Abstract
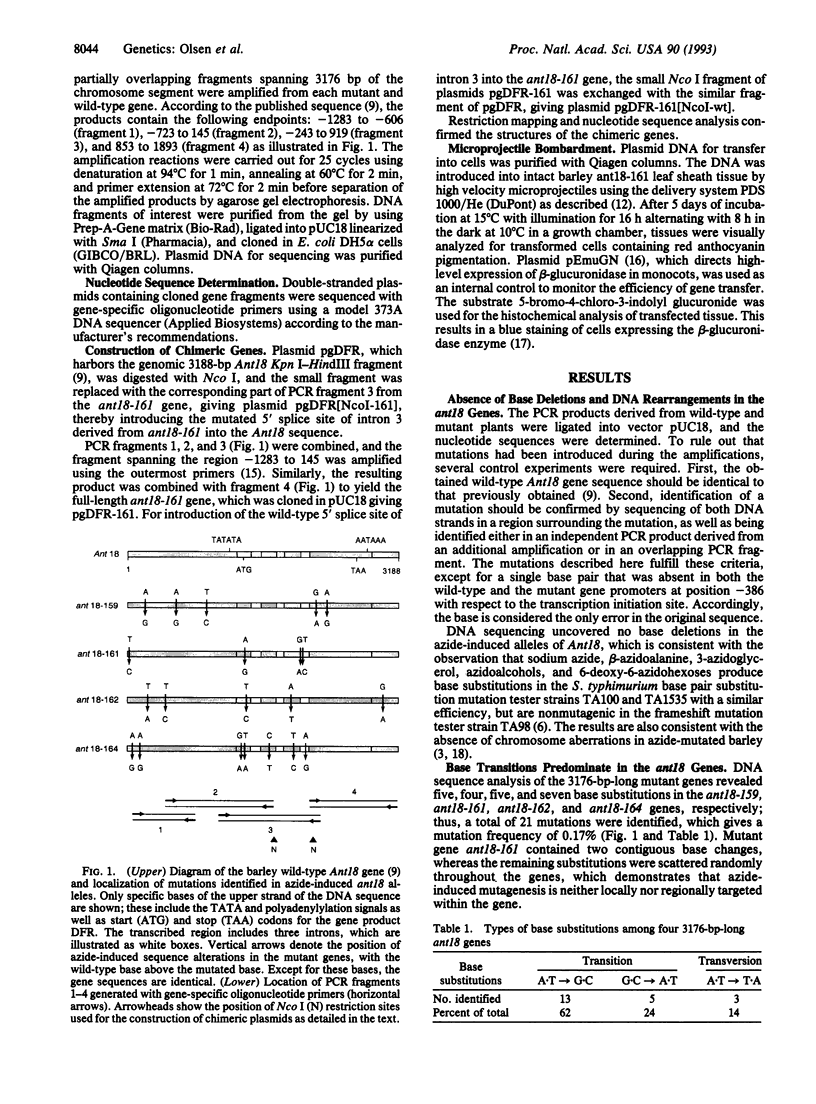
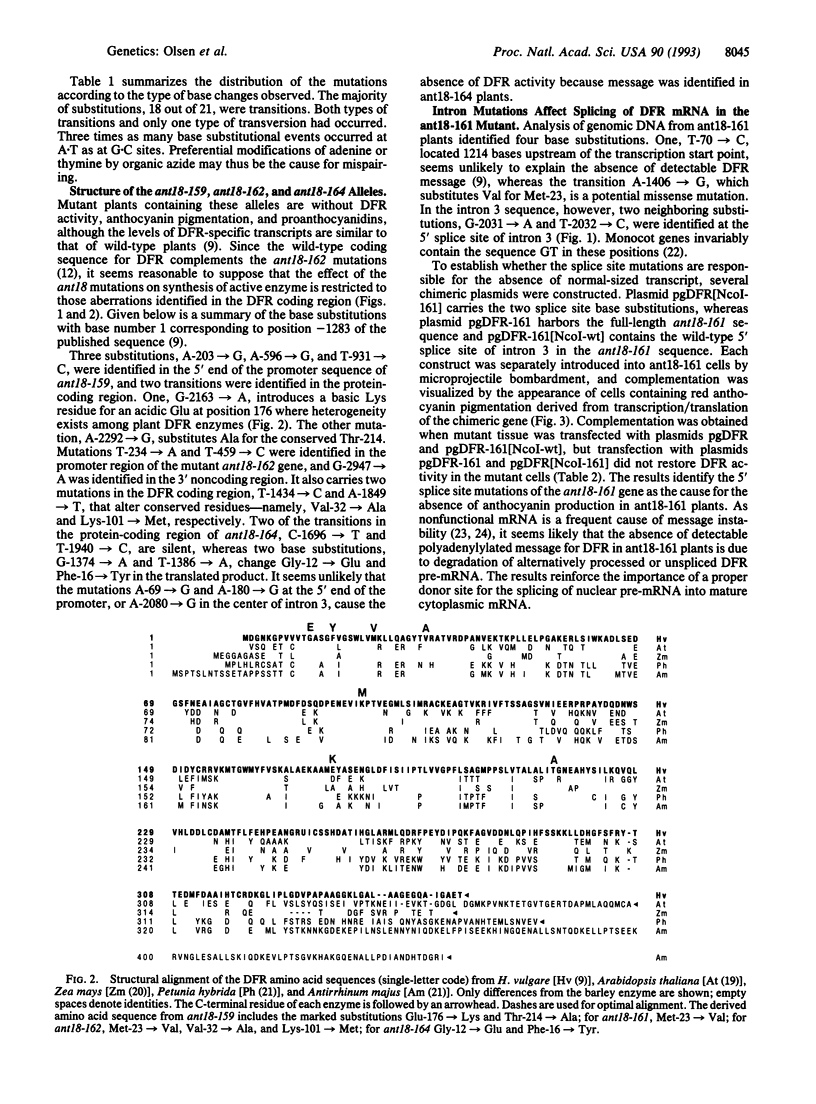
The molecular basis for the absence of anthocyanins and proanthocyanidins in four independent sodium azide-induced ant18 mutants of barley was examined by sequencing the gene encoding dihydroflavonol 4-reductase in these mutants. Sodium azide generated 21 base substitutions, which corresponds to 0.17% of the 12,704 nucleotides sequenced. Of the substitutions, 86% were nucleotide transitions, and 14% were transversions. A.T-->G.C base pair transitions were about 3 times more frequent than G.C-->A.T transitions. No deletions or mutation hot spots were found. The absence of dihydroflavonol 4-reductase activity in ant18-159, ant18-162, and ant18-164 plants is caused by missense mutations in the respective genes. By using microprojectile bombardment, a plasmid harboring the wild-type Ant18 gene was introduced into ant18-161 mutant cells and resulted in the development of anthocyanin pigmentation, which demonstrates that the mutation is corrected by expression of the introduced gene. On the other hand, a plasmid derivative with the two ant18-161-specific base transitions at the 5' splice site of intron 3 prevented complementation. It is concluded that the absence of detectable mRNA for dihydroflavonol 4-reductase in ant18-161 cells is due to the mutations in the pre-mRNA splice donor site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akasaka S., Takimoto K., Yamamoto K. G:C-->T:A and G:C-->C:G transversions are the predominant spontaneous mutations in the Escherichia coli supF gene: an improved lacZ(am) E. coli host designed for assaying pZ189 supF mutational specificity. Mol Gen Genet. 1992 Nov;235(2-3):173–178. doi: 10.1007/BF00279358. [DOI] [PubMed] [Google Scholar]
- Beld M., Martin C., Huits H., Stuitje A. R., Gerats A. G. Flavonoid synthesis in Petunia hybrida: partial characterization of dihydroflavonol-4-reductase genes. Plant Mol Biol. 1989 Nov;13(5):491–502. doi: 10.1007/BF00027309. [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991 Mar 25;19(6):1349–1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux C. N., Mis J. R., Pierce M. K., Kohalmi S. E., Kunz B. A. DNA sequence analysis of spontaneous mutations in the SUP4-o gene of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Feb;8(2):978–981. doi: 10.1128/mcb.8.