Abstract
The mycotoxin, aflatoxin B1 (AFB1) is a hepatotoxic, immunotoxic, and mutagenic contaminant of food and animal feeds. In poultry, AFB1 can be maternally transferred to embryonated eggs, affecting development, viability and performance after hatch. Domesticated turkeys (Meleagris gallopavo) are especially sensitive to aflatoxicosis, while Eastern wild turkeys (M. g. silvestris) are likely more resistant. In ovo exposure provided a controlled AFB1 challenge and comparison of domesticated and wild turkeys. Gene expression responses to AFB1 in the embryonic hepatic transcriptome were examined using RNA-sequencing (RNA-seq). Eggs were injected with AFB1 (1 μg) or sham control and dissected for liver tissue after 1 day or 5 days of exposure. Libraries from domesticated turkey (n = 24) and wild turkey (n = 15) produced 89.2 Gb of sequence. Approximately 670 M reads were mapped to a turkey gene set. Differential expression analysis identified 1535 significant genes with |log2 fold change| ≥ 1.0 in at least one pair-wise comparison. AFB1 effects were dependent on exposure time and turkey type, occurred more rapidly in domesticated turkeys, and led to notable up-regulation in cell cycle regulators, NRF2-mediated response genes and coagulation factors. Further investigation of NRF2-response genes may identify targets to improve poultry resistance.
Keywords: aflatoxin, domesticated turkey, wild turkey, embryonic, liver, RNA-seq, differential expression
1. Introduction
Domesticated turkeys (Meleagris gallopavo) are highly susceptible to the hepatotoxic and immunotoxic effects of the mycotoxin, aflatoxin B1 (AFB1). Consumption of AFB1-contaminated feed by poultry can induce liver damage, immunosuppression and poor growth [1,2,3,4]. Generated in the liver, exo-AFB1-8,9- epoxide (AFBO) is the major metabolite responsible for AFB1 toxicity [4,5,6]. AFBO can bind to DNA and other macromolecules causing mutations, impairing transcription and translation, and initiating apoptosis [4,5,6,7]. In domesticated turkeys, hepatic cytosolic alpha-class glutathione S-transferase (GSTA) enzymes lack the ability to detoxify AFBO, which is likely the most important factor underlying their extreme sensitivity [4,8,9].
Unlike their domesticated relatives, Eastern wild turkeys (M. g. silvestris) appear relatively AFB1-resistant [10]. Supporting this, AFBO-conjugating activity was demonstrated for hepatic GSTA enzymes from both the Eastern and Rio Grande (M. g. intermedia) subspecies of wild turkey [11]. More efficient GST-mediated hepatic detoxification of AFBO could be largely responsible for greater resistance, although variation in apoptotic processes, cellular regulation, immune responses, and other pathways could also contribute. To our knowledge, no in vivo experiments have compared domesticated and wild turkey responses to AFB1 and most work has utilized ad libitum feeding studies that allow variation in the dose of AFB1 ingested by each bird. Therefore, to better characterize the differences between domesticated and wild turkeys, direct comparison of their responses to a controlled dose of AFB1 is needed.
This study utilized an egg injection (in ovo) route of AFB1 challenge to examine domesticated and wild turkey responses in the liver after either 1 or 5 days of exposure. Embryonic exposure ensures a precise administration of the toxin and provides a more cost effective model for toxicity assessment than in hatched poults. During egg formation, AFB1 can transfer from the laying hen into the egg yolk and albumen [2,12,13,14,15,16,17]. Presence of AFB1 in embryonated and unfertilized eggs is of concern to the poultry industry. In ovo AFB1 exposure of developing chickens (Gallus gallus) and turkeys results in DNA damage in the liver, morphological defects, and embryonic mortality [14,18,19,20,21,22,23,24]. Direct experimental injection of eggs or embryonic exposure through maternal feeding can also compromise cellular and humoral immune functions in the hatched progeny [19,22,25,26,27].
In ovo responses to AFB1 can be compared in embryos by analysis of differential gene expression. Comparison of domesticated and wild birds could potentially identify genes or alleles associated with decreased susceptibility to the effects of AFB1. Previous work in our laboratory applied RNA-sequencing (RNA-seq) to investigate AFB1 effects on hepatic and splenic gene expression in domesticated turkey poults after dietary exposure [28,29]. In the liver, expression changes were identified in transcripts linked to apoptosis, carcinogenesis and lipid metabolism [28]. In the spleen, both innate and adaptive immune response genes were affected [29]. RNA-seq allows for characterization of expressed sequence from the entire transcriptome, without the hybridization and inclusion bias of microarrays. This study was designed to apply RNA-seq to AFB1-exposed embryonic liver tissue, which will provide insight into the mechanisms of toxicity, determine the similarity between in ovo models and live dietary exposures, and detect differences in the responses of wild and domesticated turkeys. We hypothesized that AFB1 would have differential in ovo effects on the hepatic transcriptome of wild turkeys versus domesticated birds.
2. Results
2.1. Phenotypic Effects of AFB1
Eggs containing viable domesticated turkey (DT) and wild turkey (WT) embryos were injected with either an AFB1-solution (AFB) or a sham control (CNTL) and incubated for the subsequent 1 day or 5 days (four groups in each turkey type: CNTL 1 day exposure (C1), AFB 1 day exposure (A1), CNTL 5 days exposure (C5) and AFB 5 days exposure (A5)). Weight measurements were collected at the end of exposure to characterize phenotypic effects of AFB1 toxicity. Embryonic exposure to AFB1 for 5 days (A5 versus C5) significantly reduced mean embryo weight in WT, relative liver weight in DT, and absolute liver weights in both types of turkey (Table 1 and Table S1). No significant differences were observed in either DT or WT after 1 day of AFB1 exposure (A1 versus C1). Although the CNTL groups were not significantly different across turkey types, egg and embryo weights in WTA5 were lower than in DTA5. The expected growth and maturation of the embryos over time (C5 versus C1) led to significant increases in embryo and liver weights (Table S1). However, as development progressed, mean relative liver weight only increased in DT.
Table 1.
Effect of aflatoxin B1 on egg, embryo, and liver weights in domesticated and wild turkeys.
| Type | Exposure | Treatment 1 | Number of Embryos | Egg Weight (g) | Embryo Weight (g) | Liver Weight (g) | Relative Liver Weight (g) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||||
| DT | 1 Day | CNTL | 7 | 79.56 | ±5.62 | 16.58 | ±0.97 | 0.26 | ±0.03 | 1.56 | ±0.12 |
| AFB | 7 | 81.13 | ±8.49 | 17.67 | ±0.49 | 0.28 | ±0.03 | 1.56 | ±0.17 | ||
| 5 Days | CNTL | 7 | 76.6 | ±5.71 | 30.7 | ±1.93 | 0.65 d | ±0.13 | 2.12 f | ±0.37 | |
| AFB | 7 | 82.16 a | ±5.56 | 32.60 b | ±2.86 | 0.53 d | ±0.22 | 1.64 f | ±0.22 | ||
| WT | 1 Day | CNTL | 4 | 66.77 | ±8.75 | 18.71 | ±1.09 | 0.34 | ±0.04 | 1.81 | ±0.20 |
| AFB | 4 | 74.7 | ±4.51 | 19.08 | ±1.17 | 0.33 | ±0.04 | 1.74 | ±0.26 | ||
| 5 Days | CNTL | 3 | 66.3 | ±10.52 | 29.59 c | ±4.46 | 0.64 e | ±0.07 | 2.21 | ±0.34 | |
| AFB | 4 | 60.18 a | ±3.68 | 24.83 b,c | ±0.46 | 0.46 e | ±0.06 | 1.87 | ±0.25 | ||
Matched superscript letters a,b,c,d,e,f indicate significant differences (p-value < 0.05) between AFB and CNTL or DT and WT groups. All post-hoc comparisons are provided in Table S1. Domesticated turkey (DT), wild turkey (WT), control (CNTL), aflatoxin B1 (AFB), standard deviation (SD). 1 In ovo exposure to 30% EtOH (CNTL) or 1 µg of AFB1 (AFB).
2.2. RNA-seq Datasets
Total RNA isolated from DT and WT embryonic liver samples (n = 24 and n = 15, respectively) was used for construction of individual barcoded libraries. Sequencing of all libraries (n = 39) produced over 441 M read pairs (883 M total reads) composing 89.2 Gb of raw sequence data (Table 2). Reads (in pairs) per library averaged 22.6 M (range of 17.1–29.9 M); mean library depth was higher in WT (25.5 M) than DT (20.9 M) (Table 2 and Table S2). After filtering and trimming, corrected datasets were only slightly reduced (98.3% of raw data, average of 22.3 M reads per library, range of 16.9–29.4 M) and contained both read pairs and single reads. Characteristics of the corrected datasets in DT and WT were similar, with minimal differences in read length (95.7 bp to 95.1 bp), mean quality score (36.6 to 35.9), and mean GC content (46.9% to 46.6%) (Table 2). In both DT and WT, quality scores in corrected reads were sufficiently high across all base positions (Figure S1).
Table 2.
RNA-seq datasets from domesticated and wild turkey embryonic liver.
| Type | Number of Libraries | Read Status | Mean Read Length (bp) | Mean Quality Score | Mean GC Content (%) | Read Pairs | Single Reads | Total Sequence (Gb) |
|---|---|---|---|---|---|---|---|---|
| DT | 24 | Raw | 101.0 | 35.8 | 47.2 | 250,429,631 | NA | 50.6 |
| Corrected | 95.7 | 36.6 | 46.9 | 245,205,648 | 4,829,880 | 47.4 | ||
| WT | 15 | Raw | 101.0 | 34.7 | 46.9 | 190,989,257 | NA | 38.6 |
| Corrected | 95.1 | 35.9 | 46.6 | 184,931,064 | 5,721,404 | 35.7 |
Domesticated turkey (DT), wild turkey (WT), not applicable (NA).
2.3. Mapping to MAKER Gene Set
Approximately 77% of the 83.1 Gb of corrected sequence mapped uniquely to the MAKER annotated turkey gene set (Table 2 and Table 3). This percentage was consistent across all treatment groups in both DT and WT (Table 3 and Table S2). In each corrected dataset, a higher percentage of paired reads mapped to the gene set than single reads (Table S2). As expected, the majority (73.3%) of mapped reads aligned to exons, rather than introns. Over half (57.8%) of these exonic reads spanned exon borders, illustrating the importance of splice junction mapping when investigating eukaryotic genomes. In total, 17,440 genes (95.5% of the gene set) were expressed in at least one group, with a mean depth of 941 reads/gene (Table 3). Gene expression across the genome was highly similar in DT and WT (Table S3). Most genes (96.8% in DT, 96.5% in WT) with known chromosomal locations were expressed; a lower 79.9% of unassigned genes were expressed in either turkey type.
Table 3.
Read mapping to the MAKER annotated turkey gene set.
| Type | Mapped Reads (% of Corrected) 1 | Unmapped Reads (% of Corrected) 1 | Expressed Genes (% of Total Genes) | Mean Read Depth per Gene |
|---|---|---|---|---|
| DT | 380.6 M (76.9%) | 114.6 M (23.1%) | 17,293 (94.7%) | 868 |
| WT | 289.4 M (77.0%) | 86.1 M (22.9%) | 17,211 (94.2%) | 1056 |
| Total | 670.1 M (76.9%) | 200.8 M (23.1%) | 17,440 (95.5%) | 941 |
Domesticated turkey (DT), wild turkey (WT). 1 Paired reads counted as two.
