195–1205, doi: 10.1007/s00726-010-0752-7 (2011). [DOI] [PubMed] [Google Scholar]
- Popov A. & Schultze J. L. IDO-expressing regulatory dendritic cells in cancer and chronic infection. Journal of molecular medicine 86, 145–160, doi: 10.1007/s00109-007-0262-6 (2008). [DOI] [PubMed] [Google Scholar]
- Wolf A. M. et al. Overexpression of indoleamine 2,3-dioxygenase in human inflammatory bowel disease. Clinical immunology 113, 47–55, doi: 10.1016/j.clim.2004.05.004 (2004). [DOI] [PubMed] [Google Scholar]
- Maes M., Leonard B. E., Myint A. M., Kubera M. & Verkerk R. The new ‘5-HT’ hypothesis of depression: cell-mediated immune activation induces indoleamine 2,3-dioxygenase, which leads to lower plasma tryptophan and an increased synthesis of detrimental tryptophan catabolites (TRYCATs), both of which contribute to the onset of depression. Progress in neuro-psychopharmacology & biological psychiatry 35, 702–721, doi: 10.1016/j.pnpbp.2010.12.017 (2011). [DOI] [PubMed] [Google Scholar]
- Guney Y. et al. Carnosine may reduce lung injury caused by radiation therapy. Medical hypotheses 66, 957–959, doi: 10.1016/j.mehy.2005.11.023 (2006). [DOI] [PubMed] [Google Scholar]
- Kraus D. et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262, doi: 10.1038/nature13198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen E. et al. Enhanced polyamine catabolism alters homeostatic control of white adipose tissue mass, energy expenditure, and glucose metabolism. Molecular and cellular biology 27, 4953–4967, doi: 10.1128/MCB.02034-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begriche K. et al. Beta-aminoisobutyric acid prevents diet-induced obesity in mice with partial leptin deficiency. Obesity 16, 2053–2067, doi: 10.1038/oby.2008.337 (2008). [DOI] [PubMed] [Google Scholar]
- Roberts L. D. et al. beta-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell metabolism 19, 96–108, doi: 10.1016/j.cmet.2013.12.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannt A. et al. Association of nicotinamide-N-methyltransferase mRNA expression in human adipose tissue and the plasma concentration of its product, 1-methylnicotinamide, with insulin resistance. Diabetologia 58, 799–808, doi: 10.1007/s00125-014-3490-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateuszuk L. et al. Activation of nicotinamide N-methyltrasferase and increased formation of 1-methylnicotinamide (MNA) in atherosclerosis. Pharmacological reports : PR 61, 76–85 (2009). [DOI] [PubMed] [Google Scholar]
- Ulanovskaya O. A., Zuhl A. M. & Cravatt B. F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nature chemical biology 9, 300–306, doi: 10.1038/nchembio.1204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloarec O. et al. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Analytical chemistry 77, 1282–1289, doi: 10.1021/ac048630x (2005). [DOI] [PubMed] [Google Scholar]
- Veselkov K. A. et al. Recursive segment-wise peak alignment of biological (1)h NMR spectra for improved metabolic biomarker recovery. Analytical chemistry 81, 56–66, doi: 10.1021/ac8011544 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.