Abstract
The present study functionally identified a new microRNA (microRNA ovual line 5, miRNVL5) with its target gene GhCHR from cotton (Gossypium hirsutum). The sequence of miRNVL5 precursor is 104 nt long, with a well developed secondary structure. GhCHR contains two DC1 and three PHD Cys/His-rich domains, suggesting that GhCHR encodes a zinc-finger domain-containing transcription factor. miRNVL5 and GhCHR express at various developmental stages of cotton. Under salt stress (50–400 mM NaCl), miRNVL5 expression was repressed, with concomitant high expression of GhCHR in cotton seedlings. Ectopic expression of GhCHR in Arabidopsis conferred salt stress tolerance by reducing Na+ accumulation in plants and improving primary root growth and biomass. Interestingly, Arabidopsis constitutively expressing miRNVL5 showed hypersensitivity to salt stress. A GhCHR orthorlous gene At2g44380 from Arabidopsis that can be cleaved by miRNVL5 was identified by degradome sequencing, but no confidential miRNVL5 homologs in Arabidopsis have been identified. Microarray analysis of miRNVL5 transgenic Arabidopsis showed six downstream genes (CBF1, CBF2, CBF3, ERF4, AT3G22920, and AT3G49200), which were induced by salt stress in wild-type but repressed in miRNVL5-expressing Arabidopsis. These results indicate that miRNVL5 is involved in regulation of plant response to salt stress.
Soil salinity is one of the major environmental and agronomical problems which seriously cause cellular dehydration and ionic toxicity, thus limiting the productivity of crops1. To survive the detrimental effect of salt stress, plants have developed various elaborate mechanisms to exclude excess salt from their cells or to tolerate salt within the cells2. Plant resistance to salt stress is controlled by multiple genes and is regulated at multiple levels. At the molecular level, various stress-responsive genes are involved3,4.
Recently, a large group of small RNAs, termed microRNAs (miRNAs), are reported as regulators in plant adaptation to abiotic stresses5,6,7,8. For examples, transgenic creeping bentgrass (Agrostis stolonifera) overexpressing rice miR319a exhibited enhanced salt and drought tolerance9; overexpressing miR396c and miR394 plants was hypersensitive to saline stress10,11. In cotton (Gossypium hirsutum), a group of miRNAs and their targets have been identified, and some of them respond to salt and drought stresses12,13,14,15,16,17. But to date, only a few of cotton miRNAs have been functionally characterized18.
Abiotic stress-responsive genes are largely regulated by complex networks of transcription factors (TFs)19,20,21. One of them is the zinc finger protein (ZFP) family, which bind to zinc ions through their Cystenine (Cys) and Histidine (His) and are involved in protein-DNA or protein-protein interactions in plants22. ZFP family proteins can be categorized based on the combinations of Cys and His for coordination divalent zinc ions, such as C2H2, C3HC4, C4, CCCH, C4HC3 or C2HC523. Each ZFP protein may contain one or more finger zinc motifs, which is incorporated into diverse functional proteins. The PHD (plant homeodomain) finger protein genes encode another type of zinc finger proteins whose motifs comprise ∼60 amino acids and typically show a C4HC3 signature (four Cys, one His, and three Cys); with the PHD-finger domain, PHD proteins are able to bind to a common nuclear ligand chromatin and play an important role in regulating chromatin remodeling24. In Arabidopsis, PHD domain-containing protein genes regulate diverse processes of development25,26. Importantly, some of the genes were reported to regulate plant response to salt stress27,28. In this study, we functionally identified a miRNA (designed as miRNVL5), along with its target gene from cotton (G. hirsutum). miRNVL5 and GhCHR were originally isolated from cotton ovules from our previous study13. RACE-PCR analysis showed that miRNVL5 guides the cleavage of the target harboring two DC1 and three PHD domains (Due to its containing a Cys/His-rich DC1/PHD domains, here referred as GhCHR). Our data showed that salt stress repressed the miRNVL5 expression, whereas induced the GhCHR expression in cotton. Transgenic Arabidopsis constitutively expressing miRNVL5 showed sensitivity to salt stress, while expression of GhCHR improved salt tolerance in Arabidopsis. These results indicate that miRNVL5 and GhCHR play an important role in plant response to salt stress.
Results
miRNVL5 was a cotton miRNA targeting a zinc finger protein family gene GhCHR
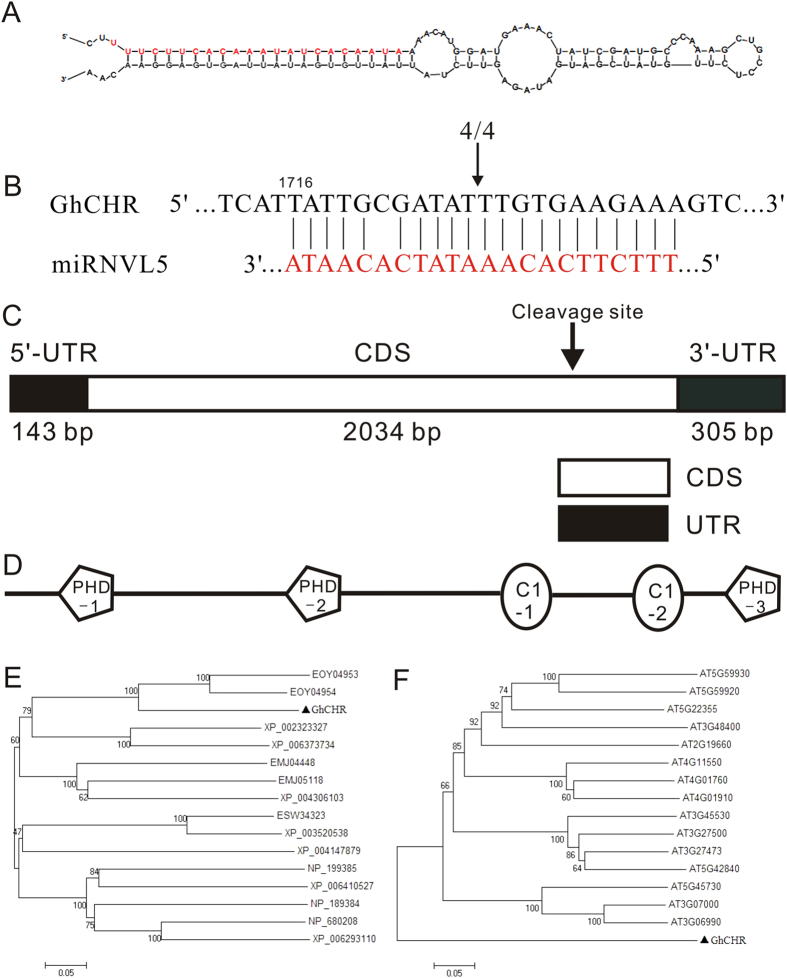
The sequence of miRNVL5 precursor is 104 nt in length, with a 22-nt mature sequence and well developed secondary structure (Fig. 1A; Data S1). Homologous search revealed that no sequences perfectly or near-perfectly match the mature miRNVL5 in Arabidopsis and other plant species. GhCHR was predicted to be the only target of miRNVL5 in G. hirsutum. 5′ complementary DNA ends (5′-RACE)-PCR analysis was performed to validate the cleavage site (Fig. 1B). The PCR products were cloned and sequenced. The four clones showed the same site of cleavage following the eleventh nucleotide from the 5′ end of miRNVL5, indicating that the GhCHR transcript is the real target of miRNVL5.
Figure 1. Basic information on miRNVL5 and GhCHR from cotton (Gossypium hirsutum).
(A) Mature (in red) and precursor sequences and the stem-loop structure of miRNVL5. (B) The GhCHR sequence with nucleotides was cleaved by miRNVL5 using 5′-RACE. (C) cDNA structure of GhCHR. (D) Predicted GhCHR protein structure with PHD and C1 domains. (E) Dendrogram showing the relationship among genes encoding PHD-type proteins in cotton and other plant species. (F) Dendrogram showing PHD-type proteins between cotton and Arabidopsis only.
