Abstract
Chemical investigation of the cultures of marine Streptomyces sp. 182SMLY led to the discovery of two new polycyclic anthraquinones, which were elucidated as N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) based on the extensive spectroscopic analysis including 2D NMR, HRESIMS, and an electronic circular dichroism (ECD) calculation. Both anthraquinones remarkably suppressed the proliferation of four different glioma cell lines with IC50 values in a range from 0.5 to 7.3 μM and induced apoptosis in the glioma cells. The ratios of IC50 for normal human astrocytes to IC50 for glioma cells were 6.4–53 for 1 and >14–31 for 2. N-acetyl-N-demethylmayamycin (1) also inhibited the growth of methicillin-resistant Staphylococcus aureus with MIC 20.0 μM.
Keywords: marine bacterium Streptomyces sp. 182SMLY, N-acetyl-N-demethylmayamycin, streptoanthraquinone A, bioactivities against glioma cells and bacteria
1. Introduction
Gliomas represent 80% of primary malignant brain tumors and remain a serious health problem despite advances in a standard treatment regimen of surgical resection followed by radiation and chemotherapy [1,2]. While chemotherapy has played an important role in the treatment and prevention of cancer, very few drugs have been approved for treating gliomas including temozolomide (TMZ), carmustine, lomustine, and bevacizumab [3]. Furthermore, only TMZ has been independently used for the treatment of gliomas, and the efficacy of TMZ and other current anti-glioma drugs remains unsatisfactory [3]. Therefore, there is an urgent need to discover lead compounds for the development of novel anti-glioma drugs. Marine-derived natural products are important sources for the discovery of new anticancer drug leads [4,5,6].
During the course of our ongoing project for the discovery of novel antibacterial and anti-tumor natural products from marine organisms [7,8,9,10,11,12], the cultures of marine bacterium strain 182SMLY isolated from a sediment sample was found to inhibit the proliferation of glioma cells. The 16S rDNA gene sequence of strain 182SMLY completely (100% identity for a 1376 bp stretch of sequence) matched those of several Streptomyces strains (Table S1), including S. griseus CB00830, S. pluricolorescens 999, S. tricolor cfcc3055, S. sporovirgulis L0801, S. sp. A15Ydz-AH, S. sp. HBUM74775, and S. sporovirgulis TGNBSA5 in the GenBank database. Previous studies showed that actinomycete S. griseus produced diversified secondary metabolites mainly including macrolides [13,14], β-lactams [15,16], quinones [17], aminoglycosides [18], diterpenoids [19], and phenazines [20], while S. pluricolorescens produced C-glycosidic quinones [21].
In this study, two novel polycyclic quinones of N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) (Figure 1) were isolated from the cultures of strain 182SMLY by using column chromatographic fractionation, followed by HPLC purification. We report herein the isolation and culture of strain 182SMLY, the structural elucidation of the isolates, and their inhibitory activities against the proliferation of glioma cells and the growth of methicillin-resistant Staphylococcus aureus and Escherichia coli.
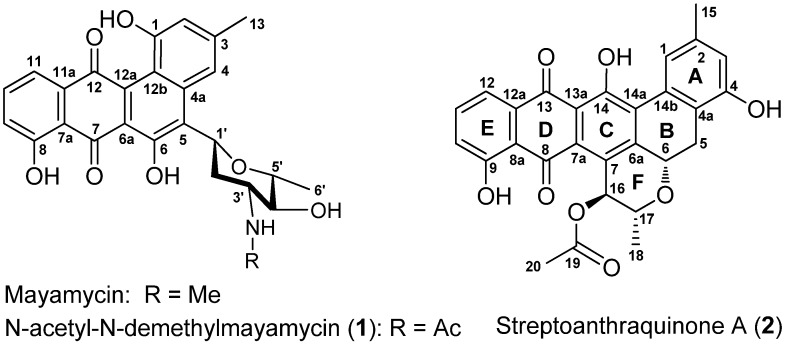
Figure 1.
Structures of compounds 1 and 2.
2. Results and Discussion
Strain 182SMLY was isolated from a sediment sample collected from the East China Sea and classified as a Streptomyces species based on the result of the 16S rDNA sequence analysis. Cultures of this bacterium were grown in the Gause’s liquid medium (30 L) and then extracted by organic solvents. The extract was fractionated by column chromatography, followed by HPLC purification, to yield two anthraquinones.