2.4. Sample Variation
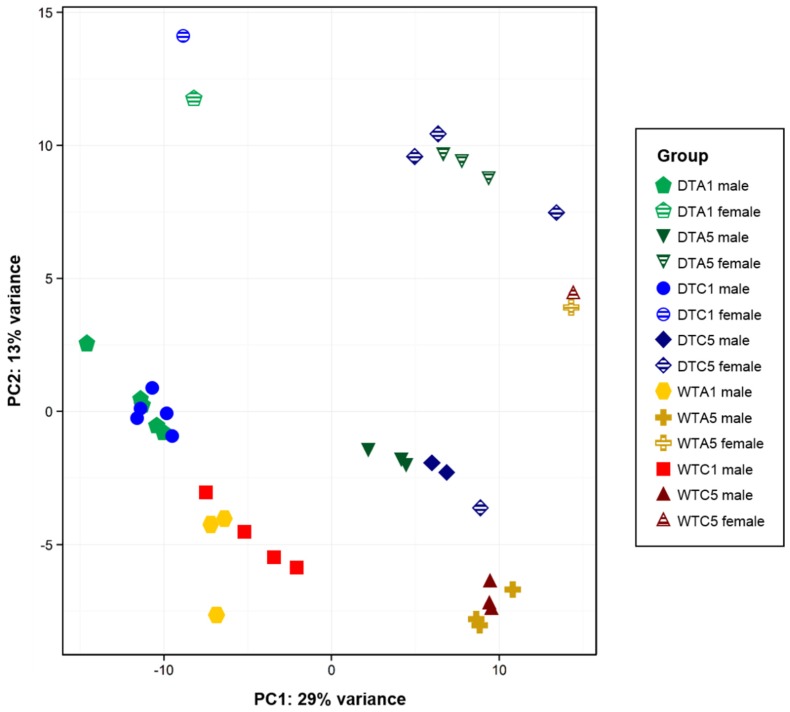
Principle component analysis (PCA) was used to evaluate variation within and between groups based on regularized log2 transformed read counts (Figure 1). The principle component 1 (PC1) axis explained the greatest amount of variation (29%) and separated transcriptomic expression profiles based on exposure time (1 day and 5 days). Further clustering occurred according to turkey type, irrespective of treatment. For example, samples from WTA1 and WTC1 intermixed but were distinct from DTA1 and DTC1. All treatment groups except WTA1 and WTC1 were also divided into two clusters along principle component 2 (PC2). Further investigation clarified that this sample distribution represented differences in the liver transcriptome due to the effects of gender. In each treatment, the female embryonic samples clustered higher along PC2 than the male; only one female (DTC5L7) did not follow this pattern. Overall, this distribution illustrates the developmental differences between samples, the distinct expression profiles of DT and WT, and the effect of gender on hepatic tissue. Hierarchical clustering of the distances between samples reiterated the relationships shown by PCA (Figure S2).
Figure 1.
Exposure time, turkey type, and gender cause variation in embryonic transcriptomes. Principle component analysis (PCA) was performed on regularized log2 transformed read counts in DESeq2 [30]. Principle component 1 (PC1) and principle component 2 (PC2) explain 42% of the variation in read counts. Samples are plotted by group: DTC1 (blue circles), DTC5 (dark blue diamonds), DTA1 (green pentagons), DTA5 (dark green inverted triangles), WTC1 (red squares), WTC5 (dark red triangles), WTA1 (gold hexagons), and WTA5 (dark gold crosses). Gender for each sample is also indicated (male = solid, female = striped). Domesticated turkey (DT), wild turkey (WT), control 1 day (C1), control 5 days (C5), aflatoxin B1 1 day (A1), aflatoxin B1 5 days (A5).
2.5. Differential Expression Analysis
After read count normalization and differential expression (DE) analysis, nearly half (48.9%, 8929) of the genes in the MAKER gene set were identified as having significant DE (q-value ≤ 0.05) in at least one pair-wise comparison between groups. However, only a small portion of these genes (17.2%, 1535) had expression changes greater than |log2 fold change (log2FC)| ≥ 1.0, illustrating the stability of embryonic expression in the liver (Table S4; Figures S3 and S4). This stability of expression profiles could be due to conservation of developmental processes across turkey types (DT versus WT), to similarities in the late stages of embryonic development (1 day versus 5 days), and to the contained environment of the egg minimizing variation and limiting AFB1 effects to direct toxicity (AFB versus CNTL). Significant genes with greatest DE (|log2FC| ≥ 1.0) also had the highest significance in each pair-wise comparison. Table S4 provides the full list of significant DE genes with |log2FC| ≥ 1.0 in at least one comparison. Of these genes, 814 (53.0%) were unique to a single pair-wise comparison, although three genes (absent in melanoma 1-like protein (AIM1L), coiled-coil domain-containing protein 135 (CCDC135), and cytochrome P450 2H2 (CYP2H2)) were identified in seven of twelve comparisons (Table S4). Analysis of each pair-wise comparison elucidates DE attributed to development, AFB1 exposure and genetic differences between DT and WT.
2.6. Developmental Effects
Comparison between the control groups at 1 day and 5 days can provide important information on gene expression during embryonic development. As development progressed in DT (DTC5 versus DTC1), 19.7% (3593) of genes in the MAKER gene set were significantly DE (q-value ≤ 0.05; log2FC from −2.8 to 3.7). Expression of the majority of these genes (82.2%) had small changes in expression and only 640 genes had |log2FC| ≥ 1.0 (Table S4; Figure S5). Significant genes with large expression changes were primarily (71.4%) up-regulated during development, with the greatest increase observed in megakaryoblastic leukemia/myocardin-like protein 1 (MKL1) and decrease in cytochrome P450 1A5 (CYP1A5) (Table S4). To best characterize affected pathways, functional analysis in Ingenuity Pathway Analysis (IPA) utilized DE data from all significant genes in the comparison. In DT, IPA assigned the highest significance to multiple signaling pathways, including “non-small cell lung cancer signaling” and “pancreatic adenocarcinoma signaling” (Table S5; Figure S6A). Genes in these pathways (including those with non-significant DE) were predominately down-regulated during development (Figure S6A) and likely reflect decreases in expression of genes involved in cellular functions and development. Exposure to AFB1 over time (DTA5 versus DTA1) reduced the significance of these pathway associations, showing that introduction of the toxin can affect normal developmental processes.
In WTC5 versus WTC1, 2313 genes (12.7% of the gene set) had significant DE (log2FC from −2.9 to 3.0), of which 316 (13.7%) had |log2FC| ≥ 1.0 (Table S4; Figure S5). More than half (61.7%) of the significant genes with large expression changes were up-regulated, with the greatest DE observed in protein phosphatase 1 regulatory subunit 3B-like (PPR3B; up-regulated) and ATP-binding cassette sub-family G member 8-like (ABCG8; down-regulated). Associations to “protein ubiquitination pathway” and “RAN signaling” had the highest significance in IPA (Table S5; Figure S6B). Similar to changes in DT, the majority of DE genes in these pathways (44.0% and 63.2%, respectively) were down-regulated in WT (Figure S6B). Introduction of AFB1 decreased or eliminated the significance of these pathway associations (WTA5 versus WTA1). Multiple genes (173) were significant and had |log2FC| ≥ 1.0 in both DT and WT comparisons of control groups, illustrating conserved changes during embryo development (Table S4; Figure S5). Examples of highly DE genes observed in both DT and WT include PPR3B, neuroligin-1 (NLGN1), ABCG8, and CYP1A5.
Direct comparisons between DT and WT were also made in the CNTL groups at each exposure time. In WTC1 versus DTC1, 66 genes had significant DE with |log2FC|≥ 1.0 (1593 total significant DE; log2FC from −1.7 to 3.2) (Table S4). Poly(U)-specific endoribonuclease (ENDOU) was the most up-regulated gene in WT, while disintegrin and metalloproteinase domain-containing protein 9 (ADAM9) was the most down-regulated. Fewer genes (1044) had significant DE (log2FC from −3.6 to 2.6) at the later developmental time point (WTC5 versus DTC5); however, 94 had |log2FC| ≥ 1.0. Glutamate receptor-interacting protein 1 (GRIP1; up-regulated) and an unannotated gene (down-regulated) had the greatest log2FC. The most significant pathway associations in WTC1 versus DTC1 were observed for “pancreatic adenocarcinoma signaling” and “coagulation system” (Table S5). Other signaling pathways had highly significant associations at the later time point, illustrating that differences in gene expression profiles in domesticated and wild turkeys changed as development progressed.
2.7. Effects of 1 Day of Exposure to AFB1
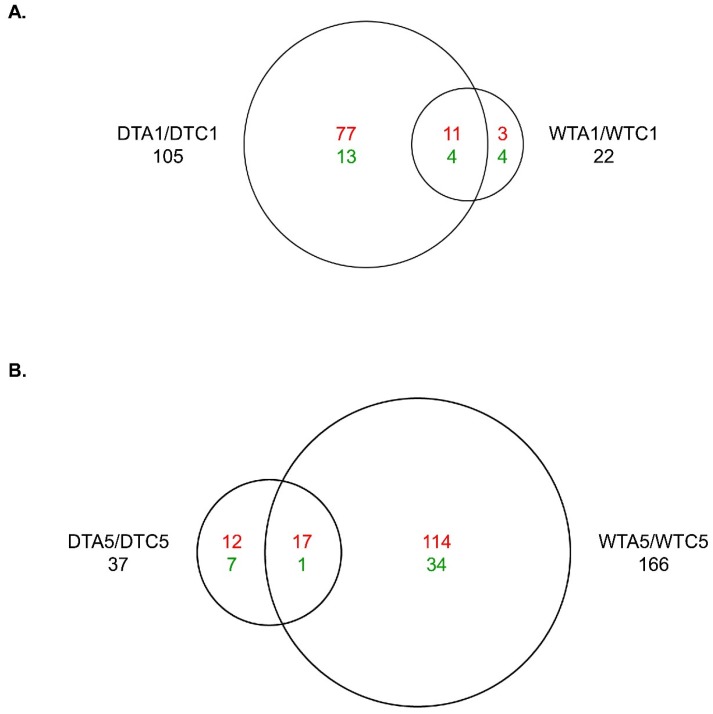
DE analysis comparing the AFB1-exposed group in DT (DTA1) versus the control (DTC1) identified 2144 genes with significant DE (q-value ≤ 0.05; log2FC from −1.8 to 4.6). In total, 105 (4.9%) had |log2FC| ≥ 1.0 (Table S4; Figure 2A); these were predominately (83.8%) up-regulated by AFB1. The greatest increases in expression were observed for cyclin dependent kinase inhibitor CIP1 (CIP1/CDKN1A/p21), ectodysokasin A2 receptor (EDA2R/TNFRSF27), S-adenosylmethionine synthase isoform type-1 (MAT1A), and E3 ubiquitin-protein ligase MDM2 (MDM2) (Table S4). Genes showing the greatest down-regulation after 1 day of exposure in DT were RNA binding protein fox-1 homolog 1 (RBFOX1) and hydrocephalus-inducing protein homolog (HYDIN).
Figure 2.
Treatment-related expression changes from aflatoxin B1 (AFB1) occurs predominately in DTA1 and WTA5. (A) A1 versus C1 in DT and WT. (B) A5 versus C5 in DT and WT. Each diagram shows the number of shared and unique genes with significant differential expression (DE) (q-value ≤ 0.05) and |log2FC| ≥ 1.0. Direction of DE (red, above = up-regulated, and green, below = down-regulated) is shown. Domesticated turkey (DT), wild turkey (WT), control 1 day (C1), control 5 days (C5), aflatoxin B1 (AFB1), aflatoxin B1 1 day (A1), aflatoxin B1 5 days (A5), log2 fold change (log2FC).