The cDNA of GhCHR was 2482 bp in length including a 2034-bp open reading frame, a 143-bp 5′ untranslated region (UTR), and a 305-bp 3′ UTR (Fig. 1C, Data S2A). The deduced GhCHR polypeptide consists of 677 residues of amino acids, with a predicted molecular weight of 23.7 kD and a pI of 6.54. A Blastp search showed that GhCHR contains three PHD (Plant Homeo Domain with Cys4-His-Cys3 motif) and two DC1 (Cys5-His) motifs capable of binding zinc (Fig. 1D, Data S2B), indicating that GhCHR transcripts encode a PHD-domain containing protein25. A phylogenetic analysis showed that GhCHR protein shared only 59% identity with a cysteine/histidine-C1 domain family protein from Cacao (Theobroma cacao; EOY04953; Fig. 1E,F). Multiple sequence alignments using BLASTx showed that the GhCHR protein also shared very low similarity with the proteins from other plant species except for PDH and C1 domains (Data S2C), suggesting that GhCHR is not a highly conserved CHC domain (Cys4-His-Cys3)-containing zinc finger protein.
Expression patterns of miRNVL5 and GhCHR under salinity stress
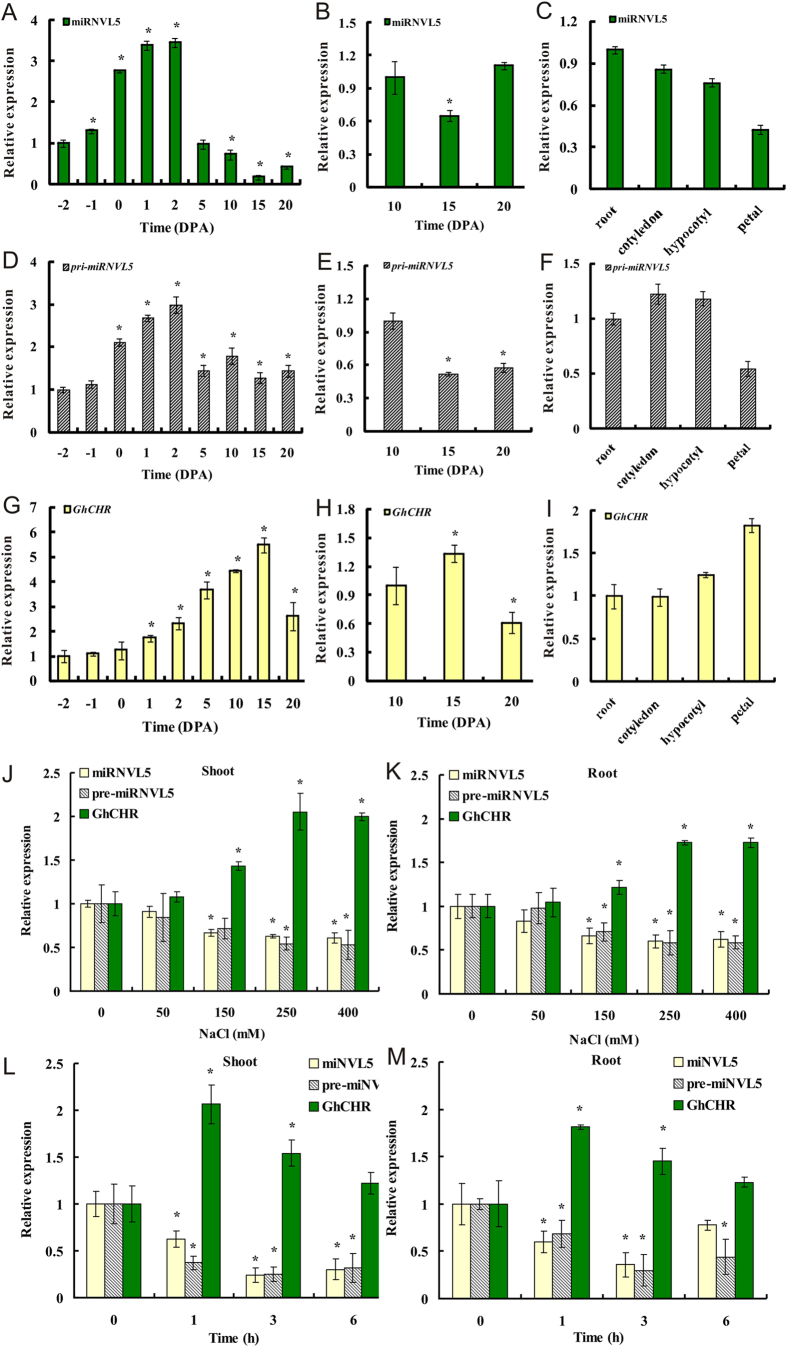
Quantitative RT-PCR (qRT-PCR) was used to analyze expression patterns of miRNVL5 and pri-miRNVL5 as well as GhCHR at different developmental stages of cotton. Both miRNVL5/pri-miRNVL5 and GhCHR were ubiquitously expressed at various developmental stages, but the transcript levels changed considerably (Fig. 2A–I). In developing ovules, miRNVL5 and pri-miRNVL5 were highly expressed at 0-2 DPA. After that, expression of miRNVL5 was decreased. In contrast, expression of GhCHR was always repressed in correlation to miRNVL5. The inverse expression pattern of miRNVL5 and GhCHR suggests that GhCHR transcript was negatively regulated by miRNVL5.
Figure 2. Expression of miRNVL5, pri-miRNVL5 and GhCHR in different tissues of cotton with or without salt treatment.
(A,D,G) Genes from ovules of −2–20 days post anthesis (DPA). (B,E,H) Genes from developing fibers of 10, 15 and 20 DPA. (C,F,I) genes from other tissues. (J–M) Two week-old cotton seedlings were treated with NaCl (0–400 mM) (J,K) for 1 h, or with 250 mM NaCl for 0–6 h (L,M). After that, total RNA was extracted and analyzed by real-time PCR. Vertical bars represent SD of the mean with three replicates. Asterisks indicate that mean values are significantly different between the treatment and control (p < 0.05).
Because recent reports indicate that PHD domain-containing protein genes are involved in plant response to abiotic stress27,28, this urged us to test the transcription of GhCHR under environmental stress. In shoots of cotton seedlings, treatment with 150–400 mM NaCl induced the expression of GhCHR but repressed the miRNVL5 expression (Fig. 2J). A similar result was found in roots (Fig. 2K). The time-course study with 250 mM NaCl also revealed that expression of miRNVL5 was decreased, whereas the expression of GhCHR was increased after 1 h treatment of NaCl (Fig. 2L,M). Compared to the control, treatment with 250 mM NaCl for 1, 3 and 6 h led to the decrease in transcripts of miRNVL5 by 37.3%, 75.6% and 70% in shoots and 39.3%, 64.3% and 22.4% in roots, respectively. These results confirmed the inverse expression pattern for miRNVL5/pri-miRNVL5 and GhCHR. The transcriptional responses of miRNVL5 and GhCHR were also tested with drought, cold and ABA, but no significant change in the transcriptional response was observed (Data S3).
Transgenic Arabidopsis response to salt stress was regulated by constitutive expression of miRNVL5 and GhCHR
In order to identify a role of GhCHR in plant response to salt stress, full-length cDNA of GhCHR was transformed into Arabidopsis wild-type (Col-0 ecotype) with cauliflower mosaic virus 35S promoter. As GhCHR mRNA generated in Arabidopsis was possibly cleaved by a putative miRNA, we simultaneously constructed an miRNVL5 cleavage-resistant version (35S::mGhCHR) by introducing seven silent mutations in the miRNVL5 binding site but without changing the protein sequences (Data S4). Five independent transgenic lines were obtained, and two of them were used for study. The transgenic lines displayed no abnormal phenotypes under normal condition. qRT-PCR analysis showed that the levels of GhCHR mRNA in 35S::mGhCHR and 35S::GhCHR transgenic lines were 72–89 and 109–132 folds higher than the wide-type, respectively (Data S4A–C). We also generated transgenic Arabidopsis plants expressing miRNVL5, because blast search on Arabidopsis genome revealed five potential orthologs of GhCHR which are possibly targeted by miRNVL5 (data to be presented below). Thus, the pri-miRNVL5 sequence was fused to the CaMV 35S promoter and transformed into the Arabidopsis Col-0. Expression analysis showed that the selected transgenic lines of 35S::miRNVL5 had high expression of miRNVL5 (Data S4D).