2.1. Structure Elucidation
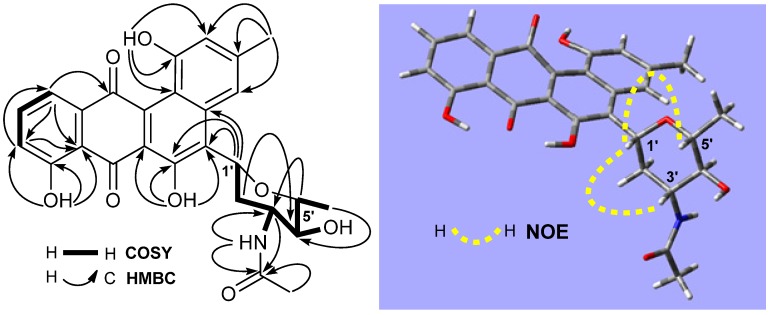
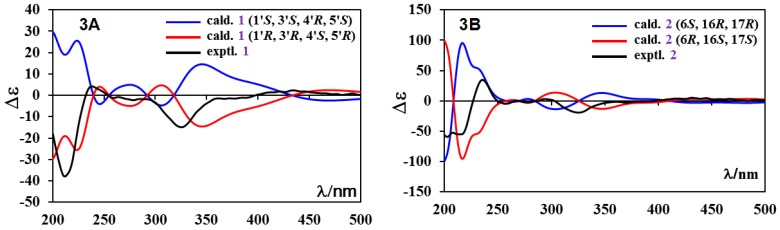
Compound 1 was obtained as a brown amorphous solid and had a molecular formula of C27H25NO8, as determined by its HRESIMS at m/z 514.1457 [M + Na]+ (calcd for C27H25NNaO8, 514.1478). Its UV absorptions at 328 and 443 nm are similar to those of mayamycin, belonging to the angucycline group [22]. The quinone group in 1 was suggested by two carbonyl signals at δ 191.6 and 184.6 (Table 1). HMBC correlations of three singlet signals at δH 12.26 (1H, OH-6), 11.36 (1H, OH-8), and 10.26 (1H, OH-1) with the three carbon signals at δC 152.1 (C-6), 160.2 (C-8), and 155.2 (C-1) indicated the presence of three hydroxy groups. The region (δ 6.5–8.0) of 1H-NMR spectrum showed five aromatic proton signals: two appeared at δ 6.64 (1H, brs) and 7.89 (1H, brs) and were assigned to H-2 and H-4; two were double doublets at δ 7.30 (1H, 7.5, 1.1 Hz) and 7.43 (1H, 7.5, 1.1 Hz) and were assigned to H-9 and H-11; and one was a triplet at δ 7.76 (1H, 7.5 Hz) and was assigned to H-10. In addition, the signals (δC 22.3; δH 2.40, 3H, s) for CH3-13 were also observed. From these NMR data, it was altogether concluded that both 1 and mayamycin had the same dehydrorabelomycin [23] as their quinone skeletons. Further NMR spectroscopic interpretation indicated that the structural difference between the two compounds was their amino sugar parts; i.e., the N-CH3 group in mayamycin was replaced by the N-acetyl group in 1. Therefore, the amino sugar in 1 was assigned as 2,3,6-trideoxy-3-acetylaminopyran, which was further confirmed by COSY correlations in combination with HMBC information, shown in Table 1 and Figure 2. The linkage of this amino sugar at C-5 was established by HMBC correlations of H-1′ (δ 5.44) with C-4a (δ 137.6), C-5 (δ 124.2), and C-6 (δ 152.1). The 3JH4′-H5′ coupling constant (9.1 Hz) and the strong NOE correlations of H-1′ (δ 5.44) with H-3′ (δ 3.85) and H-5′ (δ 3.40) (Figure 2) in the NOESY spectrum indicated the same orientation for H-1′, H-3′, and H-5′, suggesting that the relative configuration of this amino is 1′R, 3′R, 4′S, 5′R (22). In order to assign the absolute configuration, the theoretical calculation of ECD spectrum of 1 was carried out. Two preferred conformers (Figures S43 and S44) of (1′R,3′R,4′S,5′R)-1 were yielded after an OPLS (optimized potentials for liquid simulations) conformational search followed by DFT (density functional theory) optimization at the B3LYP/6-31+G(d) level. The ECD spectra of both conformers were calculated using the TDDFT (time-dependent density functional theory) method at the B3LYP-SCRF/6-31+G(d) level in methanol. The Boltzmann-weighted ECD spectrum of the conformer (1′R,3′R,4′S,5′R)-1 showed good agreement with the experimental curve of 1 (Figure 3A). Thus, the absolute configuration of 1 was assigned as (1′R,3′R,4′S,5′R). The full 1H and 13C assignments (Table 1) of 1 were made based on the COSY, HSQC, HMBC, and NOESY spectroscopic interpretations. Compound 1 was determined as N-acetyl-N-demethylmayamycin, a new analogue of mayamycin (22). Cytotoxic and antibacterial mayamycin was previously isolated from Streptomyces sp. strain HB202, a symbiotic bacterium with the marine sponge Halichondria panicea, and is the first example in the angucycline class with a C-glycoside at the C-5 of the quinone skeleton [22]. Although a robust regio- and stereocontrolled route for the synthesis of mayamycin scaffold has been recently developed [24], the total synthesis of mayamycin has not achieved yet [24,25] and is still underway [24]. To the best of our knowledge, N-acetyl-N-demethylmayamycin (1) is the first reported addition to this class of mayamycin.
Table 1.
13C- and 1H-NMR Data of N-acetyl-N-demethylmayamycin (1) (in DMSO-d6).
| No. | δC, Type | δH (J = Hz) | HMBC | No. | δC, Type | δH (J = Hz) | HMBC |
|---|---|---|---|---|---|---|---|
| 1 | 155.2, C | 12a | 137.7 a, C | ||||
| 2 | 111.5, CH | 6.64, brs | 1, 4, 12b, 13 | 12b | 115.4, C | ||
| 3 | 141.0, C | 13 | 22.3, CH3 | 2.40, s | 2, 3, 4 | ||
| 4 | 115.4, CH | 7.89, brs | 2, 5, 12b, 13 | 1′ | 71.0, CH | 5.44, dd (12.1, 2.3) | 4a, 5, 6 |
| 4a | 137.6 a, C | 2′ | 36.0, CH2 | 1.86, m; 2.10, 1H, m | 1′, 3′, 4′ | ||
| 5 | 124.2, C | 3′ | 52.5, CH | 3.85, m | 7′ | ||
| 6 | 152.1, C | 4′ | 74.2, CH | 3.20, m | 3′ | ||
| 6a | 117.6, C | 5′ | 77.7, CH | 3.40, dd (9.1, 6.1) | 3′ | ||
| 7 | 191.6, C | 6′ | 18.6, CH3 | 1.27, d (6.1) | 4′, 5′ | ||
| 7a | 115.6, C | 7′ | 169.2, C | ||||
| 8 | 160.2, C | 8′ | 22.3, CH3 | 1.80, s | 7′ | ||
| 9 | 122.9, CH | 7.30, dd (7.5, 1.1) | 7a, 8, 11 | 1-OH | 10.26, s | 1, 2, 12b | |
| 10 | 137.5, CH | 7.76, t (7.5) | 8, 11, 11a | 6-OH | 12.26, s | 5, 6, 6a | |
| 11 | 117.9, CH | 7.43, dd (7.5, 1.1) | 7a, 9, 12 | 8-OH | 11.36, s | 7a, 8, 9 | |
| 11a | 136.6, C | 3′-NH | 7.87, d (8.2) | 3′, 7′ | |||
| 12 | 184.6, C | 4′-OH | 5.02, d (5.1) |
a The data with the same label in each column may be interchanged.