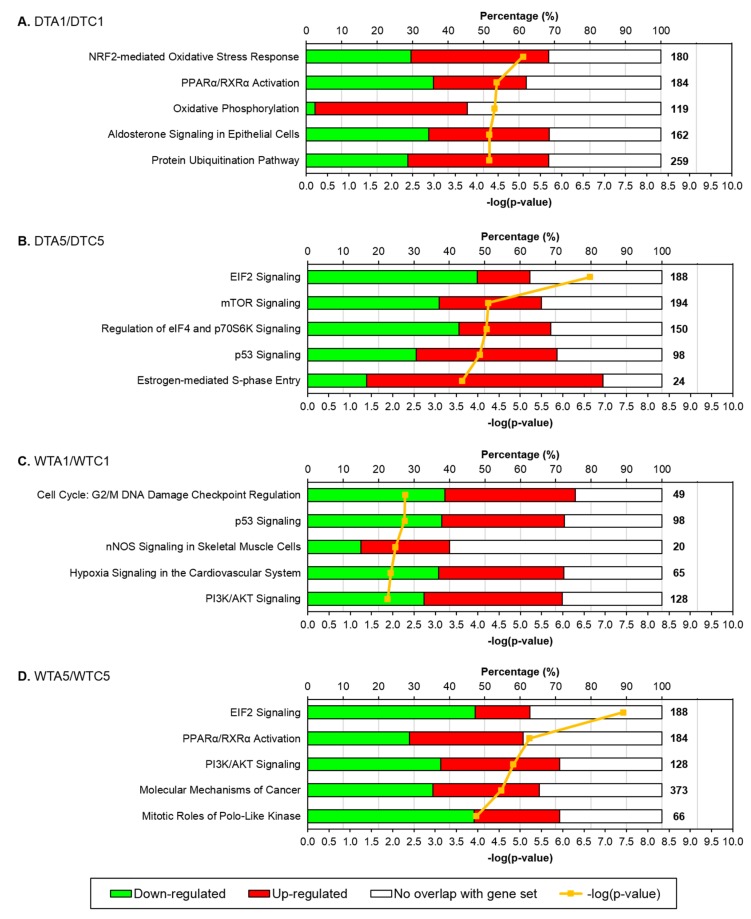
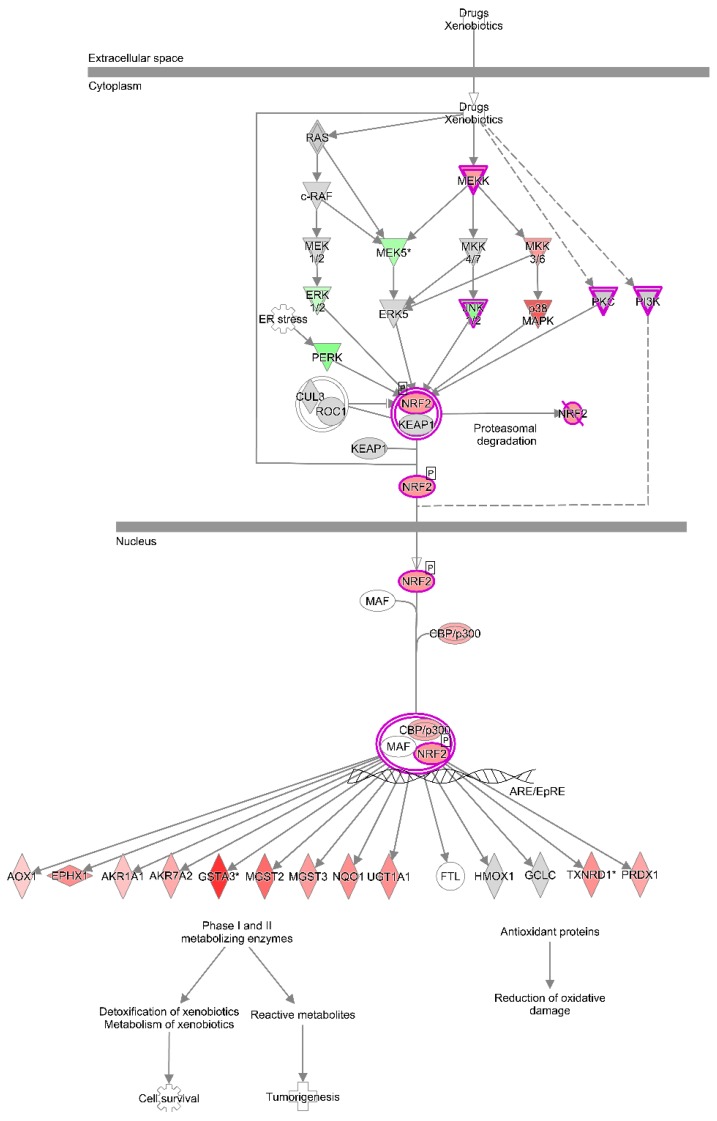
The highest IPA associations in the DTA1/DTC1 comparison were made to the “NRF2-mediated oxidative stress response”, “PPARα/RARα activation” and “oxidative phosphorylation” pathways (Table S5; Figure 3A). AFB1 exposure increased expression of many genes in these pathways (Figure 3A). For example, 38.9% of genes in the NRF2-mediated oxidative stress response pathway were up-regulated (both significant and non-significant DE) in DTA1. Although significant, nuclear factor erythroid 2-related factor 2-like (NFE2L2/NRF2) itself was only moderately up-regulated (Table S6; Figure 4). Expression of many downstream targets of this transcription factor increased in DT after 1 day exposure to AFB1 (Table 4 and Table S6; Figure 4). Highest significant up-regulation was observed in alpha-class glutathione S-transferase 3 (GSTA3), alpha-class glutathione S-transferase 4 (GSTA4), and microsomal glutathione S-transferase 2 (MGST2) (Table 4). It should be noted that three genes in the MAKER gene set are annotated to GSTA3; each representing a fragment of the known sequence [31]. This likely results from misassembly in this repetitive genomic region. Increased expression was also seen in other phase I and II metabolic enzymes and anti-oxidant proteins, including microsomal glutathione S-transferase 3 (MGST3), aflatoxin B1 aldehyde reductase 2 (AKR7A2/AFAR), epoxide hydrolase 1 (EPHX1), NAD(P)H dehydrogenase quinone 1 (NQO1), thioredoxin reductase 1 cytoplasmic (TXNRD1) and UDP-glucuronosyltransferase 1-1 (UGT1A1).
Figure 3.
Differential expression in highly significant canonical pathways in comparisons of AFB versus CNTL groups. (A) DTA1/DTC1. (B) DTA5/DTC5. (C) WTA1/WTC1. (D) WTA5/WTC5. Ingenuity Pathway Analysis (IPA) was used to identify significant effects on canonical pathways (−log(p-value) > 1.3). Percentage of genes down-regulated (green), up-regulated (red), not represented in MAKER annotated gene set (white) are shown for the 5 most significant pathways (−log(p-values) in yellow) in each pair-wise comparison; both significant and non-significant DE is included. Total numbers of genes in each pathway are in bold. Domesticated turkey (DT), wild turkey (WT), control (CNTL), control 1 day (C1), control 5 days (C5), aflatoxin B1 (AFB), aflatoxin B1 1 day (A1), aflatoxin B1 5 days (A5).
Figure 4.
Up-regulation of NRF2-mediated responses in DTA1 versus DTC1. Ingenuity Pathway Analysis (IPA) identified highly significant association of this pathway (p-value of 8.22 × 10−6). Genes were up-regulated (red), down-regulated (green), not significant (grey), or not represented in the MAKER gene set (white). Purple outlines represent groups of molecules and indicate gene annotations that occur more than once in the gene set. Nuclear factor erythroid 2-related factor 2-like (NRF2), domesticated turkey (DT), control 1 day (C1), aflatoxin B1 1 day (A1).
Table 4.
Significant DE of genes regulated by nuclear factor erythroid 2-related factor 2-like (NRF2) in AFB versus control (CNTL) groups.
| Gene ID | Gene | DTA1/DTC1 | DTA5/DTC5 | WTA1/WTC1 | WTA5/WTC5 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Log2FC | q-value | Log2FC | q-value | Log2FC | q-value | Log2FC | q-value | ||
| T_ALT_S_008999 | AKR1A1 | 0.34 | 3.18 × 10−2 | 0.07 | 7.76 × 10−1 | 0.25 | 8.07 × 10−1 | 0.69 | 5.58 × 10−5 |
| T_ALT_S_014672 | AKR7A2 | 0.47 | 1.15 × 10−2 | 0.14 | 4.91 × 10−1 | 0.34 | 5.55 × 10−1 | 0.18 | 3.37 × 10−1 |
| T_ALT_S_008024 | AOX1 | 0.28 | 3.65 × 10−2 | 0.00 | 9.96 × 10−1 | −0.02 | 9.95 × 10−1 | −0.04 | 8.86 × 10−1 |
| T_ALT_S_003995 | CAT | −0.18 | 3.15 × 10−1 | −0.44 | 3.49 × 10−4 | −0.18 | 8.54 × 10−1 | −0.30 | 1.58 × 10−1 |
| T_ALT_S_010665 | DNAJA3 | 0.40 | 1.15 × 10−3 | −0.03 | 8.79 × 10−1 | 0.07 | 9.78 × 10−1 | 0.21 | 1.66 × 10−1 |
| T_ALT_S_009842 | DNAJB11 | −0.32 | 7.44 × 10−3 | −0.08 | NA | −0.39 | 2.36 × 10−1 | 0.31 | 1.15 × 10−1 |
| T_ALT_S_008078 | DNAJC10 | −0.39 | 1.01 × 10−3 | −0.02 | 9.65 × 10−1 | −0.05 | 9.91 × 10−1 | 0.18 | 4.09 × 10−1 |
| T_ALT_S_014426 | DNAJC11 | 0.26 | 4.81 × 10−2 | −0.06 | 7.63 × 10−1 | −0.20 | 8.47 × 10−1 | 0.20 | 3.88 × 10−1 |
| T_ALT_S_007708 | DNAJC13 | 0.03 | 8.56 × 10−1 | −0.40 | 6.56 × 10−3 | 0.07 | 9.80 × 10−1 | 0.08 | 7.48 × 10−1 |
| T_ALT_S_014061 | DNAJC18 | −0.02 | 8.95 × 10−1 | −0.02 | 9.44 × 10−1 | −0.24 | 5.93 × 10−1 | −0.42 | 1.81 × 10−2 |
| T_ALT_S_002734 | EPHX1 | 0.63 | 5.74 × 10−4 | −0.06 | 8.95 × 10−1 | 0.03 | 9.93 × 10−1 | 0.57 | 2.36 × 10−3 |
| T_ALT_S_003948 | FTH1 | 0.10 | 6.77 × 10−1 | 0.38 | 4.52 × 10−2 | −0.10 | 9.41 × 10−1 | 0.12 | 7.29 × 10−1 |
| T_ALT_S_003347 | GSTA3 1 | 1.60 | 1.47 × 10−10 | 0.74 | 1.79 × 10−2 | 0.48 | 3.63 × 10−1 | 0.52 | 2.34 × 10−1 |
| T_ALT_S_003348 | GSTA3 1 | 1.52 | 2.87 × 10−9 | 0.85 | 4.31 × 10−3 | 0.39 | 5.52 × 10−1 | 0.23 | 6.61 × 10−1 |
| T_ALT_S_003346 | GSTA3 1 | 1.11 | 9.74 × 10−5 | 0.40 | 2.66 × 10−1 | 0.36 | 5.83 × 10−1 | 0.80 | 4.76 × 10−2 |
| T_ALT_S_003345 | GSTA4 | 0.82 | 8.65 × 10−7 | 0.47 | 2.82 × 10−2 | 0.35 | 6.56 × 10−1 | 0.42 | 4.09 × 10−2 |
| T_ALT_S_001023 | GSTK1 | 0.07 | 6.99 × 10−1 | −0.31 | 2.39 × 10−2 | −0.26 | 5.83 × 10−1 | −0.11 | 6.34 × 10−1 |
| T_ALT_S_009475 | GSTO1 | 0.13 | 5.87 × 10−1 | 0.06 | 8.65 × 10−1 | −0.28 | 3.27 × 10−1 | 1.07 | 4.63 × 10−3 |
| T_ALT_S_011992 | HERPUD1 | −0.25 | 3.76 × 10−1 | 0.69 | 3.75 × 10−4 | −0.20 | 9.21 × 10−1 | −0.26 | 3.11 × 10−1 |
| T_ALT_S_000662 | HMOX1 | 0.29 | 1.91 × 10−1 | −0.14 | 7.31 × 10−1 | 0.32 | 3.84 × 10−1 | 0.85 | 1.48 × 10−9 |
| T_ALT_S_005601 | MGST2 | 0.84 | 6.76 × 10−7 | 0.14 | 6.23 × 10−1 | 0.41 | 2.18 × 10−1 | 0.37 | 8.19 × 10−2 |
| T_ALT_S_008700 | MGST3 | 0.56 | 9.79 × 10−5 | 0.10 | 7.83 × 10−1 | 0.33 | 2.55 × 10−1 | 0.73 | 4.64 × 10−4 |
| T_ALT_S_012166 | NQO1 | 0.63 | 2.64 × 10−6 | 0.03 | 8.87 × 10−1 | 0.57 | 1.81 × 10−3 | 0.02 | 9.39 × 10−1 |
| T_ALT_S_004785 | NQO2 | 0.24 | 2.42 × 10−1 | 0.63 | 1.94 × 10−4 | 0.00 | 1.00 | 0.73 | 6.78 × 10−6 |
| T_ALT_S_009000 | PRDX1 | 0.49 | 2.13 × 10−3 | 0.10 | 6.39 × 10−1 | 0.10 | 9.69 × 10−1 | 0.66 | 1.03 × 10−4 |
| T_ALT_S_016734 | STIP1 | −0.03 | 8.93 × 10−1 | −0.19 | 2.64 × 10−1 | −0.14 | 8.98 × 10−1 | 0.36 | 9.48 × 10−3 |
| T_ALT_S_007072 | TXN | 0.09 | 5.73 × 10−1 | −0.62 | 2.26 × 10−3 | −0.05 | 9.80 × 10−1 | −0.07 | 6.80 × 10−1 |
| T_ALT_S_000674 | TXNRD1 1 | 0.61 | 1.32 × 10−2 | 0.15 | 7.63 × 10−1 | 0.54 | 2.55 × 10−1 | 0.45 | 2.82 × 10−1 |
| T_ALT_S_011677 | TXNRD1 1 | −0.30 | 2.07 × 10−3 | 0.00 | 9.99 × 10−1 | 0.00 | 9.99 × 10−1 | −0.16 | 3.06 × 10−1 |
| T_ALT_S_007907 | UGT1A1 | 0.62 | 2.35 × 10−5 | 0.06 | 8.19 × 10−1 | 0.15 | 9.05 × 10−1 | 0.34 | 8.16 × 10−3 |
| T_ALT_S_009396 | MRP2 | 0.16 | 3.07 × 10−1 | −0.32 | 3.68 × 10−2 | 0.24 | 4.82 × 10−1 | 0.24 | 2.59 × 10−1 |
| T_ALT_S_006174 | SOD3 | 0.33 | 3.12 × 10−1 | 0.17 | 7.08 × 10−1 | 0.07 | 9.92 × 10−1 | 0.58 | 2.25 × 10−2 |
Genes down-stream of nuclear factor erythroid 2-related factor 2-like (NRF2) were identified using Ingenuity Pathway Analysis (IPA). Log2 fold change (Log2FC) and FDR-adjusted p-values (q-values) were determined in DESeq2 [30]. Non-significant comparisons (q-value > 0.05) are shown in grey. Differential expression (DE), domesticated turkey (DT), wild turkey (WT), control (CNTL), control 1 day (C1), control 5 days (C5), aflatoxin B1 (AFB), aflatoxin B1 1 day (A1), aflatoxin B1 5 days (A5), no statistics due to low read counts (NA). 1 Multiple genes in the MAKER gene set had significant DE and annotated to the same reference.