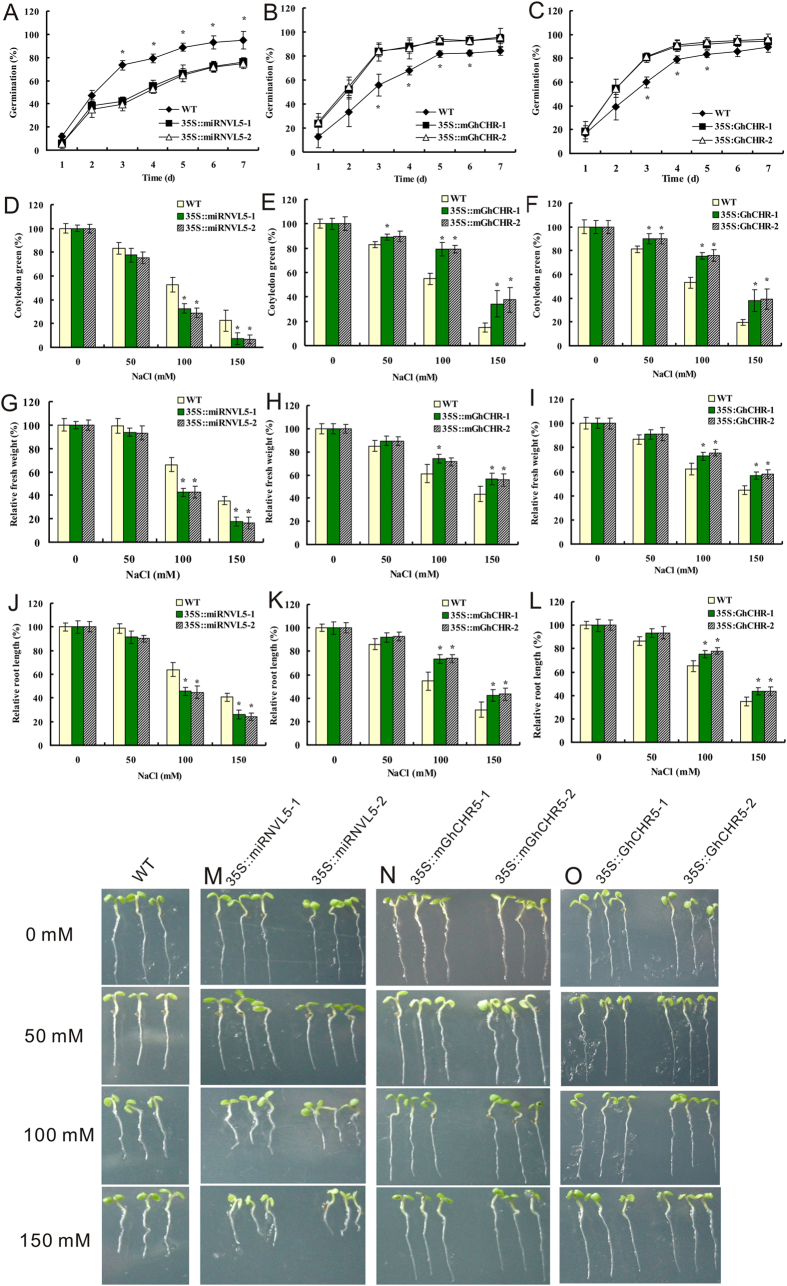
We then investigated the transgenic plants in regulating plant response to salt stress. Arabidopsis seeds were placed on the solid MS medium supplemented with 0–150 mM NaCl. The seed germination and post-germination growth responded differently to NaCl after a 1–7 d period of treatment. Following a 3-day treatment with 100 mM NaCl, the seed germination rate of 35S::miRNVL5 lines were only 53.4–57.5% of the wild-type (Fig. 3A), whereas seed germination rate of 35S::mGhCHR and 35S::GhCHR plants was 51.9–52.7% and 37.3–39.1% higher than the wild-type, respectively (Fig. 3B,C). Exposure to 100–150 mM NaCl also inhibit cotyledon greening in 35S::miRNVL5 seedlings (Fig. 3D). Under the same condition, the percentage of cotyledon greening in 35S::mGhCHR and 35S::GhCHR seedlings was higher than that of the wild-type (Fig. 3E,F). The biomass (fresh weight) of 35S::miRNVL5 and 35S::mGhCHR or 35S::GhCHR lines was also examined. Compared to wild-type, lower fresh weight of 35S::miRNVL5 seedlings was observed, whereas higher biomass of 35S::mGhCHR and 35S::GhCHR seedlings was determined (Fig. 3G–I). The root elongation of 35S::miRNVL5 lines was significantly inhibited by 100–150 mM NaCl; in contrast, the 35S::mGhCHR and 35S::GhCHR seedlings had higher tolerance to 100–150 mM NaCl stress in root growth (Fig. 3J–O). These results indicate that 35S::miRNVL5 plants were hypersensitive, whereas 35S::mGhCHR or 35S::GhCHR plants were tolerant to salt stress.
Figure 3.
Seed germination and seedling growth of transgenic Arabidopsis over-expressing miRNVL5 (35::miRNVL5) (A,D,G,J,M), miRNVL5-resistant version (35S::mGhCHR)(B,E,H,K,N) and GhCHR (35S::GhCHR)(C,F,I,L,O) under the salt stress. (A–C) Seeds were germinated on MS medium containing 100 mM NaCl. Germination rates were measured during one week. (D–F) Cotyledon greening and expansion of 7 day-old seedlings with 50–150 mM NaCl. (G–I) Fresh weight of 7 day-old seedlings exposed to 50–150 mM NaCl. (J–K) The elongation of primary root of 7 day-old seedlings exposed to 50–150 mM NaCl. M-O: phenotype of primary root growth of 7 day-old seedlings exposed to 50–150 mM NaCl. Vertical bars represent SD of the mean with three replicates. Asterisks indicate that mean values are significantly different between the transgenic plants and wild-type (WT) (p < 0.05).
35S::miRNVL5 Arabidopsis plants accumulated more Na+ than wild-type plants
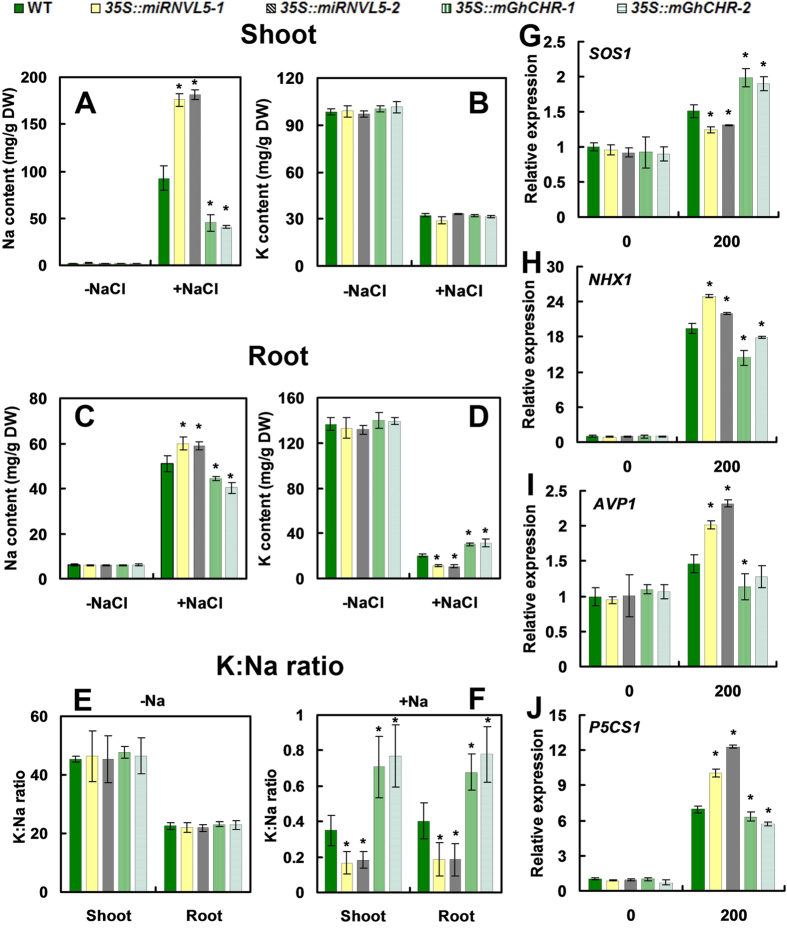
Excess Na+ is toxic to plants, whereas K+ is antagonistic to Na+ under salt stress3. To identify the potential mechanism of transgenic plants in response to saline stress, concentrations of Na+ and K+ were assessed in both transgenic and WT plants. There was no difference of Na+ and K+ concentrations in wild-type and transgenic plants under normal condition (Fig. 4A–D). However, when exposed to 200 mM NaCl, the 35::miRNVL5 plants accumulated more Na+ in shoots and roots; conversely, the concentration of Na+ in the tissues was lower in 35S::mGhCHR plants as compared to wild-type (Fig. 4A,C). In shoots, no difference of K+ concentration was observed between the transgenic plants and wild-type; but in roots, lower levels of K+ in 35::miRNVL5 plants and higher levels in 35S::mGhCHR plants were determined. The imbalance of Na+ and K+ levels led to an increase in K+/Na+ ratio in 35S::mGhCHR plants and decrease in K+/Na+ ratio in 35::miRNVL5 plants (Fig. 4E,F).