Figure 2.
1H–1H COSY, key HMBC and NOE correlations of N-acetyl-N-demethylmayamycin (1).
Figure 3.
The experimental and calculated ECD spectra of (A) N-acetyl-N-demethylmayamycin (1) and (B) streptoanthraquinone A (2).
Compound 2, a dark brown amorphous powder, had a molecular formula of C28H22O8 deduced from its HRESIMS data at m/z [M + Na]+ 509.1233 (calcd for C28H22NaO8, 509.1212) and 13C-NMR data (Table 2). The prelimirary NMR spectroscopic interpretation indicated that compound 2 contained the moieties of a quinone group (δC 193.5, 189.6), a carbonyl (δC 170.3), three hydroxyls, 18 aromatic carbons, three oxymethines, three methyls, and a methylene. These data suggested that 2 may have a dihydrobenzo[a]naphthacenequinone skeleton, which usually has five rings A, B, C, D, and E (Figure 1) [26]. More detailed NMR data from 1H, 13C, 1H–1H COSY, HSQC, HMBC, and NOESY spectra and an ECD calculation enabled the determination of the structure of 2 as described below.
Table 2.
13C- and 1H-NMR Data of streptoanthraquinone A (2) (in CDCl3, J = Hz).
| No. | δC, Type | δH (J = Hz) | HMBC | No. | Δc, Type | δH (J = Hz) | HMBC |
|---|---|---|---|---|---|---|---|
| 1 | 118.1, CH | 8.14, d (1.6) | 3, 4a, 14a, 14b, 15 | 12a | 135.3, C | ||
| 2 | 143.0, C | 13 | 189.6, C | ||||
| 3 | 119.1, CH | 7.01, d (1.6) | 1, 4, 4a, 15 | 13a | 118.9 a, C | ||
| 4 | 154.7, C | 14 | 154.3, C | ||||
| 4a | 139.1, C | 14a | 129.7, C | ||||
| 5 | 29.8, CH2 | 2.88, dd (15.6, 12.1); 3.57, dd (15.6, 3.3) |
4, 6, 6a 6, 6a, 14b |
14b | 118.8 a, C | ||
| 6 | 72.3, CH | 5.77, dd (12.1, 3.3) | 4a, 7, 14a, 17 | 15 | 22.6, CH3 | 2.50, s | 1, 2, 3 |
| 6a | 152.9, C | 16 | 73.1, CH | 5.33, d (9.5) | 6a, 7a, 17, 18, 19 |
||
| 7 | 127.7, C | 17 | 77.1, CH | 3.93, dd (9.5, 6.1) |
7 | ||
| 7a | 133.4, C | 18 | 19.0, CH3 | 1.43, d (6.1) | 16, 17 | ||
| 8 | 193.5, C | 19 | 170.3, C | ||||
| 8a | 114.9, C | 20 | 21.0, CH3 | 2.23, s | 19 | ||
| 9 | 162.2, C | 4-OH | 9.25, s | 3, 4 | |||
| 10 | 125.5, CH | 7.34, dd (7.5, 1.2) | 8a, 9, 12 | 9-OH | 11.69, s | 8a, 9, 10 | |
| 11 | 138.1, CH | 7.71, t (7.5) | 9, 12a | 14-OH | 12.84, s | 13a, 14, 14a | |
| 12 | 121.7, CH | 7.81, dd (7.5, 1.2) | 8a, 10, 13 |
a The data with the same label in each column may be interchanged.
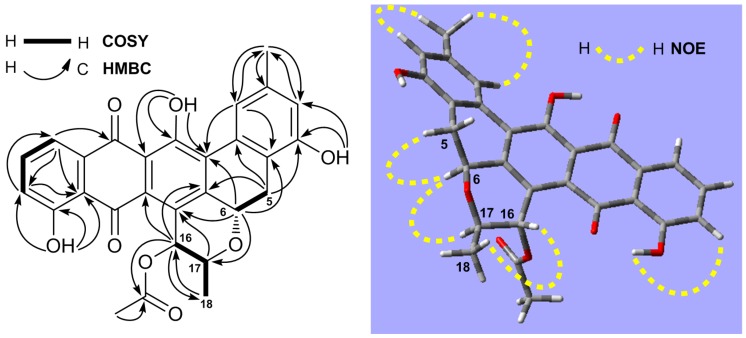
The 1H-NMR spectrum displayed five aromatic protons at δ 8.14 (d, 1.6 Hz), 7.01 (d, 1.6 Hz), 7.34 (dd, 7.5, 1.2 Hz), 7.71 (t, 7.5 Hz), and 7.81 (dd, 7.5, 1.2 Hz), which were assigned to H-1, H-3, H-10, H-11, and H-12, respectively. Three OH signals resonated at δ 12.84 (1H, s), 11.69 (1H, s), and 9.25 (1H, s), and the downfield shifts of δ 11.69 and 12.84 suggested that the protons from these two hydroxy groups had hydrogen bond relationships with carbonyls at C-8 and C-13. Further location of these three OH groups at C-4, C-9, and C-14 was made from the HMBC correlations, shown in Table 2 and Figure 4. One methyl (δC 22.6 and δH 2.50) was assigned at C-15 based on the HMBC correlations of H-15 (δ 2.50) with C-1 (δ 118.1), C-2 (δ 143.0), and C-3 (δ119.1), and both H-1 (δ 8.14) and H-3 (δ 7.01) with C-15 (δ 22.6). COSY correlations of H-5 with H-6 indicated their neighbor relationship of the methylene (δC 29.8 and δH 2.88, 3.57) at C-5 and the oxymethine at C-6 (δC 72.3 and δH 5.77). The HMBC correlations of H-5 with C-4 (δ 154.7), C-6a (δ 152.9), and C-14b (δ 118.8), and of H-6 with C-4a (δ 139.1), C-7 (δ 127.7), C-14a (δ 129.7), and C-17 (δ 77.1) demonstrated the presence of ring B. Similarly, the link sequence of the two oxymethines at C-16 (δC 73.1 and δH 5.33) and C-17 (δC 77.1 and δH 3.93) with the methyl at C-18 (δC 19.0 and δH 1.43, d, 6.1 Hz) was reduced from the COSY correlations of H-16 with H-17 and H-17 with H-18. The HMBC correlations of H-16 with C-6a, C-7a (δ 133.4), C-17, and C-18, and of H-17 with C-7, as well as a strong NOE correlation of H-17 with H-6, proved the presence of ring F. Finally, the acetyl group at C-16 was established by the HMBC correlations of H-16 with C-19 (δ 170.3) and of H-20 (δ 2.23) with C-19. The relative