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. Different effects of intron nucleotide composition and secondary structure on pre-mRNA splicing in monocot and dicot plants. EMBO J. 1991 Sep;10(9):2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser J., Seidman M. M., Sidur K., Dixon K. Sequence specificity of point mutations induced during passage of a UV-irradiated shuttle vector plasmid in monkey cells. Mol Cell Biol. 1986 Jan;6(1):277–285. doi: 10.1128/mcb.6.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebee B., Brouwer J., van de Putte P., Loman H., Retèl J. 60Co gamma-rays induce predominantly C/G to G/C transversions in double-stranded M13 DNA. Nucleic Acids Res. 1988 Aug 25;16(16):8147–8156. doi: 10.1093/nar/16.16.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juricek M., Gruz P., Veleminsky J., Stanek J., Kefurt K., Moravcova J., Jary J. Mutagenic activity of 6-azido deoxyhexoses and azido alcohols in Salmonella typhimurium and its inhibition by a structure-similar carbon source in the medium. Mutat Res. 1991 Nov;251(1):13–20. doi: 10.1016/0027-5107(91)90211-6. [DOI] [PubMed] [Google Scholar]
- Kleinhofs A., Owais W. M., Nilan R. A. Azide. Mutat Res. 1978;55(3-4):165–195. doi: 10.1016/0165-1110(78)90003-9. [DOI] [PubMed] [Google Scholar]
- Kleinhofs A., Smith J. A. Effect of excision repair on azide-induced mutagenesis. Mutat Res. 1976 Dec;41(2-3):233–240. doi: 10.1016/0027-5107(76)90096-8. [DOI] [PubMed] [Google Scholar]
- Kristiansen K. N., Rohde W. Structure of the Hordeum vulgare gene encoding dihydroflavonol-4-reductase and molecular analysis of ant18 mutants blocked in flavonoid synthesis. Mol Gen Genet. 1991 Nov;230(1-2):49–59. doi: 10.1007/BF00290650. [DOI] [PubMed] [Google Scholar]
- Mattox W., Ryner L., Baker B. S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992 Sep 25;267(27):19023–19026. [PubMed] [Google Scholar]