Figure 4. Sodium (Na+)/potassium (K+) accumulation and expression of SOS1, NHX1, AVP1 and P5CS1 in transgenic and wild-type (WT) plants exposed to NaCl.
(A,C) Na+ in shoots and roots. (B,D) K+ in shoots and roots of three week-old WT, 35S::miRNVLU5 and 35S::mGhCHR transgenic plants exposed to 200 mM NaCl exposure for 3 d. (E,F) K+/Na+ ratio in WT and the transgenic plants under −NaCl (E,F) and +NaCl (F) Condition. (G–J) qRT-PCR analysis of the indicated gene expression. Three week-old seedlings were exposed to 200 mM NaCl for 1 h. Vertical bars represent SD of the mean with three replicates. Asterisks indicate that mean values are significantly different between the transgenic plants and WT (p < 0.05).
We further assessed transcripts of several genes responsible for plant tolerance to salt stress. SOS1 (Salt Overly Sensitive 1) is a plasma membrane Na+/H+ antiporter and mediates Na+ efflux29. Compared to the control, expression of SOS1 was lower in 35::miRNVL5 plants, whereas higher in 35S::mGhCHR plants under salt stress (Fig. 4G). NHX1 (Sodium Hydrogen Exchanger 1) encodes a vacuolar Na+/H+ antiporter involved in salt resistance and transports Na+ into the vacuole by using the electrochemical gradient of protons generated by the vacuolar H+ -translocating enzyme, H+ -adenosine triphosphatase and H+ -inorganic pyrophosphatase30. AVP1 (Pyrophosphate-energized vacuolar membrane proton pump 1) encodes a H+ -translocating (pyrophosphate-energized) inorganic pyrophosphatase (H+ -PPase) located in the vacuolar membrane31. P5CS1 (Delta-1-pyrroline-5-carboxylate synthase 1) is a rate-limiting enzyme in biosynthesis of proline, whose mRNA is induced by drought, salinity, and ABA32. qRT-PCR analysis showed that expression of NHX1, AVP1 and P5CS1 were highly expressed in 35::miRNVL5 plants, whereas those in 35S::mGhCHR plants were inhibited (Fig. 4H–J).
Identification of genes targeted by miRNVL5 in Arabidopsis
Because expression of miRNVL5 in Arabidopsis led to salt sensitive phenotypes, this allowed for hypothesis that some putative targets of miRNVL5 may exist in Arabidopsis. Based on the assumption, we predicted the target genes of miRNVL5 in Arabidopsis. According to the commonly accepted criteria, five putative target genes were identified. Interestingly, all these predicted genes belong to the PHD family protein genes (Table 1). We then analyzed their transcriptional abundance of the genes from our microarray datasets (WT + S/WT-S, miR5 − S/WT-S and miR5 + S/WT + S) and found all of them were repressed in the 35S::miRNVL5 plants under salt stress (Table 1; Data S5). To confirm the genes predicted, genome-wide high throughput degradome sequencing was performed based on the recently developed approach33,34. We sequenced three libraries including WT + S (wild-type with salt), MIR5 − S (35S::miRNVL5 plants without salt treatment), and MIR5 + S (35S::miRNVL5 plants treated with salt) from Arabidopsis. A total of 24.9–30.4 million short sequencing reads representing the 5’ ends, uncapped and polyadenylated RNAs were obtained (Data S6). After removal of low quality, error and redundant reads, 4.4–6.5 million unique reads were mapped to the Arabidopsis genome, which accounted for 70.6%, 83.6%, and 72.0% of the clean reads for the WT + S, MIR5 − S, and MIR + S libraries, respectively.
Table 1. Prediction of miRNVL5 targeted genes from Arabidopsis and expression pattern in wild-type and 35S::miR5 (miR5) plants under salt stress.
| Gene ID | Expectation value | Inhibition type | WT + S/WT-S Fold Change | miR5 − S/WT-S Fold Change | miR5 + S/WT + S Fold Change |
|---|---|---|---|---|---|
| AT2G44380 | 1.5 | Cleavage | 2.20 | −1.07 | −1.21 |
| AT4G39070 | 2.5 | Cleavage | 1.31 | −1.03 | −1.01 |
| AT4G01930 | 3 | Cleavage | 1.28 | −1.06 | −1.09 |
| AT2G19260 | 3 | Translation | 1.13 | 1.12 | −1.27 |
| AT2G21850 | 3 | Translation | -1.03 | −1.11 | −1.16 |
The prediction was run according to the online tool (http://plantgrn.noble.org/psRNATarget/). The threshold was set as 3.
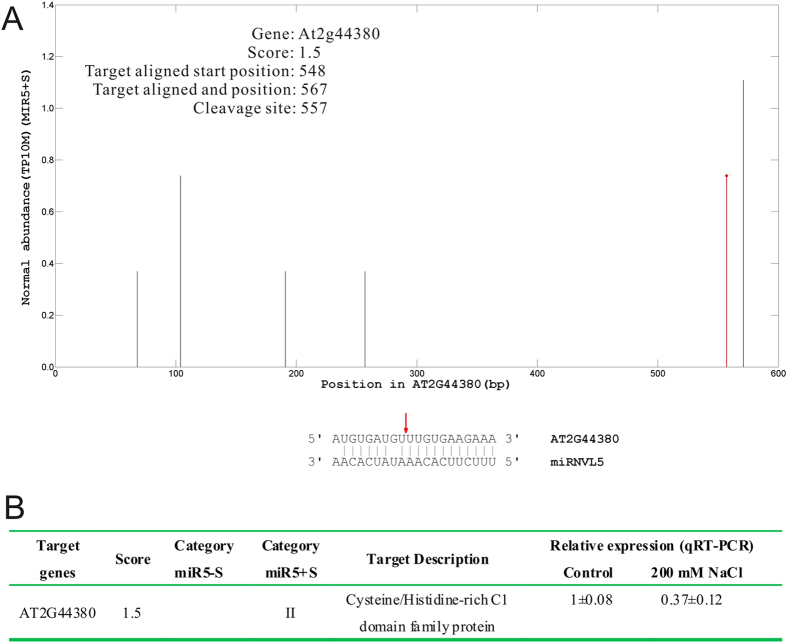
miRNVL5 target was identified by using the CleaveLand pipeline33. According to the results, AT2G44380 has been identified as the target of miRNVL5 (Fig. 5A). The t-plot graphics showed that the target transcripts fall into miRNA-guided cleavage remnants; the alignment of the sequence manifested the signature that can be produced by miRNVL5-directed cleavage (Fig. 5B). Both microarray and qRT-PCR analyses showed that AT2G44380 in wild-type seedlings was induced by NaCl, but repressed by in 35S::miRNVL5 plants under salt stress (Table 1).
Figure 5. Identification of miRNVL5 target from Arabidopsis through degradome sequencing.
(A) The red line with a dot indicates the significant signature. The arrow indicates the signature produced by miRNA-directed cleavage. (B) Target for miRNVL5 identified from degradome of 35S::miRNVL5 Arabidopsis exposed to minus salt (miR5 − S) and plus salt (miR5 + S). Three week-old Arabidopsis seedlings were treated with 0 (control) and 200 mM NaCl for 1 h and the treated seedlings were used for degradome sequencing and qRT-PCR.