stereochemistry of 2 was proposed based on 3J coupling constants of protons and NOESY experiment. The 3J coupling constants of 12.1 Hz for αH-5/βH-6 and 9.5 Hz for αH-16/βH-17 indicated axial orientations for αH-5/βH-6 and αH-16/βH-17. NOESY spectrum showed strong NOE correlations of βH-6 (δ 5.77) with βH-5 (δ 3.57) and βH-17 (δ 3.93), and of αH-16 (δ 5.33) with αH-18 (δ 1.43), but no NOE for βH-6 with αH-5 (δ 2.88) or weak NOE for αH-16 with βH-17 were observed (Figure 4 and Figures S40). All of the above evidence suggested the orientations of βH-6, αH-16, and βH-17. The absolute configuration of 2 was proposed by a quantum chemical calculation of its ECD spectrum using the same method as that of 1. Considering the rigid structure [27,28,29] of 2, one preferred conformer (Figure S45) of (6S,16R,17R)-2 was afforded by conformational analysis and geometrical optimization. The ECD spectrum was then calculated using the TDDFT method. As shown in Figure 3B, the experimental ECD curve of 2 was similar to the calculated ECD spectrum of (6S,16R,17R)-2 rather than of (6R,16S,17S)-2. Thus, the absolute configuration of 2 was proposed to be 6S, 16R, 17R. The full 1H and 13C assignments (Table 2) of 2 were achieved by extensive NMR spectral analysis. Compound 2 was elucidated as streptoanthraquinone A, a novel dihydrobenzo[a]naphthacenequinone [26] with a rare oxygen-contained hexatomic ring F at C-6 and C-7 positions. The skeleton of dihydrobenzo[a]naphthacenequinone, such as benaphthamycin [30], benanomicins, ericamycin, pradimicins, and WS-79089A [26], usually has five rings, A, B, C, D, and E. Benaphthamycin, ericamycin, and WS-79089A have an additional ring F at C-2 and C-3 positions, while compound 2 had its sixth ring F at C-6 and C-7 positions, which is unique. To the best of our knowledge, such a structure is the first that has ever been found in the class of dihydrobenzo[a]naphthacenequinones.
Figure 4.
1H–1H COSY, key HMBC and NOE correlations of streptoanthraquinone A (2).
2.2. Biological Activities
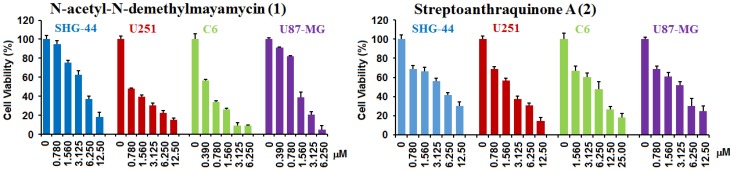
The compounds 1 and 2 were assayed for their activities inhibiting the proliferation of glioma C6, U251, U87-MG, and SHG-44 cells using the sulforhodamine B (SRB) assay. The SRB assay is a method that measures total cellular protein content for evaluating the activity of tested compounds against the proliferation of tumor cells. Doxorubicin (DOX) was used as a positive control. Glioma cells were treated with tested compounds for 72 h at different concentrations. The results (Table 3 and Figure 5) showed that N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) significantly inhibited the proliferation of four different tested glioma cell lines with IC50 values in a range from 0.5–3.9 μM for 1 and 3.3–7.3 μM for 2. The control DOX displayed activities with IC50 0.9–9.0 μM. N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) were also assayed for their activity against normal human astrocytes (HA). The initial assay was conducted on the concentrations from 0.35 to 7.0 μM for 1 and on those from 1.65 to 16.5 μM for 2. The cell viability of HA cells from each tested concentration of both compounds was 100%. The results from the second assay in higher concentrations showed (Table 3) that the IC50 values of compounds 1 and 2 against HA cells were 25 μM for 1 and >100 μM for 2. The ratios of IC50 for human astrocytes (IC50HA) to IC50 for glioma cells (IC50gc) were 6.4–53 for 1 and >14–31 for 2.
Table 3.
Activity of N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) inhibiting the proliferation of cells (IC50: μM).
| Compounds | U251 | U87-MG | SHG-44 | C6 | HA |
|---|---|---|---|---|---|
| N-acetyl-N-demethylmayamycin (1) | 0.7 ± 0.2 | 1.4 ± 0.1 | 3.9 ± 0.4 | 0.5 ± 0.1 | 25 ± 1.3 |
| Ratios of IC50HA/IC50gc | 35 | 18 | 6.4 | 53 | |
| Streptoanthraquinone A (2) | 3.3 ± 0.3 | 4.6 ± 0.3 | 6.5 ± 1.1 | 7.3 ± 1.4 | >100 |
| Ratios of IC50HA/IC50gc | >31 | >22 | >16 | >14 | |
| Doxorubicin (DOX) | 6.7 ± 1.1 | 0.9 ± 0.1 | 9.0 ± 0.8 | 1.0 ± 0.1 | NT |
NT: No testing.