In contrast to the DT, 1 day exposure to AFB1 in WT (WTA1 versus WTC1) had limited effects on gene expression; only 201 genes (1.1%) in the MAKER gene set had significant DE (log2FC from −1.7 to 2.8). Among these, 22 (10.9%) had |log2FC| ≥ 1.0 and 8 were down-regulated (Table S4; Figure 2A). Similar to DT, the most up-regulated gene in WT was MDM2 (Table S4). The most significant IPA associations in the WTA1/WTC1 comparison were to “G2/M DNA damage checkpoint regulation” and “p53 signaling”, illustrating misregulation of the cell cycle after AFB1 exposure in WT (Table S5; Figure 3C). Despite the vast difference in the numbers of significant genes, 15 genes were in common between the DT and WT and shared even the direction of expression changes following exposure (Figure 2A). For instance, MDM2, E3 ubiquitin-protein ligase MYCBP2 (MYCBP2), RBFOX1, and thioredoxin-like protein AAED1 (AAED1) were affected in both types of turkey (Table S4). Therefore, even with the lower AFB1 sensitivity of WT, some expression changes in the initial response to AFB1 were conserved with DT.
2.8. Effect of 5 Days of Exposure to AFB1
After 5 days of exposure to AFB1 in DT (DTA5 versus DTC5), 1036 genes had significant DE (q-value ≤ 0.05; log2FC from −1.8 to 3.9), but only 37 had |log2FC| ≥ 1.0 (Table S4, Figure 2B). AFB1 increased expression of 78.4% (29) of the genes with |log2FC| ≥ 1.0 and many of these genes were also up-regulated by 1 day exposure (e.g., AAED1, CIP1, MAT1A, MDM2 and EDA2R). As in DTA1, CIP1 had the greatest increase in expression after 5 days exposure (DTA5), although the log2FC was slightly lower (Table S4). “EIF2 signaling”, “mTOR signaling”, and other signaling pathways were the most significantly associated IPA pathways after 5 days of exposure in DT (Table S5; Figure 3B).
The number of significant genes (1904; log2FC from −1.9 to 4.6) in WTA5 verses WTC5 was greater than that observed in the DTA5/DTC5 comparison. Nearly 4.5 times as many genes in WT (166) had |log2FC| ≥ 1.0 (Table S4, Figure 2B). The largest log2FC values were observed for EDA2R, MAT1A, CIP1 and cytochrome P450 2C45 (CYP2C45) (Table S4). Consistent with the other comparisons involving the AFB group, 78.9% of genes with significant DE and |log2FC| ≥ 1.0 were up-regulated (Table S4, Figure 2B). Pathway associations with the highest significance in WT after 5 days of exposure were “EIF2 signaling” and “PPARα/RARα activation” (Table S5; Figure 3D), both of which were highly significant in other AFB to CNTL group comparisons. Eighteen significant and highly DE genes were identified in both WTA5/WTC5 and DTA5/DTC5, including the three with the greatest DE (EDA2R, MAT1A and CIP1) (Table S4, Figure 2B).
2.9. Gender Effects
Gender also warranted further examination in DE analysis since it contributed to the clustering of samples observed in PCA (Figure 1). Since gender was unequally represented in most comparisons, only DTA5 versus DTC5 was evaluated using gender as a second factor in DESeq2. When incorporating gender, the number of genes with significant DE in DTA5/DTC5 slightly increased (1102 genes; log2FC from −1.8 to 3.9). However, the number of significant DE genes (39) with |log2FC| ≥ 1.0 was consistent with the original analysis (Tables S4 and S7). Most (36) of the significant genes with |log2FC| ≥ 1.0 were identified irrespective of gender as a factor. Among the 980 genes with significant DE in both analyses, gender only determined whether three genes met the log2FC threshold. Furthermore, only one gene (annexin A4 (ANXA4)) had significant DE and |log2FC| ≥ 1.0 in the two-factor analysis, but did not have significance in the original comparison (q-value of 1.86 × 10−1).
Thirty genes (76.9%) were found in the two-factor analysis to be significantly up-regulated (significant DE and |log2FC| ≥ 1.0) by AFB1 exposure (Table S7). Three genes, (CIP1, EDA2R, and MAT1A) had the largest positive log2FC values, while tubulointerstitial nephritis antigen-like (TINAG) was the most down-regulated (Table S7). This is in complete agreement with the original comparison of DTA5/DTC5. Incorporating gender into the analysis also had limited effect on the canonical pathway associations identified by IPA (Table S5). Overall, the identity and functions of significant genes were highly consistent in both analyses, demonstrating that gender had minimal effects on AFB1-induced expression changes despite its role in the transcriptome as a whole. Therefore, due to the unbalanced distribution across groups and the congruence of both DTA5/DTC5 comparisons, gender was not included as a secondary factor in the overall DE analysis.
2.10. Comparison of AFB1 Effects
Differences in transcriptome responses to AFB1 exposure were examined in direct comparisons between WT and DT. In WTA1 versus DTA1, 716 genes had significant DE (log2FC from −1.9 to 2.8; 45 with |log2FC| ≥ 1.0), while 2050 were significant (log2FC from −3.3 to 2.9; 248 with |log2FC| ≥ 1.0) in WTA5 versus DTA5 (Table S4). Some of the detected differences between WT and DT occurred irrespective of treatment (CNTL or AFB). For example, similar to the CNTL comparisons, ENDOU was one of the most highly up-regulated genes in the WTA1/DTA1 and WTA5/DTA5 comparisons. A large increase in GRIP1 was also observed in both AFB and CNTL comparisons, but only at the later time point in AFB-exposed birds (WTA5 versus DTA5). However, pathway analysis identified greater effects on the “oxidative phosphorylation” and “mitochondrial dysfunction” pathways in the WTA5/DTA5 comparison than WTC5/DTC5 (Figure S7). In WTA1 versus DTA1, “DNA double-strand break repair by non-homologous end joining” had a significant pathway association not observed in other comparisons between turkey types.
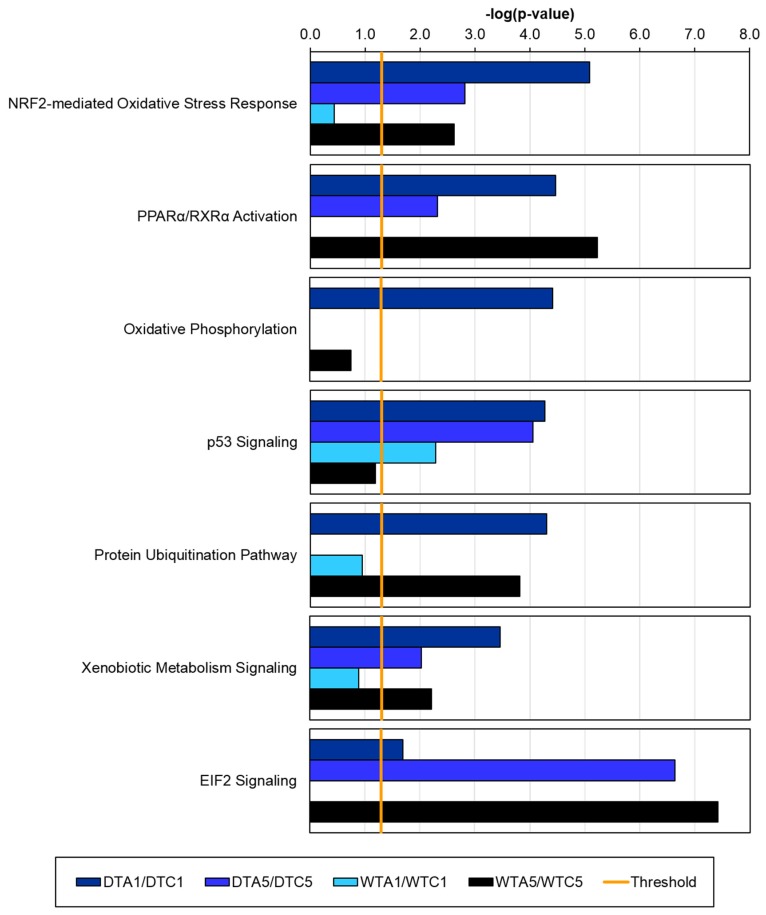
Differences in the pathways affected by AFB1 in DT and WT were also highlighted by comparative pathway analysis in IPA (Figure 5). For example, “EIF2 signaling” was significant after 5 days of exposure to AFB1 in both DT and WT, due to similar levels of down-regulation of genes in this pathway (47.9% and 47.3%, respectively) (Figure 3 and Figure 5). The “PPARα/RARα activation” and “protein ubiquitination” pathways occurred in the comparisons with greatest AFB1 effects (DTA1/DTC1 and WTA5/WTC5). Significant associations were made to the “NRF2-mediated oxidative stress response” pathway in all AFB versus CNTL comparisons except WTA1/WTC1 (Figure 5). In these comparisons (DTA1/DTC1, DTA5/DTC5, and WTA5/WTC5), there were similar levels of up-regulated (37.8%–38.9%) and down-regulated (28.3%–30.0%) genes in the NRF2-response pathway. Interestingly, the gene encoding NRF2 (NFE2L2) was up-regulated in both DT time points and in WTA1, but not in the WTA5 group (Table S6). The significance of DE in downstream targets of NRF2 was even more variable (Table 4 and Table S6). After 5 days of exposure, GSTA3, homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein (HERPUD1), and dehydrogenase quinone 2 (NQO2) were most up-regulated in DT, while GSTA3, heme oxygenase 1-like (HMOX1), MGST3, and NQO2 increased most in WT. In WTA1 versus WTC1, NQO1 was the only downstream gene with significant DE (up-regulated).