Identification of downstream genes of AT2G44380 in Arabidopsis
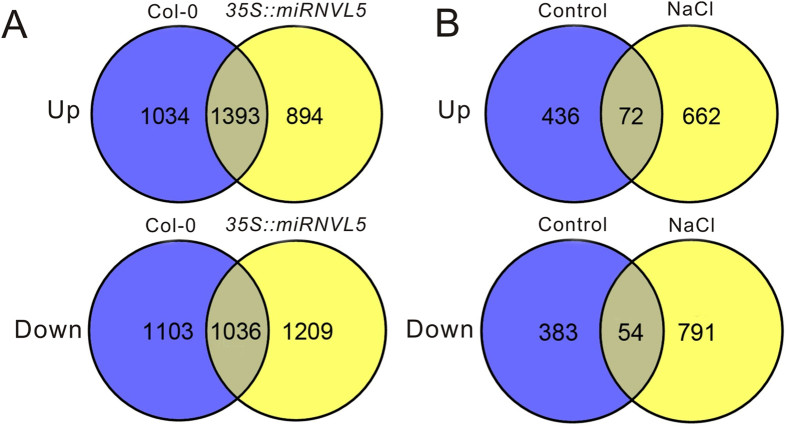
The expression of miRNVL5 and GhCHR in Arabidopsis generating distinct phenotypes suggests that downstream genes responsible for salt stress tolerance could be regulated. To identify putative downstream genes in Arabidopsis under salt stress, we analyzed the global transcripts in 35S::miRNVL5 and wild-type plants under salt stress by generating an Agilent GeneChip with 43,803 distinct probes. By profiling the microarray, 1034 genes were induced and 1103 were repressed in wild-type, while 894 genes were induced and 1209 genes were repressed in 35S::miRNVL5 plants under salt stress (Fig. 6A; Data S5). Comparative analysis of transcripts between 35S::miRNVL5 and WT plants showed that 436 genes were induced and 383 genes were repressed under control condition, whereas 662 genes were induced and 791 genes were repressed under salt stress (Fig. 6B; Data S7).
Figure 6. Differentially expressed genes in Arabidopsis wild-type (Col-0) and 35S::miRNVL5 seedlings under salt stress.
Three weeks-old Arabidopsis seedlings were treated with 200 mM NaCl for 1 h. (A) Venn diagram showing up- and down-regulated genes in Col-0 and 35S::miRNVL5 seedlings under salt stress. (B) Venn diagram showing up- and down-regulated genes in 35S::miRNVL5 seedlings relative to Col-0 grown on the control (0 mM) and 200 mM NaCl media.
The commonly repressed or induced genes were examined by screening the microarray datasets (WT + S/WT-S, miR5 − S/WT-S and miR5 + S/WT + S). Only 6 genes matched the criteria, including CBF1, CBF2, CBF3/DREB1A, ERF4, AT3G22920 and AT3G49200, and all of the genes were repressed (Table 2). CBF1/2/3 proteins are APETALA2/ethylene-responsive transcription factors that bind to DRE and activate the transcription of target stress-inducible genes to confer plant tolerance to abiotic stresses35. ERF4 encodes a ethylene-responsive transcription factor 436. AT3G22920 and AT3G49200 encode peptidylprolyl isomerase and O-acyltransferase (WSD1-like) family protein, respectively. Our data showed that all six genes in WT were induced under NaCl exposure but repressed in 35S::miRNVL5 plants under either normal or NaCl stress condition. These results indicate that the genes may be directly or indirectly mediated by a putative component, which is in turn negatively regulated by miRNVL5 in Arabidopsis. Of the six genes, three were randomly selected for qRT-PCT validation, and all of them were well confirmed (Data S8).
Table 2. List of genes that are commonly induced in Arabidopsis Col-0 (wild-type, WT) and 35S::miRNVL5 (miR5) seedlings under salt (S, 200 mM NaCl) stress.
| AGI ID | Description | WT + S/WT-S | miR5 − S/WT-S | miR5 + S/WT + S |
|---|---|---|---|---|
| CBF1 | Dehydration-responsive element-binding protein 1B | 2.26 | −3.93 | −1.07 |
| CBF2 | Dehydration-responsive element-binding protein 1C | 2.37 | −2.67 | −1.10 |
| CBF3 (DREB1A) | Dehydration-responsive element-binding protein 1A | 20.75 | −2.18 | −1.04 |
| ERF4 | Ethylene-responsive transcription factor 4 | 3.18 | −3.53 | −1.32 |
| AT3G22920 | Peptidylprolyl isomerase | 2.76 | −2.47 | −1.32 |
| AT3G49200 | O-acyltransferase (WSD1-like) family protein | 2.97 | −6.57 | −1.23 |
Genes presented showed a fold change more than 2.0. Transcript abundance in Col-0 and 35S::miRNVL5 seedlings based on microarray analysis.
A group of salt-responsive genes were co-expressed with AT2G44380
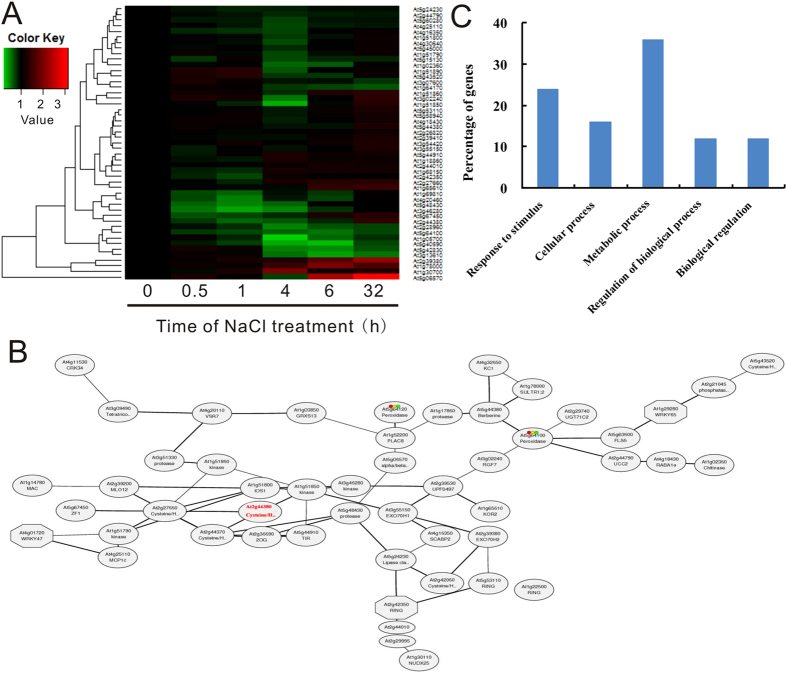
Recent studies have shown that many genes may have a similar expression pattern under a certain environmental stimuli, and in this case, they are usually working in a similar pathway37,38. By extraction of tightly co-expressed genes, one may find important or even novel genes associated with specific pathways. All salt-responsive genes were searched based on the Gene Orthology (GO) annotation in the Arabidopsis database (The Arabidopsis Information Resource, TAIR, http://www.arabidopsis.org/). A total of 49 genes that can co-express with AT2G44380 were identified (Data S9). These genes were further compared to the microarray datasets with salt stress39 (Fig. 7A). A strict screening criterion was set up, by which only genes with strong salt stress response could be considered as tightly co-expressed genes. Two genes (At2g42060 and At1g14780) were identified as core (or guide) genes that connect the remaining 56 genes. We took an advantage of the database ATTED-II, which used a unique co-expression measure, namely Mutual Rank (MR) of the Pearson’s correlation coefficient, to compare the co-expression strength for the guide gene AT2G4438040. With a strict cutoff (MR < 50), a gene network from the 50 co-expression genes was generated (Fig. 7B). These genes were found to be well associated. For instance, two genes At2g27660 and At2g44370 were predicted to interact with AT2G44380. The GO and KEGG (Kyoto Encyclopedia of Genes and Genomes) analyses revealed four diverse pathways with salt stress (Fig. 7C). Genes belonging to the Response to stimulus (24%) and metabolic process (36%) comprise the majority of the genes.
Figure 7. Co-expression, network and Gene Orthology (GO) analysis of salt-inducible genes in Arabidopsis.
(A) Differentially expression profiles of salt-responsive genes that are co-expressed with AT2g44380 in Arabidopsis treated with 100 mM NaCl for 0–32 h. Hierarchial clustering display performed using Euclidean distance according to expression pattern with the MeV software. The intensities of the color represent the relative magnitude of fold changes in log values. The raw transcriptional data that the heatmap used were obtained from Li et al. (2009) and normalized using RMA algorithm (Dinneny, et al., 2008) with Expression Console software (Affymetrix Technologies). (B) The hypothetic network under salt stress. (C) Gene Orthology analysis.