Figure 5.
N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) inhibited the proliferation of glioma SHG-44, U251, C6, and U87-MG cells. Glioma cells were treated with 1 or 2 for 72 h at different concentrations. Values are means ± S.D. from five independent experiments.
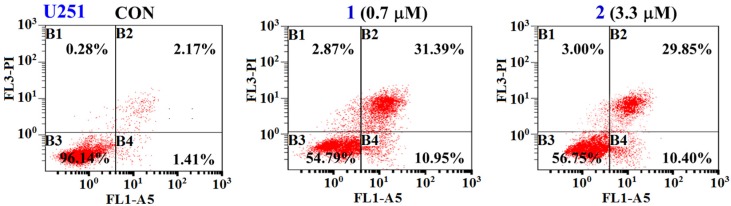
N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) were further tested for their ability to induce apoptosis in glioma U251 cells by flow cytometry using Annexin V-FITC/PI double staining. U251 cells were treated with N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) in their IC50 concentrations of 0.7 μM for 1 and 3.3 μM for 2 for 36 h, stained with annexin-V FITC and PI, and then analyzed by using flow cytometry. It was found that the total apoptotic cells including early and late apoptotic cells were increased by 38.76% for 1 and 36.67% for 2 when compared to the control (CON, 3.58%, Figure 6). These data demonstrated that N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) significantly induced apoptosis in the glioma U251 cells.
Figure 6.
N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) induced apoptosis in the glioma U251 cells quantified by cytometric analysis. U251 cells were treated with 1 (0.7 μM) or 2 (3.3 μM) for 36 h and then stained with Annexin-V FITC and PI double staining (B1: necrotic cells; B2: late apoptotic cells; B3: normal glioma cells; B4: early apoptotic cells).
Compounds 1 and 2 were also determined for their activities against methicillin-resistant Staphylococcus aureus ATCC 43300 and Escherichia coli ATCC 25922. The result indicated that only N-acetyl-N-demethylmayamycin (1) inhibited the growth of S. aureus with MIC 20.0 μM. The positive control norfloxacin inhibited the growth of both S. aureus and E. coli with MIC values of 62.6 μM and 31.3 μM, respectively. However, both compounds showed no activity against E. coli.
3. Experimental Section
3.1. General Experimental Procedures
Optical rotations were measured on a JASCO DIP-370 digital polarimeter. CD spectra were recorded on a JASCO J 715 spectropolarimeter. IR spectra were recorded on an AVATAR 370 FT-IR spectrometer (Thermo Nicolet, Madison, WI, USA). NMR spectra were acquired on a Bruker 500 spectrometer using standard pulse programs and acquisition parameters. Chemical shifts were expressed in δ (ppm) and referred to the NMR solvent used. HRESIMS data were acquired on an Agilent 6230 TOF LC/MS spectrometer. Octadecyl-functionalized silica gel (ODS, Cosmosil 75C18-Prep, Nacalai Tesque Inc., Kyoto, Japan) was used for column chromatography. HPLC purification was performed on an Agilent 1260 HPLC system with DAD detector. HPLC and analytic grade solvents used for this study were purchased from the Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China). Human glioma U251, U87-MG, and SHG-44 cells, and rat glioma C6 cells, were obtained from the Cell Bank of the Chinese Academy of Sciences. Normal human astrocytes (HA, Cat. No. 1800) were obtained from ScienCell. The methicillin-resistant Staphylococcus aureus ATCC 43300 and Escherichia coli ATCC 25922 were gifts from Professor Zhongjun Ma and Dr. Pinmei Wang. Nutrient Broth (NB), Mueller Hinton Broth (MHB), and Gause’s-agar were purchased from Hangzhou Microbial Reagent Co. Ltd. (Hangzhou, China), Thermo Fisher Scientific Inc. (Waltham, MA, USA), and Guangdong Huankai Microbial Science and Technology Co. Ltd. (Guangzhou, China), respectively. Doxorubicin (DOX, >98.0%) was obtained from Sigma-Aldrich and norfloxacin (98%) from Saen Chemical Technology Co. Ltd. (Shanghai, China).
3.2. Isolation and Taxonomic Identity of Marine Streptomyces sp. 182SMLY
Strain 182SMLY was derived from a sample of marine sediment at a 3.6 m depth, which was collected from the East China Sea, close to Zhoushan City, Zhejiang Province, China in August 2013. Briefly, the marine sediments (3.0 g) were air-dried for five days in a sterile centrifuge tube. The dried sample was diluted into 0.01 g/mL with seed broth (1.5% glucose, 1.5% glycerol, 1.5% malt extract, 2.5% yeast extract, 0.5% casamino acids, and 0.1% calcium carbonate). The diluted sample (200 μL) was dispersed across a Bacto-agar plate and then incubated at room temperature for 10 days. Bactria colonies were picked with sterile needles and transferred to Bacto-agar plates. After another seven days of growth at room temperature, the single colony (strain 182SMLY) that grew well was transferred onto Gause’s synthetic agar media. Working stocks were prepared on Gause’s synthetic agar slants and stored at 4 °C until use.
16S rDNA analysis was used to determine the taxonomic identity of strain 182SMLY, and the DNA sequence using BLAST (nucleotide sequence comparison) was compared to the GenBank database. The 16S rDNA sequence of strain 182SMLY has been deposited in GenBank (accession number: KT899860). A voucher strain (Streptomyces sp. 182SMLY) of this actinomycete was preserved at the Laboratory of Institute of Marine Biology, Ocean College, Zhejiang University, China.