Figure 5.
Significance of pathway associations vary in AFB versus CNTL comparisons. In each pair-wise comparison, Ingenuity Pathway Analysis (IPA) assigned p-values to canonical pathways based on differential expression (DE). Bar plot provides 6 example pathways with variable significance between the DTA1/DTC1 (dark blue), DTA5/DTC5 (bright blue), WTA1/WTC1 (light blue), and WTA5/WTC5 (black) comparisons. Pathway associations must have a -log(p-value) > 1.3 (threshold, vertical yellow line) to be considered significant. Domesticated turkey (DT), wild turkey (WT), control (CNTL), control 1 day (C1), control 5 days (C5), aflatoxin B1 (AFB), aflatoxin B1 1 day (A1), aflatoxin B1 5 days (A5).
Toxicological function analysis in IPA also identified significant associations to hepatotoxic outcomes that reflect the known phenotypic effects of AFB1 exposure. In all pair-wise comparisons of AFB versus CNTL, “liver hyperplasia” had highest significance (Figure S8). The strength of the associations also mirrored the overall effects of AFB1 on expression, with highest p-values in DTA1/DTC1 and WTA5/WTC5 and lowest in the WTA1/WTC1 comparison. Outcomes such as “hepatocellular carcinoma” and “liver steatosis” also had significant associations in most comparisons. These potential hepatotoxic outcomes fit with expected tissue-wide effects, while canonical pathways and individual expression changes illustrate differential responses to AFB1 exposure in WT and DT.
3. Discussion
This study utilized an in ovo exposure model to characterize gene expression responses to AFB1 in the hepatic transcriptome of domesticated and wild turkeys. Responsible for phase II detoxification, the liver is also the primary site of AFB1 activation into AFBO and its resulting toxicity [4,5,6]. Consequently, in ovo AFB1 exposure of chickens and domesticated turkeys induces dose-related DNA damage in the embryonic liver [24] and can reduce embryo viability [14,18,19,20,21,22,23,24]. Although no embryo mortality was seen in this study, in ovo AFB1 exposure had phenotypic effects and altered expression in the hepatic transcriptome.
3.1. Responses to AFB1
Dietary AFB1 exposure in poultry leads to lipid accumulation in the liver, which causes hepatomegaly and increased liver weight relative to body weight in both chickens and domesticated turkeys [1,2,3,32,33,34,35,36,37,38,39]. In the present study, embryonic exposure to AFB1 decreased the relative weight in domesticated turkey embryos. That liver mass was not increased in AFB1-exposed embryos is not surprising given the short duration of exposure. A short-term decline in relative liver weight has been reported in broilers fed dietary aflatoxins; as the study progressed, both lipids and relative liver weights increased [35]. In this study, embryo weights also significantly decreased after in ovo exposure in wild turkeys, but not domesticated turkeys. Both AFB1 injection and maternal feeding have been shown to reduce embryo weight in chickens [14,20,21]. In addition to the observed phenotypic changes, hepatic transcriptome analysis identified significant effects on genes involved in cell cycle regulation and NRF2-mediated responses.
Genes linked to the cell cycle and apoptosis were consistently among the most significant and up-regulated genes in AFB1-exposed groups. For example, expression of MDM2, CIP1 and MAT1A were significantly increased in domesticated and wild turkey embryos in AFB versus CNTL comparisons (CIP1 and MAT1A were not significant in WTA1/WTC1). AFB1-induced expression changes in these genes have been identified in multiple species, suggesting that AFB1 effects on the cell cycle are conserved. Up-regulation of MDM2 after AFB1 exposure has been observed in human hepatocellular carcinoma (HCC) cells [40], rats [41], swine [42], and in our previous study of turkey poults [28]. Increased hepatic expression of CIP1 in response to AFB1 has been seen in human primary hepatocytes or HCC cell lines [40,43,44], mice [45], rats [41], swine [42], and turkeys [28]. Interestingly, MAT1A expression has been shown to decrease in swine [42] and human HCC cells [40] after AFB1 exposure.
Inhibition of apoptotic pathways or misregulation of the cell cycle by these genes could facilitate the development of hepatic lesions in AFB1-exposed poultry, including vaoculation of hepatocytes, necrotic loci, focal hemorrhages, biliary hyperplasia, fibrosis and nodular tissue regeneration [1,2,33,34,36,46,47,48,49]. MDM2 is an E3 ubiquitin-protein ligase known to mark the tumor suppressor p53 for degradation by the proteasome [50,51]. However, overexpression of MDM2 can downregulate p53 in cells that should undergo apoptosis, causing aberrant cell cycle progression. CIP1 encodes a p53-activated inhibitor of cyclin-dependent kinase activity that regulates cell cycle progression. Overexpression of CIP1 could lead to cell cycle arrest and apoptosis [42], contrary to the proliferative effects of MDM2-mediated p53 loss. Increased expression of these regulatory genes during short-term embryonic exposure in the turkey suggests that the apoptotic and hyper-proliferative phenotypes seen in the liver begin to develop in the early stages of aflatoxicosis.
Expression of S-adenosylmethionine synthase could affect liver health and sulfur metabolism. In mammals, MAT1A and S-adenosylmethionine synthase isoform type-2 (MAT2A) encode interchangeable subunits that metabolize methionine for use in DNA methylation [52,53,54]. MAT enzymes can also regulate hepatocyte proliferation and apoptosis [52,53] and affect glutathione production [54]. Mammalian MAT1A is the normal hepatic form of the enzyme, while MAT2A is expressed extrahepatically, as well as in fetal liver and HCC [52,53,54]. In a recent study from our laboratory, MAT2A was also up-regulated in AFB1-fed domesticated turkey poults [28]. Conversely, up-regulation of MAT1A after AFB1 exposure in this study could occur as part oxidative stress responses; increased MAT expression could feed additional S-adenosylmethionine into glutathione synthesis. However, three genes in the turkey MAKER gene set used in this study annotate as MAT synthases (two to MAT1A and one to MAT2A). Further clarification of their identity, normal expression patterns, and the functional consequences of AFB1-induced differential expression is needed to fully understand the role of these enzymes in poultry.
Based on expression changes in the embryonic liver, cellular responses to oxidative stress and xenobiotics were also initiated by AFB1 exposure. NFE2L2 encodes the NRF2 leucine zipper transcription factor central to many of these responses; this gene was up-regulated after 1 day of exposure to AFB1 in both domesticated and wild turkey embryos. After activation by phosphorylation, NRF2 binds anti-oxidant response elements (AREs) in nuclear DNA and activates transcription of metabolic/detoxifying enzymes, anti-oxidants and anti-apoptotic factors [55,56,57,58]. Mammalian NRF2 can be stabilized in its active form by interacting with CIP1 [56,59]. In this study, significant up-regulation of NFE2L2 and CIP1 was observed in the same comparisons of embryonic exposure to AFB1, suggesting a similar interaction could be possible in poultry.
GSTs are NRF2-response genes with a critical role in domesticated turkey susceptibility to AFB1 toxicity [4,8,9]. In embryonic turkey liver, GSTA3 and GSTA4 were significantly up-regulated in all AFB versus CNTL comparisons except WTA1/WTC1. In mice, efficient glutathione conjugation by murine GSTA3 causes high resistance to aflatoxicosis [6,60,61]. Hepatic GST enzymes in domesticated turkey are essentially unable to detoxify AFBO in vitro, whereas wild turkey GST enzymes retain anti-AFBO activity [4,8,9,11]. However, as the greatest up-regulation in GSTA genes in this study was observed in DTA1/DTC1, the lack of AFBO detoxification by hepatic GSTs does not appear to reflect an inability to respond to AFB1 exposure. Rather post-translational modifications may be responsible for the inactivity of GSTAs against AFBO in domesticated turkeys [8]. It is important to note that while hepatic GST enzymes are produced in turkey embryos [62]; their AFB1-conjugating activity has not been examined. Furthermore, since AFB1 exposure can lead to oxidative stress and lipid peroxidation [63,64,65], up-regulation of GSTA genes may reflect antioxidant functions of glutathione instead of AFBO detoxification.
Many other genes regulated by NRF2 also encode phase I and II enzymes that could be involved in the metabolism of AFB1 in turkey liver. In WTA5/WTC5, omega-class glutathione S-transferase 1 (GSTO1) was the most up-regulated NRF2-response gene. Up-regulation of AFAR was unique to DTA1/DTC1, while EPHX1 was significant in both DTA1 and WTA5. Mammalian homologs for both of these enzymes can metabolize AFB1 or one of its metabolites into AFB1-dihydrodiol [6,66,67,68]. AFAR activity has been demonstrated in turkey liver [69,70]. The role of EPHX1 is uncertain in both birds and mammals. NRF2 also regulates genes that encode antioxidant enzymes, including NQO1 and NQO2 [71]; expression of these genes was time point dependent, with significant DE in both turkey types for NQO1 after 1 day of exposure and for NQO2 after 5 days. Differences in the response of GSTs and other NRF2-response genes between domesticated and wild turkeys could play a role in determining their resistance to AFB1 toxicity.
3.2. Time of Response in WT and DT
One of the overall differences in AFB1 treatment between wild and domesticated turkey embryos is the time-dependency of response. More significant DE was observed after 5 days of exposure in wild turkey embryos, while expression in domesticated turkeys changed quickly (1 day exposure). After only 4 hours of exposure to AFB1, liver DNA damage has been seen in chicken and domesticated turkey embryos; however, less damage was observed in the longer 4 days of exposure [24]. The rapid effects of AFB1 on domesticated turkey embryonic expression in this study could be explained by effective production of AFBO or insufficient hepatic GST detoxification. The slower expression response in wild turkey embryos could be due to a delay in toxicity from either lower levels of AFBO production or better detoxification. We are currently investigating P450-mediated activation of AFB1 in wild versus domesticated turkey liver to better understand AFB1 metabolism in both types of turkey. Other response pathways could also vary in the wild birds. Further investigation of phenotypic or biochemical measures of toxicity, such as hepatic lesions, DNA damage or AFB1-adduct production, alongside expression effects will be needed to clarify the differences in wild turkeys.
It should be noted that the effects of longer exposure (5 days) were more detrimental to the growth of wild turkey than domesticated turkey embryos. Feeding AFB1 to both domesticated and wild turkey poults has been previously shown to decrease growth [1,3,10]. Embryonic exposure to AFB1 can impair developmental processes identified in the CNTL groups (DTC5/DTC1 and WTC5/WTC1), as shown by decreased pathway associations in AFB-treated groups (DTA5/DTA1 and WTA5/WTA1). Furthermore, pathway analysis identified differences in oxidative phosphorylation and mitochondrial dysfunction in the direct comparison of WTA5 versus DTA5. Differences in the oxidative capabilities of the mitochondria in muscle have been shown to effect growth in chickens [72,73] and AFB1 is known to adversely affect hepatic mitochondrial enzymes and oxidative phosphorylation [74,75]. Therefore, if AFB1 effects on mitochondrial functions extend throughout the embryo, these changes may explain the decreased growth of the wild turkeys during development.
Expression changes during development may also contribute to the greater effects of AFB1 seen after 1 day of exposure in domesticated turkeys. For example, cytochrome P450 1A5 (CYP1A5) was significantly down-regulated in developmental comparisons (except WTA5 versus WTA1 due to an outlier). Cytochrome P450 1A5 (and 3A37) can efficiently activate AFB1 into AFBO in vitro [76,77,78,79]. The activity of hepatic P450 enzymes and therefore AFB1 sensitivity is inversely related to age in turkey poults [1,34,69,76]. An active protein from the CYP1A family has been identified in embryonic liver [62], suggesting that hepatotoxicity in turkey embryos may also be driven by P450 1A5. In this study, decreased expression of CYP1A5 over time may indicate that domesticated turkeys are more susceptible to toxicity earlier in development.