Discussion
Although there are substantial miRNAs isolated from higher plants recently, only a few of them have been functionally characterized as regulators of plant response to salt stress8,10,11,34. In this study, we isolated and functionally characterized a new miRNA and its target from cotton. Sequence analysis revealed that the miRNVL5 precursor can fold into a perfect secondary hairpin structure, with higher negative minimal free energies (MFEs) and minimal free energy index (MFEIs)41. The target of miRNVL5 was predicted and experimentally confirmed by 5′-RACE analysis. In silico analysis revealed that GhCHR belongs to the Cys/His-rich DC1/PHD domains-containing protein gene family. Although miRNVL5 was isolated from cotton ovules, both mature and primary miRNVL5 are ubiquitously expressed in cotton plants. GhCHR expressed in the same tissues where miRNVL5 expressed, but its expression level was lower, indicating that GhCHR is constantly under the control of miRNVL5. miRNVL5-regulated GhCHR expression was also detected under salt stress, but both miRNVL5 and pri-miRNVL5 expression was repressed in cotton with concomitant higher expression levels of GhCHR in the same tissues. Such an expression pattern suggests that GhCHR may serve as a positive regulator of plant salt stress response.
Our data shows that ectopic expression of GhCHR in Arabidopsis promoted seed germination, seedling growth and primary root elongation under salt stress. Furthermore, 35S::mGhCHR accumulated less Na+ in shoots as compared to wild-type. Although there was no change of K+ concentration in shoots, the K+/Na+ ratio was higher in 35S::mGhCHR shoots. The higher ratio of K+/Na+ was maintained in shoots possibly owning to the exclusion mechanism, because SOS1 responsible for Na+ pumping-out was activated in the 35S::mGhCHR plants. In contrast, expression of NHX1 which is encoding Na+/H+ antiporter in vacuoles was repressed in 35S::mGhCHR plants, suggesting that sequestration of higher Na+ in vacuoles under the condition may not be the mechanism for salt tolerance. A similar expression pattern for AVP1 was observed in the 35S::mGhCHR plants. P5CS1 can be considered as a marker gene responding to salt stress42. The lower level of P5CS1 suggests that expression of GhCHR in Arabidopsis attenuated degree of salt stress.
One striking observation in this study is that the expression of miRNVL5 in Arabidopsis led to the sensitive phenotypes of 35S::miRNVL5 plants to sodium ions. Such an observation allowed for assumption that there are putative transcripts of genes possibly targeted by miRNVL5 in Arabidopsis. To identify the putative target of miRNVL5, degradome from wild-type and 35::miRNVL5 plants under control and salt stress was profiled. Degradome is a newly developed and highly efficient approach for identifying global authentic miRNA targets, which has been applied to many plant species33,34,43. Our analysis showed that among the several candidate targets, At2g44380 is most likely the target of miRNVL5. Several lines of evidence are given as follows: (1) there is a perfect target sit of miRNVL5 in At2g44380 mRNA; (2) At2g44380 encodes a cysteine/histidine-rich C1 domain family protein similar to GhCHR; and (3) Co-expression analysis showed that At2g44380 acts as a core gene capable of connecting several other genes in the networks.
We attempted to identify orthologs of miRNVL5 in Arabidopsis. A query search was made for all miRNAs from Arabidopsis and other plant species that have been so far discovered in miRNA database (http://www.mirbase.org/) and NCBI (http://www.ncbi.nlm.nih.gov/). Unfortunately, no confident miRNAs from Arabidopsis compatible to miRNVL5 have been detected. However, we cannot rule out the possibility of existing of putative miRNAs, because our studies have shown that although there was a similar phenotype of 35S::GhCHR and 35S::mGhCHR plants, and both transgenic plants have a similar level of GhCHR transcripts, the relative lower level of GhCHR transcripts was observed in 35S::GhCHR plants. This suggests that some putative miRNAs may regulate the GhCHR abundance in a negative way. Gossypium hirsutum is a tetraploid crop with four copies of chromosomes. The extra copies of genetic materials and/or increased levels of heterozygosity in two or more sets of chromosome are predicted to be advantageous for selection, adaption and domestication44. Polyploidy induces non-additive expression of homologous genes through mechanisms of transcriptional regulation via chromatin modifications and DNA methylation45,46,47 or posttranscriptional regulation involving small RNA regulation48. The transcriptional or posttranscriptional regulation divergence between GhCHR and At2g44380 may be a good example of the miRNA-mediated mechanisms for evolutionary differences of duplicate genes in polyploids. Meanwhile, miRNVL5 in Arabidopsis might be lost during the course of evolution.
Our microarray analysis showed that miRNVL5 expression in Arabidopsis altered expression of many genes under normal and salt stress conditions. It was interesting to identify the downstream genes of At2g44380 in Arabidopsis. There were more down-regulated genes detected in 35S::miRNVL5 seedlings than those in wild-type. An array of genes could be repressed by miRNVL5 expression in Arabidopsis under salt stress. We further narrowed down the number of genes differentially expressed in the three datasets (WT + S/WT-S, miR5 − S/WT-S and miR5 + S/WT + S). Only 6 genes (CBF1, CBF2, CBF3, ERF4, AT3G22920 and AT3G49200) remained after most of genes were filtered out. All these genes are stress-responsive genes. CBFs encode transcriptional activators that control the expression of genes with the CRT/DRE-responsive element in their promoters and are rapidly induced in response to low temperature49. Evidence also demonstrates that the CBFs-family genes play an important role in activating gene expression during salt stress35,50. Overexpression of the DREB1/CBF family genes increased tolerance to freezing, drought, and high salinity in transgenic Arabidopsis plants51. Ethylene-responsive transcription factor 4 (ERF4) is known as one of ethylene-responsive element binding proteins (EREBPs) that interacts with the GCC-box sequence with the core GCCGCC motif 52. Expression of AtERF4 can be induced by ethylene, jasmonic acid (JA), ABA and salt stress36,39. Overexpression of ERF4 from Brissica rapa increased tolerance to salt and drought stresses in Arabidopsis53. In this study, CBF1, CBF2, CBF3 and ERF4 were found to be repressed in the transgenic Arabidopsis plants expressing miRNVL5, which shows a phenotype of NaCl sensitivity. These results suggest that miRNVL5-regulated repression of CBF1, CBF2, CBF3 and ERF4 should be responsible for plant sensitivity to the salinity stress.
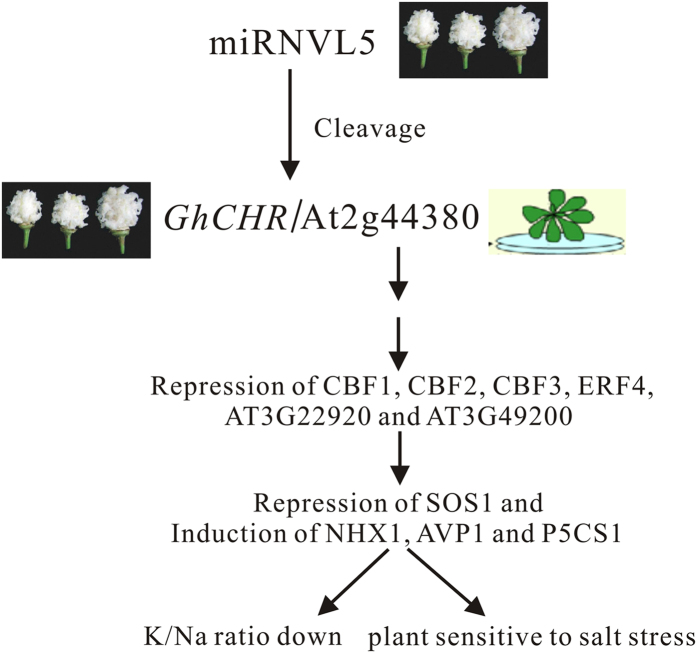
Taken together, the present study showed that miRNVL5 and GhCHR were involved in plant response to salt stress. Ectopic expression of miRNVL5 resulted in NaCl sensitive phenotype in Arabidopsis, whereas a tolerance phenotype was observed in the transgenic Arabidopsis expressing GhCHR. By degradome analysis, a GhCHR orthorlous gene At2g44380 has been identified in Arabidopsis as a target of miRNVL5. Whether At2g44380 is involved in regulation of plant tolerance to salt stress remains to be further investigated. Based on our observation, a simple model could be proposed of how miRNVL5 regulates plant response to salt stress (Fig. 8).
Figure 8. The model for cotton miRNVL5 regulating its target genes GhCHR (cotton) and At2g44380 (Arabidopsis).
The schematic model reflects that transformation of miRNVL5 into Arabidopsis results in decrease in K/Na ratio and seedling sensitivity to salt stress possibly through repressing At2g44380, and several other downstream genes CBF1, CBF2, CBF3, ERF4, AT3G22920, AT3G49200 and others.