3.3. Culture of Strain Streptomyces sp. 182SMLY
Colonies of the strain growing on Gause’s synthetic agar media were inoculated into a 500 mL Erlenmeyer flask containing 200 mL of a liquid medium (20 g/L starch, 1.0 g/L KNO3, 0.5 g/L K2HPO4, 0.5 g/L MgSO4·7H2O, 0.5 g/L NaCl, 0.01 g/L FeSO4·7H2O), and the colonies were then incubated at 28 °C for 5 days on a rotary shaker (180 rpm) to produce seed broth. The seed broth (5 mL) was then inoculated into a 500 mL Erlenmeyer flask that contained 250 mL of liquid Gause’s synthetic media. The flask was incubated at 28 °C for 10 days on a rotary shaker (180 rpm). A total of 30 L fermentation was made for this study.
3.4. Extraction and Isolation of Compounds
The fermentation broth (30 L) of the isolated marine Streptomyces sp. 182SMLY was filtered with a filter press to give filtrate and mycelia. The filtrate was extracted with EtOAc three times (each 10.0 L), and the mycelia were extracted with MeOH. The EtOAc and MeOH phases were combined and then dried in vacuo to afford a crude extract (5.98 g). This crude extract was fractionated by column chromatography of ODS (10 × 150 cm) successively eluting with 70%, 85%, and 100% MeOH to yield three fractions (Frs. 1–3). Fr. 3 was further separated by HPLC using an Agilent column (Zorbax SB-C18, 250 × 9.4 mm, 5 μm) with an isocratic mobile phase of MeOH and H2O (80:15) at a flow rate of 1.0 mL/min and UV detection wavelength of 256 nm to give N-acetyl-N-demethylmayamycin (1, 4.5 mg, tR 16.46 min) and streptoanthraquinone A (2, 2.3mg, tR 21.45 min).
N-acetyl-N-demethylmayamycin (1): dark brown amorphous powder; molecular formula C27H25NO8; tR 16.48 min (85% MeOH in H2O); + 63.67 (c 0.50, DMSO); UV (MeOH) λmax (log ε) 217 (4.70), 236 (4.69), 328 (4.36), 443 (4.06) nm; ECD (10 mg/L, MeOH) λmax (Δε) 212 (−38.1), 238 (+4.0), and 325 (−14.8) nm; IR (KBr) νmax 3426, 2925, 2853, 1732, 1712, 1629, 1598, 1458, 1375, 1066, 1024 cm−1; 1H (500 MHz, in DMSO-d6) and 13C (125 MHz, in DMSO-d6) NMR data, see Table 1; HRESIMS m/z [M + Na]+ 514.1457 (calcd for C27H25NNaO8, 514.1478).
Streptoanthraquinone A (2): dark brown amorphous powder; molecular formula C28H22O8; tR 21.45 min (85% MeOH in H2O); + 51.90 (c 0.50, DMSO); UV (MeOH) λmax (log ε) 220 (4.70), 330 (3.40), 445 (3.06) nm; ECD (10 mg/L, MeOH) λmax (Δε) 215 (−59.2), 236 (+39.1, and 325 (−20.0) nm; IR (KBr) νmax 3435, 2925, 2853, 1738, 1712, 1639, 1459, 1381, 1024 cm−1; 1H (500 MHz, in DMSO-d6) and 13C (125 MHz, in DMSO-d6) NMR data, see Table 2; HRESIMS m/z [M + Na]+ 509.1233 (calcd for C28H22NaO8, 509.1212).
3.5. Computational Methods
Conformational analysis was performed with a MacroModel employing an OPLS force field. Geometrical optimization and energy calculations were performed applying the DFT method at the B3LYP/6-31+G(d) level with the Gaussian 09 program package [31]. The TDDFT calculation was run at the same level and the ECD spectra were generated by the program SpecDis v1.53 and were summed according to the Boltzmann-weighting of each individual conformer.
3.6. Cells Culture
Human glioma U251 and rat glioma C6 cells were cultured in DMEM (Dulbecco’s Modified Eagle Medium, Gibco, Thermo Fisher Scientific Inc., Waltham, MA, USA) with 10% FBS (Fetal Bovine Serum, PAA Laboratories Inc., Dartmouth, MA, USA), human glioma SHG-44 in RPMI-1640 Medium (Roswell Park Memorial Institution 1640 Medium, Gibco, Thermo Fisher Scientific Inc., Waltham, MA, USA), human glioma U87-MG cells in MEM (Minimum Essential Medium, Gibco), and normal human astrocytes (HA) in AM (Astrocyte Medium, ScienCell, Cat. No. 1801). All cells were incubated at 37 °C in a humidified incubator with 5% CO2. Cells after the third generation were used for experiment.
3.7. Sulforhodamine B (SRB) Assay
The SRB assay [8,9] was used to evaluate the activity of the isolated compounds in inhibiting the proliferation of glioma U87-MG, U251, SHG-44, and C6 cells. Doxorubicin (DOX) was used as the positive control. Briefly, glioma cells were plated in a 96-well plate and then treated with different concentrations of tested compound after cells adhesion for 24 h. After 72 h of the treatment, cells were fixed with 50 μL of a 10% cold TCA solution for 1 h at 4 °C, washed with distilled water five times, and then dried at room temperature. The dried cells were stained with 50 μL of 0.4% SRB for ten minutes and rinsed with a 1% acetic acid solution five times. After dried, dye was dissolved in a 10 mM Tris buffer and measured at 515 nm on a microplate reader (BioTech, Winooski, VT, USA).
3.8. Antimicrobial Assay
The micro broth dilution method as described in the previous report [12] was used to determine the antimicrobial activity of the isolated compounds against the methicillin-resistant Staphylococcus aureus ATCC 43300 and Escherichia coli ATCC 25922. Norfloxacin, a broad-spectrum antibiotic against both Gram-positive and Gram-negative bacteria, was used as the positive control.