3.3. In ovo Exposure as Model of Aflatoxicosis
Another objective of this study was to evaluate whether in ovo injection of AFB1 could be used to model expression changes induced by dietary exposure in poults. Our laboratory has previously characterized transcriptome responses in the liver and spleen of young domesticated turkeys fed AFB1 [28,29]. AFB1 had a predominately up-regulatory effect on gene expression in both the embryonic and poult AFB1 exposures. Significant expression changes were identified in both hepatic RNA-seq experiments for genes or transcripts involved in apoptosis, signaling, and cell cycle regulation [28]. Direct numerical comparisons of expression level between previous studies on poults and the present study cannot be made since different study designs and techniques were utilized (de novo assembly of predicted transcripts versus mapping to an annotated gene set). However, MDM2, CIP1, EDA2R, growth differentiation factor 15-like (GDF15), keratin, type II cytoskeletal cochleal-like (K2CO) and serine/threonine-protein kinase RIO3 (RIOK3) were significantly differentially expressed after AFB1 exposure in both the previous experiment [28] and in DTA1 and WTA5 in this study. Less genes shared significant expression changes in DTA5 (MDM2, CIP1, EDA2R, and GCF15) and WTA1 (MDM2, GDF15, and RIOK3), likely due to the reduced effects of AFB1 on these groups.
Embryonic exposure in the spleen did not recapitulate the effects of dietary AFB1 on the splenic transcriptome previously observed in poults [29]. Matched embryonic spleens were collected and the RNA sequenced alongside the hepatic samples described in this study. However, only 8 significant genes with |log2FC| ≥ 1.0 were identified in the spleen and only for the WTA1/WTC1 comparison [80]. This same group comparison had minimal expression changes in the liver. Birds rely principally on maternally transferred antibodies until immune system development is complete after hatch [81,82]. Thus, the lack of response in the spleen is likely attributable to the immature status of the organ. Therefore, effects of in ovo AFB1 exposure on immune gene expression would be better investigated after hatch.
4. Experimental Section
4.1. Embryos and Toxin Preparation
Embryonic AFB1 exposures were performed according to protocols approved by the Institutional Biosafety Committee (IBC) at the University of Minnesota (IBC Protocol Number: 1302-30324H). Willmar Poultry Co. (Willmar, MN, USA) generously provided fertilized commercial turkey eggs at day 14 of incubation. Eastern wild turkey eggs (Meleagris gallopavo silvestris) at day 0 of incubation were purchased from Stromberg’s Chicks and Game Birds (Pine River, MN, USA). Throughout the experiment, eggs were incubated at 37.0 ± 0.5 °C with approximately 40% humidity and rotation every 2 h. AFB1-solution was made by directly suspending AFB1 (Sigma-Aldrich, St. Louis, MO, USA) in 100% EtOH and diluting to a final concentration of 5 μg/mL in 30% EtOH.
4.2. In ovo AFB1 Exposure
Eggs were candled prior to the start of the exposure period to verify viability. Fewer embryos were used for wild turkey (WT) (N = 15) than domesticated turkey (DT) (N = 28) in this experiment as a result of lower fertilization rates in the wild birds. Viable eggs were randomly divided into 4 treatment groups (7 domesticated eggs/group and 4 wild eggs/group): control 1 day exposure (C1), AFB1 1 day exposure (A1), control 5 days exposure (C5), and AFB1 5 days exposure (A5). Due to a shorter incubation period in DT than WT, control (CNTL) and AFB1 (AFB) treatments were performed on developmentally equivalent days. Thus on day 17 (DT) and day 19 (WT), 0.2 mL of 30% EtOH was sterilely injected into the air sac of each egg in the C1 and C5 groups, while eggs in the A1 and A5 groups received an injection of 0.2 mL of AFB1-solution (1 μg of AFB1/egg). After 1 day of exposure, DT (day 18) and WT (day 20) in the A1 and C1 groups embryos were sacrificed. Egg, embryo, and liver weights were measured and liver tissue was collected directly into RNAlater (Ambion, Inc., Austin, TX, USA). Tissue samples were perfused overnight at 4 °C and stored at −20 °C to preserve RNA. Only 3 WT eggs could be processed for group A1 due to the death of one embryo prior to the start of the exposure period. After 5 days of exposure (day 22 and day 24 for DT and WT, respectively), eggs in the A5 and C5 groups were processed as described for the 1 day exposure groups. Three-way ANOVA and simultaneous tests for general linear hypotheses (multiple comparisons tests) were performed on weight measurements in R using the multcomp package [83,84].
4.3. RNA Isolation and Sequencing
Total RNA was isolated from each liver sample by TRIzol extraction (Ambion, Inc., Austin, TX, USA), DNase-treated (Turbo DNA-freeTM Kit, Ambion, Inc., Austin, TX, USA), and stored at −80 °C. Spectrophotometry (Nanodrop 1000, Nanodrop Technologies, Wilmington, DE, USA) was used for an initial assessment of RNA concentration and quality. RNA samples were submitted for QC, library preparation and sequencing at the University of Minnesota Genomics Center (UMGC). Each sample was fluorometrically quantified by RiboGreen Assay (Invitrogen Corp., Carlsbad, CA, USA) and RNA integrity was confirmed on the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) or the LabChip GX (Caliper Life Sciences, Inc., Hopkinton, MA, USA). Samples from 6 DT embryos/group (n = 24) with the highest RNA quality were selected for sequencing. All WT liver RNA samples were utilized (n = 15) due to the smaller sample sizes. Each sequenced sample possessed an RNA Integrity Number (RIN) between 5.9 and 7.4 or an RNA Quality Score (RQS) between 5.9 and 7.9. All RNA samples had clear separation of the 18S and 28S peaks on the electropherograms. Indexed libraries were constructed with 1 μg of total RNA/sample with the TruSeq RNA Sample Preparation Kit version 2 (Illumina, Inc., San Diego, CA, USA), and size selected for approximately 200 bp inserts. Libraries were multiplexed (see Table S2 for lane and flow cell for each sample) and sequenced on the HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) to produce 100-bp paired-end reads.
4.4. Read Filtering, Trimming, and Dataset QC Analysis
De-multiplexed RNA-seq datasets have been accessioned as part of SRA project ID: SRP067990 (see Table S2 for individual dataset IDs). Raw reads were filtered and trimmed using CLC Genomics Workbench 7.5 (CLCGWB, CLC Bio, Cambridge, MA, USA) according to the following protocol. TruSeq adapter sequences were removed and 4 bp were trimmed from the 3’ end of each read to reduce end-quality dips. Any reads less than 40 bp were discarded. Reads were rechecked for adapter sequence and trimmed for low sequence quality (limit 0.05 error probability and maximum of 2 ambiguities) and passed through a final length filter (discarded reads <40 bp). Quality of each dataset before and after processing was measured with CLCGWB and FastQC [85].
4.5. Read Mapping to MAKER Gene Set
Corrected reads were mapped onto a gene set [86] created on the turkey genome (UMD 5.0) using the MAKER pipeline [87]. Summaries of the MAKER gene set are provided in Table 5 and Table S2. Genes in the MAKER gene set were identified using BLAST alignment to the NCBI RefSeq and Uniprot SwissProt databases [88]. Mapping to the annotated genome was performed in CLCGWB with the standard parameters (length fraction and similarity fraction of 80%, mismatch cost of 2, insertion cost of 3 and deletion cost of 3). Paired read distances were calculated by CLCGWB and each read in a pair was counted separately (pair = 2) to allow direct comparison to single reads and inclusion of broken pairs (reads separated during mapping). Both exonic and intronic matches were counted in order to include reads that map to unannotated exons or genes with incorrect exon borders. However, only reads that mapped uniquely to a single gene were included in counts; reads that mapped to multiple locations were ignored, as their actual source could not be determined.
Table 5.
MAKER gene set for the turkey genome (UMD 5.0).
| Statistic | Number of Genes per Chromosome 1 | % Annotated Genes per Chromosome 1 | Number of Exons per Gene | Genomic Region (Including Introns) | Exons Only |
|---|---|---|---|---|---|
| Min | 2 | 87.40% | 1 | 6 | 6 |
| Mean | 553 | 93.90% | 10 | 24,805 | 2298 |
| Max | 2228 | 100.00% | 141 | 1,524,779 | 55,896 |
| N50 | N/A | N/A | N/A | 57,428 | 3654 |
| Total | 18,265 | 93.70% | 180,541 | 453,055,619 | 41,981,535 |
1 Gene distribution and BLAST annotation across the genome are shown in Table S3.
4.6. Differential Expression and Functional Analysis
Read counts were used for pair-wise comparisons between treatment groups in the R package DESeq2 following the standard workflow [30]. Read counts were first fit to a model based on a negative binomial distribution and normalized by size-scaling for differences in library sequencing depth. Empirical Bayes shrinkage estimates of dispersion and log2 fold change (log2FC) were employed by DESeq2 to prevent over-dispersion, equalize the dynamic range of read counts, handle variable sample sizes, and make log2FC reproducible. Differential expression (DE) of genes between groups was then evaluated with Wald inference tests using these shrinkage estimates and normalized read counts. Genes must have a q-value (FDR adjusted p-value based on the Benjamin-Hochberg procedure) ≤ 0.05 to be considered statistically significant DE in each pair-wise comparison. Significant transcripts were also filtered for a minimum |log2FC| ≥ 1.0. To evaluate the effects of gender, DTA5 versus DTC5 was re-analyzed in DESeq2 using the same parameters, except a second factor was incorporated (design = ~ gender + treatment). Gender of embryos was determined by PCR using sex-specific primer sets [89,90].
Principle component analysis (PCA), MA plots and Venn diagrams were created in R to visualize the expression data and the results of significance testing as previously described in Monson et al. [28,29]. Hierarchical clustering of CNTL or AFB samples (based on Euclidean sample distances) was performed in R using regularized log2 transformed reads counts [30]. For all significant DE genes in each pair-wise comparison (including those with |log2FC| < 1.0), gene pathways and toxicological functions were investigated using Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA, USA).
5. Conclusions
Pathways and genes with significant differential expression identified in this study provide the first direct comparison of the responses of domesticated and wild turkeys to AFB1. Transcriptome responses to AFB1 occurred more rapidly in domesticated than in wild embryos, perhaps as a result of differences in AFB1 metabolism. The most significant effects observed in the AFB1-exposed groups highlighted conserved responses of cell cycle regulators, such as MDM2 and CIP1. Although most evident in the early response of domesticated birds, NRF2-mediated responses to AFB1 were present in both domesticated and wild turkey embryos with varied effects in down-stream detoxifying and anti-oxidant enzymes. Further investigation of these NRF2-response genes may help identify underlying differences in AFB1 sensitivity between domesticated and wild turkeys. Overall, in ovo exposure to AFB1 successfully induced expression changes in the liver, recapitulated hepatic expression effects observed in poults fed AFB1, and identified similarities and differences in transcriptome response between domesticated and wild turkeys.
Acknowledgments
The authors gratefully acknowledge support from the United States Department of Agriculture, Agriculture and Food Research Initiative Grant from the USDA National Institute of Food and Agriculture Animal Genome Program (2013-01043). Support from the University of Minnesota College of Veterinary Medicine Hatch Formula Fund and the Minnesota and Utah Agricultural Experiments Stations is also acknowledged. Analyses were conducted in part using the computing resources at the University of Minnesota Supercomputing Institute. The authors thank Brian McComb and Willmar Poultry Co. (Willmar, MN, USA) for supplying domesticated turkey eggs, Cristian Flores Figueroa (University of Minnesota, Willmar, MN, USA) for instruction on egg injection techniques, Michael Campbell and Mark Yandell (University of Utah, Salt Lake City, UT, USA) for sharing the MAKER gene set, Robert Settlage (Virginia Tech, Blacksburg, VA, USA) for the BLAST annotation, and Juan Abrahante-Llorens (University of Minnesota Informatics Institute, Minneapolis, MN) for assisting with CLCGWB.
Supplementary Materials
The following are available online at www.mdpi.com/2072-6651/8/1/16/s1.