- McBride T. J., Preston B. D., Loeb L. A. Mutagenic spectrum resulting from DNA damage by oxygen radicals. Biochemistry. 1991 Jan 8;30(1):207–213. doi: 10.1021/bi00215a030. [DOI] [PubMed] [Google Scholar]
- Mo J. Y., Maki H., Sekiguchi M. Mutational specificity of the dnaE173 mutator associated with a defect in the catalytic subunit of DNA polymerase III of Escherichia coli. J Mol Biol. 1991 Dec 20;222(4):925–936. doi: 10.1016/0022-2836(91)90586-u. [DOI] [PubMed] [Google Scholar]
- Owais W. M., Kleinhofs A. Metabolic activation of the mutagen azide in biological systems. Mutat Res. 1988 Feb;197(2):313–323. doi: 10.1016/0027-5107(88)90101-7. [DOI] [PubMed] [Google Scholar]
- Owais W. M., Ronald R. C., Kleinhofs A., Nilan R. A. Synthesis and mutagenicity of the two stereoisomers of an azide metabolite (azidoalanine). Mutat Res. 1986 Nov;175(3):121–126. doi: 10.1016/0165-7992(86)90109-0. [DOI] [PubMed] [Google Scholar]
- Saling E., Blücher U., Sander H. Equipment for the recording of tractive power in vaccum extractions. J Perinat Med. 1973;1(2):142–144. [PubMed] [Google Scholar]
- Schaaper R. M., Dunn R. L. Spontaneous mutation in the Escherichia coli lacI gene. Genetics. 1991 Oct;129(2):317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Shepherd N., Tacke E., Gierl A., Rohde W., Leclercq L., Mattes M., Berndtgen R., Peterson P. A., Saedler H. Influence of transposable elements on the structure and function of the A1 gene of Zea mays. EMBO J. 1987 Feb;6(2):287–294. doi: 10.1002/j.1460-2075.1987.tb04752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B. W., Hanley S., Goodman H. M. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 1992 Mar;4(3):333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris E. G., Argyrakis M. Chemical alterations induced in DNA and DNA components by the mutagenic agent azide. Biochim Biophys Acta. 1974 Nov 6;366(4):367–373. doi: 10.1016/0005-2787(74)90034-3. [DOI] [PubMed] [Google Scholar]
- Wakamatsu N., Kobayashi H., Miyatake T., Tsuji S. A novel exon mutation in the human beta-hexosaminidase beta subunit gene affects 3' splice site selection. J Biol Chem. 1992 Feb 5;267(4):2406–2413. [PubMed] [Google Scholar]