4. Conclusions
Two new anthraquinones were isolated from the culture of Streptomyces sp. 182SMLY, which was isolated from a marine sediment sample. Their structures were determined to be N-acetyl-N-demethylmayamycin (1) and streptoanthraquinone A (2) based on the extensive spectroscopic analysis including 2D NMR, HRESIMS and the electronic circular dichroism (ECD) calculation. Both new compounds significantly inhibited the proliferation of four different glioma cell lines with IC50 values of 0.5 to 7.3 μM and induced apoptosis in the glioma cells. N-acetyl-N-demethylmayamycin (1) also showed strong activity against the growth of methicillin-resistant Staphylococcus aureus with MIC 20.0 μM.
Acknowledgments
This study was supported by a grant from the National Natural Science Foundation of China (No. 81273428). We appreciate Kai Chen (Laboratory of Computational Chemistry and Drug Design, Laboratory of Chemical Genomics, Peking University Shenzhen Graduate School, China) for the ECD calculation. Authors also thank Jianyang Pan (Pharmaceutical Informatics Institute of Zhejiang University, China) for performing the NMR spectrometry, and Zhongjun Ma and Pinmei Wang (Ocean College, Zhejiang University) for their gifts of Staphylococcus aureus and Escherichia coli.
Abbreviations
- DFT
Density functional theory
- DOX
Doxorubicin
- ECD
Electronic circular dichroism
- ODS
Octadecyl-functionalized silica gel
- OPLS
Optimized potentials for liquid simulations
- SRB
Sulforhodamine B
- TDDFT
Time-dependent density functional theory
- TMZ
Temozolomide
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/14/1/10. UV, NMR, and HRESIMS spectra of compounds 1 and 2 as well as other supporting data.
Author Contributions
Ying Liang performed the isolation and structural elucidation of compounds; Xin Xie, Lu Chen, Xuewei Ye and Komal Anjum conducted the bioactive assay; Shilun Yan analyzed the data; Zhizhen Zhang, Xiaoyuan Lian and Haocai Huang designed the experiments and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ostrom Q.T., Gittleman H., Farah P., Ondracek A., Chen Y., Wolinsky Y., Stroup N.E., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl. 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patil S.A., Hosni-Ahmed A., Jones T.S., Patil R., Pfeffer L.M., Miller D.D. Novel approaches to glioma drug design and drug screening. Expert Opin. Drug Discov. 2013;8:1135–1151. doi: 10.1517/17460441.2013.807248. [DOI] [PubMed] [Google Scholar]
- 3.Chamberlain M.C. Temozolomide: Therapeutic limitations in the treatment of adult high-grade gliomas. Expert Rev. Neurother. 2010;10:1537–1544. doi: 10.1586/ern.10.32. [DOI] [PubMed] [Google Scholar]
- 4.Schumacher M., Kelkel M., Dicato M., Diederich M. Gold from the sea: Marine compounds as inhibitors of the hallmarks of cancer. Biotechnol. Adv. 2011;29:531–547. doi: 10.1016/j.biotechadv.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Petit K., Biard J.F. Marine natural products and related compounds as anticancer agents: An overview of their clinical status. Anticancer Agents Med. Chem. 2013;13:603–631. doi: 10.2174/1871520611313040010. [DOI] [PubMed] [Google Scholar]
- 6.Newman D.J., Cragg G.M. Marine-sourced anti-cancer and cancer pain control agents in clinical and late preclinical development. Mar. Drugs. 2014;12:255–278. doi: 10.3390/md12010255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin W.X., Ye X.W., Yu S.R., Lian X.Y., Zhang Z.Z. New capoamycin-type antibiotics and polyene acids from marine Streptomyces fradiae PTZ0025. Mar. Drugs. 2012;10:2388–2402. doi: 10.3390/md10112388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S.R., Ye X.W., Chen L., Lian X.Y., Zhang Z.Z. Polyoxygenated 24,28-epoxyergosterols inhibiting the proliferation of glioma cells from sea anemone Anthopleura midori. Steroids. 2014;88:19–25. doi: 10.1016/j.steroids.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Yu S.R., Ye X.W., Huang H.C., Peng R., Su Z.H., Lian X.Y., Zhang Z.Z. Bioactive sulfated saponins from sea cucumber Holothuria moebii. Planta Med. 2015;81:152–159. doi: 10.1055/s-0034-1383404. [DOI] [PubMed] [Google Scholar]
- 10.Chen L., Liang L., Song T.F., Anjum K., Wang W.L., Yu S.R., Huang H.C., Lian X.Y., Zhang Z.Z. Synthesis and bioactivity of tripolinolate A from Tripolium vulgare and its analogs. Bioorg. Med. Chem. Lett. 2015;25:2629–2633. doi: 10.1016/j.bmcl.2015.04.091. [DOI] [PubMed] [Google Scholar]
- 11.Yu S.R., Ye X.W., Chen L., Xie X., Zhou Q., Lian X.Y., Zhang Z.Z. Cytotoxic and anti-colorectal tumor effects of sulfated saponins from sea cucumber Holothuria moebii. Phytomedicine. 2015;22:1112–1119. doi: 10.1016/j.phymed.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Ye X.W., Anjum K., Song T.F., Wang W.L., Yu S.R., Huang H.C., Lian X.Y., Zhang Z.Z. A new curvularin glycoside and its cytotoxic and antibacterial analogues from marine actinomycete Pseudonocardia sp. HS7. Nat. Prod. Res. 2015 doi: 10.1080/14786419.2015.1047775. [DOI] [PubMed] [Google Scholar]