Author Contributions
Melissa S. Monson and Kent M. Reed conceived and designed the experiments as part of a PhD thesis; Melissa S. Monson and Carol J. Cardona performed the experiments; Melissa S. Monson and Kent M. Reed analyzed and interpreted the data; Melissa S. Monson took the lead on writing the manuscript; Melissa S. Monson, Kent M. Reed, Roger A. Coulombe and Carol J. Cardona edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Giambrone J.J., Diener U.L., Davis N.D., Panangala V.S., Hoerr F.J. Effects of aflatoxin on young turkeys and broiler chickens. Poult. Sci. 1985;64:1678–1684. doi: 10.3382/ps.0641678. [DOI] [PubMed] [Google Scholar]
- 2.Pandey I., Chauhan S.S. Studies on production performance and toxin residues in tissues and eggs of layer chickens fed on diets with various concentrations of aflatoxin AFB1. Br. Poult. Sci. 2007;48:713–723. doi: 10.1080/00071660701713534. [DOI] [PubMed] [Google Scholar]
- 3.Rauber R.H., Dilkin P., Giacomini L.Z., Araújo de Almeida C.A., Mallmann C.A. Performance of turkey poults fed different doses of aflatoxins in the diet. Poult. Sci. 2007;86:1620–1624. doi: 10.1093/ps/86.8.1620. [DOI] [PubMed] [Google Scholar]
- 4.Rawal S., Kim J.E., Coulombe R., Jr. Aflatoxin B1 in poultry: Toxicology, metabolism and prevention. Res. Vet. Sci. 2010;89:325–331. doi: 10.1016/j.rvsc.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Bedard L.L., Massey T.E. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006;241:174–183. doi: 10.1016/j.canlet.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 6.Eaton D.L., Gallagher E.P. Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1994;34:135–172. doi: 10.1146/annurev.pa.34.040194.001031. [DOI] [PubMed] [Google Scholar]
- 7.Corrier D.E. Mycotoxicosis: Mechanisms of immunosuppression. Vet. Immunol. Immunopathol. 1991;30:73–87. doi: 10.1016/0165-2427(91)90010-A. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.E., Bunderson B.R., Croasdell A., Coulombe R.A., Jr. Functional characterization of alpha-class glutathione S-transferases from the turkey (Meleagris gallopavo) Toxicol. Sci. 2011;124:45–53. doi: 10.1093/toxsci/kfr212. [DOI] [PubMed] [Google Scholar]
- 9.Klein P.J., Buckner R., Kelly J., Coulombe R.A., Jr. Biochemical basis for the extreme sensitivity of turkeys to aflatoxin B1. Toxicol. Appl. Pharmacol. 2000;165:45–52. doi: 10.1006/taap.2000.8926. [DOI] [PubMed] [Google Scholar]
- 10.Quist C.F., Bounous D.I., Kilburn J.V., Nettles V.F., Wyatt R.D. The effect of dietary aflatoxin on wild turkey poults. J. Wildl. Dis. 2000;36:436–444. doi: 10.7589/0090-3558-36.3.436. [DOI] [PubMed] [Google Scholar]
- 11.Kim J.E., Bunderson B.R., Croasdell A., Reed K.M., Coulombe R.A., Jr. Alpha-class glutathione S-transferases in wild turkeys (Meleagris gallopavo): Characterization and role in resistance to the carcinogenic mycotoxin aflatoxin B1. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aly S.A., Anwer W. Effect of naturally contaminated feed with aflatoxins on performance of laying hens and the carryover of aflatoxin B1 residues in table egg. Pakistan J. Nutr. 2009;8:181–186. [Google Scholar]
- 13.Azzam A.H., Gabal M.A. Aflatoxin and immunity in layer hens. Avian Pathol. 1998;27:570–577. doi: 10.1080/03079459808419386. [DOI] [PubMed] [Google Scholar]
- 14.Khan W.A., Khan M.Z., Khan A., Hassan Z.U., Rafique S., Saleemi M.K., Ahad A. Dietary vitamin E in White Leghorn layer breeder hens: A strategy to combat aflatoxin B1-induced damage. Avian Pathol. 2014;43:389–395. doi: 10.1080/03079457.2014.943691. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira C.A., Kobashigawa E., Reis T.A., Mestieri L., Albuquerque R., Corrêa B. Aflatoxin B1 residues in eggs of laying hens fed a diet containing different levels of the mycotoxin. Food Addit. Contam. 2000;17:459–462. doi: 10.1080/02652030050034037. [DOI] [PubMed] [Google Scholar]
- 16.Oliveira C.A.F., Rosmaninho J.F., Castro A.L., Butkeraitis P., Alves Reis T., Corrêa B. Aflatoxin residues in eggs of laying Japanese quail after long-term administration of rations containing low levels of aflatoxin B1. Food Addit. Contam. 2003;20:648–653. doi: 10.1080/0265203031000119025. [DOI] [PubMed] [Google Scholar]
- 17.Wolzak A., Pearson A.M., Coleman T.H., Pestka J.J., Gray J.I. Aflatoxin deposition and clearance in the eggs of laying hens. Food Chem. Toxicol. 1985;23:1057–1061. doi: 10.1016/0278-6915(85)90052-3. [DOI] [PubMed] [Google Scholar]
- 18.Celik I., Oguz H., Demet O., Boydak M., Donmez H.H., Sur E., Nizamlioglu F. Embryotoxicity assay of aflatoxin produced by Aspergillus parasiticus NRRL 2999. Br. Poult. Sci. 2000;41:401–409. doi: 10.1080/713654961. [DOI] [PubMed] [Google Scholar]
- 19.Dietert R.R., Qureshi M.A., Nanna U.C., Bloom S.E. Embryonic exposure to aflatoxin-B1: Mutagenicity and influence on development and immunity. Environ. Mutagen. 1985;7:715–725. doi: 10.1002/em.2860070510. [DOI] [PubMed] [Google Scholar]
- 20.Edrington T.S., Harvey R.B., Kubena L.F. Toxic effects of aflatoxin B1 and ochratoxin A, alone and in combination, on chicken embryos. Bull. Environ. Contam. Toxicol. 1995;54:331–336. doi: 10.1007/BF00195101. [DOI] [PubMed] [Google Scholar]
- 21.Oznurlu Y., Celik I., Sur E., Ozaydın T., Oğuz H., Altunbaş K. Determination of the effects of aflatoxin B1 given in ovo on the proximal tibial growth plate of broiler chickens: Histological, histometric and immunohistochemical findings. Avian Pathol. 2012;41:469–477. doi: 10.1080/03079457.2012.712673. [DOI] [PubMed] [Google Scholar]
- 22.Qureshi M.A., Brake J., Hamilton P.B., Hagler W.M., Jr., Nesheim S. Dietary exposure of broiler breeders to aflatoxin results in immune dysfunction in progeny chicks. Poult. Sci. 1998;77:812–819. doi: 10.1093/ps/77.6.812. [DOI] [PubMed] [Google Scholar]
- 23.Sur E., Celik I. Effects of aflatoxin B1 on the development of the bursa of Fabricius and blood lymphocyte acid phosphatase of the chicken. Br. Poult. Sci. 2003;44:558–566. doi: 10.1080/00071660310001618352. [DOI] [PubMed] [Google Scholar]
- 24.Williams J.G., Deschl U., William G.M. DNA damage in fetal liver cells of turkey and chicken eggs dosed with aflatoxin B1. Arch. Toxicol. 2011;85:1167–1172. doi: 10.1007/s00204-011-0653-x. [DOI] [PubMed] [Google Scholar]
- 25.Neldon-Ortiz D.L., Qureshi M.A. Effects of AFB1 embryonic exposure on chicken mononuclear phagocytic cell functions. Dev. Comp. Immunol. 1992;16:187–196. doi: 10.1016/0145-305X(92)90018-8. [DOI] [PubMed] [Google Scholar]
- 26.Sur E., Celik I., Oznurlu Y., Aydin M.F., Oguz H., Kurtoglu V., Ozaydin T. Enzyme histochemical and serological investigations on the immune system from chickens treated in ovo with aflatoxin B1 (AFB1) Rev. Méd. Vét. 2011;162:443–448. [Google Scholar]
- 27.Ul-Hassan Z., Khan M.Z., Khan A., Javed I. Immunological status of the progeny of breeder hens kept on ochratoxin A (OTA)- and aflatoxin B1 (AFB1)-contaminated feeds. J. Immunotoxicol. 2012;9:381–391. doi: 10.3109/1547691X.2012.675365. [DOI] [PubMed] [Google Scholar]
- 28.Monson M.S., Settlage R.E., McMahon K.W., Mendoza K.M., Rawal S., El-Nezami H.S., Coulombe R.A., Reed K.M. Response of the hepatic transcriptome to aflatoxin B1 in domestic turkey (Meleagris gallopavo) PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monson M.S., Settlage R.E., Mendoza K.M., Rawal S., El-Nezami H.S., Coulombe R.A., Reed K.M. Modulation of the spleen transcriptome in domestic turkey (Meleagris gallopavo) in response to aflatoxin B1 and probiotics. Immunogenetics. 2015;67:163–178. doi: 10.1007/s00251-014-0825-y. [DOI] [PubMed] [Google Scholar]
- 30.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.E., Bauer M.M., Mendoza K.M., Reed K.M., Coulombe R.A., Jr. Comparative genomics identifies new alpha class genes within the avian glutathione S-transferase gene cluster. Gene. 2010;452:45–53. doi: 10.1016/j.gene.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Chen X., Horn N., Applegate T.J. Efficiency of hydrated sodium calcium aluminosilicate to ameliorate the adverse effects of graded levels of aflatoxin B1 in broiler chicks. Poult. Sci. 2014;93:2037–2047. doi: 10.3382/ps.2014-03984. [DOI] [PubMed] [Google Scholar]
- 33.Giambrone J.J., Diener U.L., Davis N.D., Panangala V.S., Hoerr F.J. Effects of purified aflatoxin on broiler chickens. Poult. Sci. 1985;64:852–858. doi: 10.3382/ps.0640852. [DOI] [PubMed] [Google Scholar]
- 34.Giambrone J.J., Diener U.L., Davis N.D., Panangala V.S., Hoerr F.J. Effects of purified aflatoxin on turkeys. Poult. Sci. 1985;64:859–865. doi: 10.3382/ps.0640859. [DOI] [PubMed] [Google Scholar]
- 35.Huff W.E., Kubena L.F., Harvey R.B., Corrier D.E., Mollenhauer H.H. Progression of aflatoxicosis in broiler chickens. Poult. Sci. 1986;65:1891–1899. doi: 10.3382/ps.0651891. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira C.A., Rosmaninho J.F., Butkeraitis P., Corrêa B., Reis T.A., Guerra J.L., Albuquerque R., Moro M.E. Effect of low levels of dietary aflatoxin B1 on laying Japanese quail. Poult. Sci. 2002;81:976–980. doi: 10.1093/ps/81.7.976. [DOI] [PubMed] [Google Scholar]
- 37.Verma J., Johri T.S., Swain B.K. Effect of varying levels of aflatoxin, ochratoxin and their combinations on the performance and egg quality characteristics in laying hens. Asian Aust. J. Anim. Sci. 2003;16:1015–1019. doi: 10.5713/ajas.2003.1015. [DOI] [Google Scholar]
- 38.Yarru L.P., Settivari R.S., Antoniou E., Ledoux D.R., Rottinghaus G.E. Toxicological and gene expression analysis of the impact of aflatoxin B1 on hepatic function of male broiler chicks. Poult. Sci. 2009;88:360–371. doi: 10.3382/ps.2008-00258. [DOI] [PubMed] [Google Scholar]
- 39.Yarru L.P., Settivari R.S., Gowda N.K., Antoniou E., Ledoux D.R., Rottinghaus G.E. Effects of turmeric (Curcuma longa) on the expression of hepatic genes associated with biotransformation, antioxidant, and immune systems in broiler chicks fed aflatoxin. Poult. Sci. 2009;88:2620–2627. doi: 10.3382/ps.2009-00204. [DOI] [PubMed] [Google Scholar]
- 40.Jennen D.G., Magkoufopoulou C., Ketelslegers H.B., van Herwijnen M.H., Kleinjans J.C., van Delft J.H. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol. Sci. 2010;115:66–79. doi: 10.1093/toxsci/kfq026. [DOI] [PubMed] [Google Scholar]
- 41.Merrick B.A., Phadke D.P., Auerbach S.S., Mav D., Stiegelmeyer S.M., Shah R.R., Tice R.R. RNA-Seq profiling reveals novel hepatic gene expression pattern in aflatoxin B1 treated rats. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0061768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rustemeyer S.M., Lamberson W.R., Ledoux D.R., Wells K., Austin K.J., Cammack K.M. Effects of dietary aflatoxin on the hepatic expression of apoptosis genes in growing barrows. J. Anim. Sci. 2011;89:916–925. doi: 10.2527/jas.2010-3473. [DOI] [PubMed] [Google Scholar]