Materials and Methods
Plant materials and stress treatment
Upland cotton (Gossypium hirsutum, cv. Xuzhou 142, wild type, WT) plants were grown in the field at the Nanjing Agricultural University Experimental Station. Flowers were tagged and ovules were sampled and dissected from −2 to 20 DPA (day of anthesis) ovaries in the early morning as described in our previous report13. The ovules were frozen immediately in liquid nitrogen and stored at −80 °C till to analysis.
For stress treatments, cotton seeds were germinated on 1/2 MS medium under a 16 h light/8 h dark cycle at 28 °C. Two or three week-old seedlings were treated by different concentrations of NaCl (0–400 mM) in 1/2 MS liquid medium for 0–6 h54. After treatment, shoots and roots of seedlings were separately harvested for RNA isolation. Arabidopsis thaliana ecotype Col-0 seeds were surface sterilized and germinated on 1/2 MS medium. Seeds were first stratified for 3d at 4 °C in the dark (to break dormancy) and then germinated in a growth chamber (22 °C, 16 h photoperiod). After 2 weeks, seedlings were used for NaCl treatment and analysis as described as the same for cotton treatments.
GhCHR cloning and identification of miRNVL5 cleavage site in GhCHR sequence
Two pairs of specific primers were designed for GhCHR cDNA cloning and used for amplification based on the consensus sequence from cotton leaves (Data S10). The PCR thermocycling conditions were as follow: 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 57 °C 30s, and 72 °C for 2 min, and a final extension at 72 °C for 10 min. The PCR product was separated by electrophoresis on a 1% agarose gel stained with ethidium bromide, purified using the Quiquick gel extraction kit (BioDev Biotech, Beijing, China). The product was then cloned into the PBI121 vector, and transformed into E. coli in JM109. The predicted target GhCHR of miRNVL5 was validated with the 5′-RACE assay using the FirstChoice RLM-RACE kit (Ambion)55. RNAs were prepared from cotton ovules. The RNA was ligated with a 5′ RNA adapter and a RT-PCR was performed. The resulting cDNA was used as template for PCR amplification. The amplified products were gel purified, cloned and sequenced.
RT-PCR analysis
RNA was extracted using the plant RNAout Kit (Tiandz) according to our previous reports13,56. All RNA samples were quantified and examined by a Nanodrop ND 1000 spectrophotometer. The first strand cDNA was synthesized from 1.0 μg total RNA by Moloney Murine Leukemia Virus Reverse Transcriptase (Promega) using oligo(dT) primers. The qRT-PCR was performed with a MyiQ Single Color Real-time PCR system (Bio-Rad) in a final volume of 20 μL containing 2 μL of a 1/10 dilution of cDNA in water, 10 μL of the 2 × SYBR Premix Ex Taq (TaKaRa) and 200 nM of forward and reverse primers. The thermal cycling conditions were 40 cycles of 95 °C for 5 s for denaturation and 60 °C for 30 s for annealing and extension. All reactions were run in triplicate. PCR efficiency was determined by a series of 2-fold dilutions of cDNAs. The calculated efficiency of all primers was 0.9 to 1.0. Cotton gene gbpolyubiquitin-2 (EE592464) was used as a normalizer and relative expression levels of genes were presented by 2−△CT (Data S10).
Expression of GhCHR and pri-miRNVL5 was detected by quantitative real-time reverse transcriptase RT-PCR (qRT-PCR) using the fluorescent intercalating dye SYBR-Green in a detection system (MJ Research, Opticon 2) according to a previous report57. A two-step qRT-PCR procedure was performed. First, total RNA samples (1.0 μg per reaction) from roots, cotyledons, hypocotyls, petals, ovules and fibers were reversely transcribed into cDNAs. Then, the cDNAs were used as templates in qRT-PCR with gene-specific primers (Data S10). Semi-quantitative RT-PCR was also used to analyze gene expression of GhCHR. The gene specific primer pairs (Data S10) were used for PCR reactions under the following conditions: pre-denaturation at 94 °C for 5 min, followed by 30 cycles of 30 s at 94 °C, 30 s at a specific annealing temperature (57 °C), and 30 s at 72 °C. Cotton actin2, an internal control for constitutive expression, was uniformly expressed in all tissues examined. As an internal control and to exclude genomic contamination, Actin2 was amplified (same cycling conditions as above for 28 cycles) from the same cDNA samples.
For qRT-PCR analysis of mature miRNVL5, total RNA isolated by TRIzol reagents (Invitrogen) was pre-treated with DNase following thein manufacturer’s instructions. Mature miRNVL5 were detected by stem-loop qRT-PCR58. The precursor miRNVL5 (pri-miRNVL5) was analyzed by the methods of Song et al.11. Transcript analysis of its target gene was performed also using qRT-PCR. The primers used for qRT-PCR are presented in Supplementary Data S10.
Transformation of GhCHR and pri-miRNVL5 into Arabidopsis
The GhCHR and Pri-miRNVL5 genomic sequences were inserted into downstream of the CaMV35s promoter in the binary vector pBI12111. The constructs were then transferred into Agrobacterium tumefaciens for Arabidopsis transformation by the floral dip method11. Transgenic lines were selected on the 1/2 MS medium with 50 mg/L kanamycin. Independent transgenic lines were obtained, and PCR was performed to verify the presence of 35S::(m)GhCHR and 35S::miRNVL5 in these transgenic Arabidopsis plants.
Degradome sequencing and data analysis in 35::miRNVL5 Arabidopsis
Degradome libraries were constructed based on a previous method34. Briefly, poly (A) RNA was extracted from total RNA sample using the Oligotex kit (Qiagen). Polyadenylated transcripts possessing 5′ monophosphates were ligated to RNA adapters consisting of a MmeI recognition site at its 3′ end. After ligation, the first-strand cDNA was generated using oligo d(T) and amplified using five PCR cycles. The PCR product was purified and digested with MmeI. The digested PCR product was then ligated to a 3′ double DNA adapter, amplified by 20 PCR cycles, and gel-purified for Illumina sequencing.
Sequenced tags with 18–21 nucleotides long were normalized after trimming sequence adapters and filtering the low quality tags. The sliced miRNA targets were identified and classified based on the CleaveLand pipeline33. The unique reads were normalized to give reads per million and subsequently mapped to annotated cDNA sequences from the Gene Index database (ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/Brassica_napus) (BnGI release 5.0).
Microarray hybridization and data analysis
Three week-old Arabidopsis seedlings were treated with 200 mM NaCl for 1 h and harvested. Total RNA was extracted using mirVanaTM RNA Isolation Kit (Applied Biosystem p/n AM1556). The extracted RNA was quantified by NanoDrop ND-2000 (Thermo Scientific). RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies). Total RNA was dephosphorylated, denaturated and labeled with Cyanine-3-CTP. After purification, the labeled RNA was hybridized onto the microarray (Agilent Technologies). The arrays were scanned with the Agilent Scanner G2505C (Agilent Technologies). Feature Extraction software (version10.7.1.1, Agilent Technologies) was used to analyze array images to get raw data. The Genspring software (version 12.5; Agilent Technologies) was employed to analyze raw data, which were normalized with the quantile algorithm. The probes that at least 1 condition out of 2 conditions has flags in “Detected” were chosen for further data analysis. Differentially expressed genes were identified through fold change as well as P value calculated using t-test. The threshold set for up- and down-regulated genes was a fold change ≥ 2.0 and a P value ≤ 0.05.
Measurement of Na+ and K+ concentrations
Three week-old Arabidopsis seedlings of transgenic and wild-type plants grown on 1/2 MS medium containing 200 mM NaCl for 3 d were harvested and dried at 80 °C, and digested with the mixture of nitric acid and hydrogen peroxide using microwave system (MARS, CEM). The digested samples were used to quantify Na+ and K+ using inductively coupled plasma-atomic emission spectrometry (ICP-AES) (Optimal 2100DV, Perkin Elmer Instruments).
Construction of Arabidopsis gene co-expression networks relevant to salt stress
Gene co-expression networks were constructed with the analytical tool of ATTED-II (http://atted.jp/), from which the relationship of genes from Arabidopsis was analyzed based on all microarray datasets38,40. Distance relationship was defined by MR (Mutual Rank) value. The co-expression of genes was analyzed with CoExSearch in ATTED-II. Usually, when MR is <50, the correlation of genes is considered as “close”; however, when MR is >1000, the relationship between two genes is very “weak”. In this study, genes with MR <1000 were considered as co-expression genes.