- 13.Werner G., Hagenmaier H., Drautz H., Baumgartner A., Zähner H. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. 1984;37:110–117. doi: 10.7164/antibiotics.37.110. [DOI] [PubMed] [Google Scholar]
- 14.Nair M.G., Putnam A.R., Mishra S.K., Mulks M.H., Taft W.H., Keller J.E., Miller J.R., Zhu P.P., Meinhart J.D., Lynn D.G. Faeriefungin: A new broad-spectrum antibiotic from Streptomyces griseus var. autotrophicus. J. Nat. Prod. 1989;52:797–809. doi: 10.1021/np50064a022. [DOI] [PubMed] [Google Scholar]
- 15.Harada S., Shinagawa S., Nozaki Y., Asai M., Kishi T. C-19393 S2 and H2, new carbapenem antibiotics. II. isolation and structures. J. Antibiot. 1980;33:1425–1430. doi: 10.7164/antibiotics.33.1425. [DOI] [PubMed] [Google Scholar]
- 16.Harada S., Nozaki Y., Shinagawa S., Kitano K. C-19393 E5, a new carbapenem antibiotic. Fermentation, isolation and structure. J. Antibiot. 1982;35:957–962. doi: 10.7164/antibiotics.35.957. [DOI] [PubMed] [Google Scholar]
- 17.Otani T., Yamawaki I., Matsumoto H., Minami Y., Yamada Y., Marunaka T., Qi C., Tian T., Zhang R. New antibiotics 4181-A and B from Streptomyces griseus; taxonomy, fermentation, isolation and characterization. J. Antibiot. 1988;41:275–281. doi: 10.7164/antibiotics.41.275. [DOI] [PubMed] [Google Scholar]
- 18.Tohma S., Kondo H., Yokotsuka J., Iwamoto J., Matsuhashi G., Ito T., Seto H. Ashimycins A and B, new streptomycin analogues. J. Antibiot. 1989;42:1205–1212. doi: 10.7164/antibiotics.42.1205. [DOI] [PubMed] [Google Scholar]
- 19.Xie P., Ma M., Rateb M.E., Shaaban K.A., Yu Z., Huang S.X., Zhao L.X., Zhu X., Yan Y., Peterson R.M., et al. Biosynthetic potential-based strain prioritization for natural product discovery: A showcase for diterpenoid-producing actinomycetes. J. Nat. Prod. 2014;77:377–387. doi: 10.1021/np401063s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosoya Y., Adachi H., Nakamura H., Nishimura Y., Naganawa H., Okami Y., Takeuchi T. The structure of diphenazithionin, a novel antioxidant from Streptomyces griseus ISP 5236. Tetrahedron Lett. 1996;37:9227–9228. doi: 10.1016/S0040-4039(96)02190-9. [DOI] [Google Scholar]
- 21.Byrne K.M., Gonda S.K., Hilton B.D. Largomycin FII chromophore component 4, a new pluramycin antibiotic. J. Antibiot. 1985;38:1040–1049. doi: 10.7164/antibiotics.38.1040. [DOI] [PubMed] [Google Scholar]
- 22.Schneemann I., Kajahn I., Ohlendorf B., Zinecker H., Erhard A., Nagel K., Wiese J., Imhoff J.F. Mayamycin, a cytotoxic polyketide from a Streptomyces strain isolated from the marine sponge Halichondria panicea. J. Nat. Prod. 2010;73:1309–1312. doi: 10.1021/np100135b. [DOI] [PubMed] [Google Scholar]
- 23.Gould S.J., Cheng X.C., Halley K. Biosynthesis of dehydrorabelomycin and PD 116740: Prearomatic deoxygenation as evidence for different polyketide synthases in the formation of benz[a]anthraquinones. J. Am. Chem. Soc. 1992;114:10066–10068. doi: 10.1021/ja00051a052. [DOI] [Google Scholar]
- 24.Mitra P., Behera B., Maiti T.K., Mal D. Angucycline C5 glycosides: Regio- and stereocontrolled synthesis and cytotoxicity. J. Org. Chem. 2013;78:9748–9757. doi: 10.1021/jo4013892. [DOI] [PubMed] [Google Scholar]
- 25.Wu K., Wang M., Yao Q., Zhang A. Synthesis study toward mayamycin. Chin. J. Chem. 2013;31:93–99. doi: 10.1002/cjoc.201201084. [DOI] [Google Scholar]
- 26.Fernekorn U., Heinze T., Schlegel B., Dahse H.M., Gräfe U. Synthesis of a new polycyclic quinone by reduction of a dihydrobenzo. J. Antibiot. 2001;54:191–192. doi: 10.7164/antibiotics.54.191. [DOI] [PubMed] [Google Scholar]
- 27.Berova N., di Bari L., Pescitelli G. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem. Soc. Rev. 2007;36:914–931. doi: 10.1039/b515476f. [DOI] [PubMed] [Google Scholar]
- 28.Gao Y.P., Shen Y.H., Zhang S.D., Tian J.M., Zeng H.W., Ye J., Li H.L., Shan L., Zhang W.D. Incarvilleatone, a new cyclohexylethanoid dimer from Incarvillea younghusbandii and its inhibition against nitric oxide (NO) release. Org. Lett. 2012;14:1954–1957. doi: 10.1021/ol3004639. [DOI] [PubMed] [Google Scholar]
- 29.Yan S.L., Su Y.F., Chen L., Que M., Gao X.M., Chang J.B. Polygonumosides A–D, stilbene derivatives from processed roots of Polygonum multiflorum. J. Nat. Prod. 2014;77:397–401. doi: 10.1021/np400720y. [DOI] [PubMed] [Google Scholar]
- 30.Ritzau M., Vettermann R., Fleck W.F., Gutsche W., Dornberger K. Benaphthamycin, a new dihydrobenzo[a]naphthacenequinone antibiotic from Streptomyces sp. HKI-0057. J. Antibiot. 1997;50:791–793. doi: 10.7164/antibiotics.50.791. [DOI] [PubMed] [Google Scholar]
- 31.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., et al. Gaussian 09. Gaussian, Inc.; Wallingford, CT, USA: 2009. Revision E.01. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.