Statistical analysis
Experiments were independently performed in triplicate. Each result shown in the figures was the mean of three biological replicates and each treatment contained 12–40 seedlings. All transgenic (T4 homozygous) seedlings were used in the study. The significant differences between treatments were statistically evaluated by standard deviation and one-way analysis of variance (ANOVA). The data between differently treated groups were compared statistically by ANOVA followed by the least significant difference (LSD) test (P < 0.05).
Additional Information
How to cite this article: Gao, S. et al. A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 6, 19736; doi: 10.1038/srep19736 (2016).
Supplementary Material
Acknowledgments
This research was financially supported by the National Basic Research Program of China, 863 Project of China (2008AA10Z128), the Priority Academic Program Development of Jiangsu Higher Education Institution (200910) and the Annoroad Gene Technology Co., Ltd, Beijing, China.
Footnotes
Author Contributions Z.M.Y. and B.Z. conceived, designed the experiments and revised the manuscript; S.G., L.Y. and H.Q.Z. carried out the experiments, prepared samples for sequencing and microarray; Z.S.Z. and H.L. analyzed the data and finalized the manuscript; D.S. and F.X. performed qRT-PCR analysis, literature survey and statistical analysis. All authors have read and approved the manuscript.
References
- Türkan I. & Demiral T. Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 67, 2–9 (2009). [Google Scholar]
- Munns R. & Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu J. K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6, 441–445 (2003). [DOI] [PubMed] [Google Scholar]
- De Costa W., Zorb C., Hautung W. & Schubert S. Salt resistance is determined by osmotic adjustment and abscisic acid in newly developed maize hybrids in the first phase of salt stress. Physiol. Plant 131, 311–321 (2007). [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W. & Bartel D. P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell, 14, 787–799 (2004). [DOI] [PubMed] [Google Scholar]
- Phillips J. R., Dalmay T. & Bartels D. The role of small RNAs in abiotic stress. FEBS Lett. 581, 3592–3597 (2007). [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K. & Zhu J. H. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta. 1819, 137–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. H. MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M. & Luo H. Role of microRNA319 in creeping bentgrass salinity and drought stress response. Plant Signal. Behav. 9, e28700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P. et al. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 231, 991–1001 (2010). [DOI] [PubMed] [Google Scholar]
- Song J. B. et al. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biology 13, 210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdurakhmonov I. Y. et al. Small RNA regulation of ovule development in the cotton plant, G. hirsutum L. BMC Plant Biol. 8, 93 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak P. B., Wang Q. Q., Chen X. S., Qiu C. X. & Yang Z. M. Enrichment of a set of microRNAs during the cotton fiber development. BMC Genomics 10, 457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M. et al. Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biol. 10, R122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan M. B., Zhao Y. T., Meng Z. H., Wang X. H. & Yang W. C. Conserved miRNA analysis in Gossypium hirsutum through small RNA sequencing. Genomics 94, 263–268 (2009). [DOI] [PubMed] [Google Scholar]
- Wang M., Wang Q. & Zhang B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 530, 26–32 (2013). [DOI] [PubMed] [Google Scholar]
- Xie F., Jones D. C., Wang Q., Sun R. & Zhang B. Small RNA sequencing identifies miRNA roles in ovule and fibre development. Plant Biotechnol. J. 13, 355–369 (2015). [DOI] [PubMed] [Google Scholar]
- Guan X. et al. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 5, 3050 (2014). [DOI] [PubMed] [Google Scholar]
- Li G. et al. Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Mol. Biol. 74, 437–452 (2010). [DOI] [PubMed] [Google Scholar]
- Li C. et al. TaCHP: a wheat zinc finger protein gene down-regulated by absicisic acid and salinity stress plays a positive role in stress tolerance. Plant Physiol. 154, 211–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi S. J. S. R. et al. Identification of abiotic stress miRNA transcription factor binding motifs (TFBMs) in rice. Gene 531, 15–22 (2013). [DOI] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger protein, the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078 (1999). [DOI] [PubMed] [Google Scholar]
- Li W. T., He M., Wang J. & Wang Y. P. Zinc finger protein (ZFP) in plants-A review. Plant Omics 6, 474–480 (2013). [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 31, 35–40 (2006). [DOI] [PubMed] [Google Scholar]
- Sung S., Schmitz R. & Amasino R. M. A. PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Gene Dev. 20, 3244–3248 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T. et al. The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 17, 73–78 (2007). [DOI] [PubMed] [Google Scholar]
- Wei W. et al. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS One 4, e7209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. et al. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol. 11, 178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C. & Zhu J. K. The Arabidopsis thalinia salt tolerance gene SOS1 encodes aputative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse M. P., Aharon G. S., Snedden W. A. & Blmwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258 (1999). [DOI] [PubMed] [Google Scholar]
- Gaxiola R. A., Li J., Undurraga S., Dang L. M. & Allen G. J. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+ -pump. Proc. Natl. Acad. Sci. USA 98 11444–11449 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406 (2001). [DOI] [PubMed] [Google Scholar]
- Addo-Quaye C., Eshoo T. W., Bartel D. P. & Axtell M. J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. S., Song J. B. & Yang Z. M. Genome-wide identification of Brassica napus microRNAs and their targets reveals their differential regulation cadmium. J. Exp. Bot. 59, 3443–3452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M. et al. DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J. Genet. 91, 385–395 (2012). [DOI] [PubMed] [Google Scholar]
- Yang Z., Tian L., Latoszek-Green M., Brown D. & Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596 (2005). [DOI] [PubMed] [Google Scholar]
- Fu F. F. & Xue H. W. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang L. & Yang Z. M. Co-expression analysis reveals a group of genes potentially involved in regulation of plant response to iron-deficiency. Gene 554, 16–24 (2015). [DOI] [PubMed] [Google Scholar]
- Dinneny J. R. et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945 (2008). [DOI] [PubMed] [Google Scholar]
- Obayashi T. & Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 16, 249–260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. H., Pan X., Cannon C. H., Cobb G. P. & Anderson T. A. Conservation and divergence of plant microRNA genes. Plant J. 46, 243–259 (2006). [DOI] [PubMed] [Google Scholar]
- Szekely G. et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28 (2008). [DOI] [PubMed] [Google Scholar]
- Liu N. et al. Small RNA and degradome profiling reveals a role for miRNAs and their targets in the developing fibers of Gossypium barbadense. Plant J. 80, 331–344 (2014). [DOI] [PubMed] [Google Scholar]
- Leitch A. R. & Leitch I. J. Genomic plasticity and the diversity of polyploidy plants. Science 320, 481–483 (2008). [DOI] [PubMed] [Google Scholar]
- Chen Z. J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58, 377–406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Ha M., Lackey E., Wang J. & Chen Z. J. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics 178, 1845–1858 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Ng D. W., Li W. H. & Chen Z. J. Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 21, 590–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. W., Lu J. & Chen Z. J. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr. Opin Plant Biol. 15, 154–161 (2012). [DOI] [PubMed] [Google Scholar]
- Stockinger E. J., Gilmour S. J. & Thomashow M. F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F., Medina J., Rodriguez-Franco M., Neuhaus G. & Salinas J. Genetic analysis reveals a complex regulatory network modulating CBF gene expression and Arabidopsis to abiotic stress. J. Exp. Bot. 63, 293–304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O. & Thomsshow M. F. Arabidopsis CBF1 overexpression induceds COR genes and enhances freezing tolerance. Science 280, 104–106 (1998). [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M. & Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y. J. et al. Overexpresion of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol. Cell 30, 271–277 (2010). [DOI] [PubMed] [Google Scholar]
- Chen J. Y. & Dai X. F. Cloning and characterization of the Gossypium hirsutum major latex protein gene and functional analysis in Arabidopsis thaliana. Planta 231, 861–873 (2010). [DOI] [PubMed] [Google Scholar]
- Huang S. Q. et al. A set of miRNAs from Brassica napus in response to sulfate-deficiency and cadmium stress. Plant Biotechnol. J. 8, 887–899 (2010). [DOI] [PubMed] [Google Scholar]
- Wang Q. Q. et al. Transcriptome profiling of early developing cotton fibre by deep-sequencing reveals significantly differential expression of genes in a fuzzless/lintless mutant. Genomics 96, 369–376 (2010). [DOI] [PubMed] [Google Scholar]
- Li X. B., Fan X. P., Wang X. L., Cai L. & Yang W. C. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17, 859–875 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R. M., Wood M., Walton E. F. & Hellens R. P. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.