- 43.Josse R., Dumont J., Fautrel A., Robin M.A., Guillouzo A. Identification of early target genes of aflatoxin B1 in human hepatocytes, inter-individual variability and comparison with other genotoxic compounds. Toxicol. Appl. Pharmacol. 2012;258:176–187. doi: 10.1016/j.taap.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 44.Ricordy R., Gensabella G., Cacci E., Augusti-Tocco G. Impairment of cell cycle progression by aflatoxin B1 in human cell lines. Mutagenesis. 2002;17:241–249. doi: 10.1093/mutage/17.3.241. [DOI] [PubMed] [Google Scholar]
- 45.Ranchal I., González R., Bello R.I., Ferrín G., Hidalgo A.B., Linares C.I., Aguilar-Melero P., González-Rubio S., Barrera P., Marchal T., et al. The reduction of cell death and proliferation by p27Kip1 minimizes DNA damage in an experimental model of genotoxicity. Int. J. Cancer. 2009;125:2270–2280. doi: 10.1002/ijc.24621. [DOI] [PubMed] [Google Scholar]
- 46.Chen K., Fang J., Peng X., Cui H., Chen J., Wang F., Chen Z., Zuo Z., Deng J., Lai W., Zhou Y. Effect of selenium supplementation on aflatoxin B1-induced histopathological lesions and apoptosis in bursa of Fabricius in broilers. Food Chem. Toxicol. 2014;74:91–97. doi: 10.1016/j.fct.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Cova L., Wild C.P., Mehrotra R., Turusov V., Shirai T., Lambert V., Jacquet C., Tomatis L., Trépo C., Montesano R. Contribution of aflatoxin B1 and hepatitis B virus infection in the induction of liver tumors in ducks. Cancer Res. 1990;50:2156–2163. [PubMed] [Google Scholar]
- 48.Cullen J.M., Marion P.L., Sherman G.J., Hong X., Newbold J.E. Hepatic neoplasms in aflatoxin B1-treated, congenital duck hepatitis B virus-infected, and virus-free Pekin ducks. Cancer Res. 1990;50:4072–4080. [PubMed] [Google Scholar]
- 49.Klein P.J., Van Vleet T.R., Hall J.O., Coulombe R.A., Jr. Dietary butylated hydroxytoluene protects against aflatoxicosis in turkeys. Toxicol. Appl. Pharmacol. 2002;182:11–19. doi: 10.1006/taap.2002.9433. [DOI] [PubMed] [Google Scholar]
- 50.Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez M.S., Desterro J.M., Lain S., Lane D.P., Hay R.T. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell Biol. 2000;20:8458–8467. doi: 10.1128/MCB.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García-Trevijano E.R., Latasa M.U., Carretero M.V., Berasain C., Mato J.M., Avila M.A. S-adenosylmethionine regulates MAT1A and MAT2A gene expression in cultured rat hepatocytes: A new role for S-adenosylmethionine in the maintenance of the differentiated status of the liver. FASEB J. 2000;14:2511–2518. doi: 10.1096/fj.00-0121com. [DOI] [PubMed] [Google Scholar]
- 53.Lu S.C., Mato J.M. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. J. Gastroenterol. Hepatol. 2008;23:S73–S77. doi: 10.1111/j.1440-1746.2007.05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mato J.M., Martínez-Chantar M.L., Lu S.C. S-adenosylmethionine metabolism and liver disease. Ann. Hepatol. 2013;12:183–189. [PMC free article] [PubMed] [Google Scholar]
- 55.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niture S.K., Khatri R., Jaiswal A.K. Regulation of Nrf2—An update. Free Radic. Biol. Med. 2014;66:36–44. doi: 10.1016/j.freeradbiomed.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Apopa P.L., He X., Ma Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008;22:63–76. doi: 10.1002/jbt.20212. [DOI] [PubMed] [Google Scholar]
- 58.Jowsey I.R., Jiang Q., Itoh K., Yamamoto M., Hayes J.D. Expression of the aflatoxin B1-8,9-epoxide-metabolizing murine glutathione S-transferase A3 subunit is regulated by the Nrf2 transcription factor through an antioxidant response element. Mol. Pharmacol. 2003;64:1018–1028. doi: 10.1124/mol.64.5.1018. [DOI] [PubMed] [Google Scholar]
- 59.Chen W., Sun Z., Wang X.J., Jiang T., Huang Z., Fang D., Zhang D.D. Direct interaction between Nrf2 and p21Cip1/WAF1 upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayes J.D., Judah D.J., Neal G.E., Nguyen T. Molecular cloning and heterologous expression of a cDNA encoding a mouse glutathione S-transferase Yc subunit possessing high catalytic activity for aflatoxin B1-8,9-epoxide. Biochem. J. 1992;285 Pt 1:173–180. doi: 10.1042/bj2850173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ilic Z., Crawford D., Vakharia D., Egner P.A., Sell S. Glutathione-S-transferase A3 knockout mice are sensitive to acute cytotoxic and genotoxic effects of aflatoxin B1. Toxicol. Appl. Pharmacol. 2010;242:241–246. doi: 10.1016/j.taap.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perrone C.E., Ahr H.J., Duan J.D., Jeffrey A.M., Schmidt U., Williams G.M., Enzmann H.H. Embryonic turkey liver: Activities of biotransformation enzymes and activation of DNA-reactive carcinogens. Arch. Toxicol. 2004;78:589–598. doi: 10.1007/s00204-004-0580-1. [DOI] [PubMed] [Google Scholar]
- 63.Chen J., Chen K., Yuan S., Peng X., Fang J., Wang F., Cui H., Chen Z., Yuan J., Geng Y. Effects of aflatoxin B1 on oxidative stress markers and apoptosis of spleens in broilers. Toxicol. Ind. Health. 2013 doi: 10.1177/0748233713500819. [DOI] [PubMed] [Google Scholar]
- 64.Eraslan G., Akdogan M., Yarsan E., Sahindokuyucu F., Essiz D., Altintas L. The effects of aflatoxins on oxidative stress in broiler chickens. Turk J. Vet. Anim. Sci. 2005;29:701–707. [Google Scholar]
- 65.Shen H.M., Shi C.Y., Lee H.P., Ong C.N. Aflatoxin B1-induced lipid peroxidation in rat liver. Toxicol. Appl. Pharmacol. 1994;127:145–150. doi: 10.1006/taap.1994.1148. [DOI] [PubMed] [Google Scholar]
- 66.Guengerich F.P., Cai H., McMahon M., Hayes J.D., Sutter T.R., Groopman J.D., Deng Z., Harris T.M. Reduction of aflatoxin B1 dialdehyde by rat and human aldo-keto reductases. Chem. Res. Toxicol. 2001;14:727–737. doi: 10.1021/tx010005p. [DOI] [PubMed] [Google Scholar]
- 67.Judah D.J., Hayes J.D., Yang J.C., Lian L.Y., Roberts G.C., Farmer P.B., Lamb J.H., Neal G.E. A novel aldehyde reductase with activity towards a metabolite of aflatoxin B1 is expressed in rat liver during carcinogenesis and following the administration of an anti-oxidant. Biochem. J. 1993;292:13–18. doi: 10.1042/bj2920013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelly E.J., Erickson K.E., Sengstag C., Eaton D.L. Expression of human microsomal epoxide hydrolase in Saccharomyces cerevisiae reveals a functional role in aflatoxin B1 detoxification. Toxicol. Sci. 2002;65:35–42. doi: 10.1093/toxsci/65.1.35. [DOI] [PubMed] [Google Scholar]
- 69.Klein P.J., Van Vleet T.R., Hall J.O., Coulombe R.A., Jr. Biochemical factors underlying the age-related sensitivity of turkeys to aflatoxin B1. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2002;132:193–201. doi: 10.1016/S1532-0456(02)00065-0. [DOI] [PubMed] [Google Scholar]
- 70.Klein P.J., Van Vleet T.R., Hall J.O., Coulombe R.A., Jr. Effects of dietary butylated hydroxytoluene on aflatoxin B1-relevant metabolic enzymes in turkeys. Food Chem. Toxicol. 2003;41:671–678. doi: 10.1016/S0278-6915(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 71.Jaiswal A.K. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 2000;29:254–262. doi: 10.1016/S0891-5849(00)00306-3. [DOI] [PubMed] [Google Scholar]
- 72.Toyomizu M., Kikusato M., Kawabata Y., Azad M.A., Inui E., Amo T. Meat-type chickens have a higher efficiency of mitochondrial oxidative phosphorylation than laying-type chickens. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2011;159:75–81. doi: 10.1016/j.cbpa.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 73.Bottje W., Pumford N.R., Ojano-Dirain C., Iqbal M., Lassiter K. Feed efficiency and mitochondrial function. Poult. Sci. 2006;85:8–14. doi: 10.1093/ps/85.1.8. [DOI] [PubMed] [Google Scholar]
- 74.Ramachandra Pai M., Jayanthi Bai N., Venkitasubramanian T.A. Effect of aflatoxins on oxidative phosphorylation by rat liver mitochondria. Chem. Biol. Interact. 1975;10:123–131. doi: 10.1016/0009-2797(75)90106-4. [DOI] [PubMed] [Google Scholar]
- 75.Sajan M.P., Satav J.G., Bhattacharya R.K. Activity of some respiratory enzymes and cytochrome contents in rat hepatic mitochondria following aflatoxin B1 administration. Toxicol. Lett. 1995;80:55–60. doi: 10.1016/0378-4274(95)03256-K. [DOI] [PubMed] [Google Scholar]
- 76.Rawal S., Yip S.S., Coulombe R.A., Jr. Cloning, expression and functional characterization of cytochrome P450 3A37 from turkey liver with high aflatoxin B1 epoxidation activity. Chem. Res. Toxicol. 2010;23:1322–1329. doi: 10.1021/tx1000267. [DOI] [PubMed] [Google Scholar]
- 77.Rawal S., Coulombe R.A., Jr. Metabolism of aflatoxin B1 in turkey liver microsomes: The relative roles of cytochromes P450 1A5 and 3A37. Toxicol. Appl. Pharmacol. 2011;254:349–354. doi: 10.1016/j.taap.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Reed K.M., Mendoza K.M., Coulombe R.A., Jr. Structure and genetic mapping of the cytochrome P450 gene (CYP1A5) in the turkey (Meleagris gallopavo) Cytogenet. Genome Res. 2007;116:104–109. doi: 10.1159/000097426. [DOI] [PubMed] [Google Scholar]
- 79.Yip S.S., Coulombe R.A., Jr. Molecular cloning and expression of a novel cytochrome P450 from turkey liver with aflatoxin B1 oxidizing activity. Chem. Res. Toxicol. 2006;19:30–37. doi: 10.1021/tx050233+. [DOI] [PubMed] [Google Scholar]
- 80.Monson M.S. Ph.D. Thesis. University of Minnesota; St. Paul, MN, USA: 2015. Hepatotoxic and Immunomodulatory Transcriptome Responses to Aflatoxin B1 in the Turkey (Meleagris gallopavo) [Google Scholar]
- 81.Friedman A., Elad O., Cohen I., Bar Shira E. The gut associated lymphoid system in the post-hatch chick: Dynamics of maternal IgA. Israel J. Vet. Med. 2012;67:75–81. [Google Scholar]
- 82.Hamal K.R., Burgess S.C., Pevzner I.Y., Erf G.F. Maternal antibody transfer from dams to their egg yolks, egg whites, and chicks in meat lines of chickens. Poult. Sci. 2006;85:1364–1372. doi: 10.1093/ps/85.8.1364. [DOI] [PubMed] [Google Scholar]
- 83.Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 84.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. [(accessed on 4 January 2016)]. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org/ [Google Scholar]
- 85.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. [(accessed on 6 October 2011)]. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 86.Campbell M. ((University of Utah, Salt Lake City, UT, USA)), Yandell M. ((University of Utah, Salt Lake City, UT, USA)). Personal communication. 2015.
- 87.Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Settlage R. ((Virginia Tech, Blacksburg, VA, USA)). Personal communication. 2015.
- 89.Granevitze Z., Blum S., Cheng H., Vignal A., Morisson M., Ben-Ari G., David L., Feldman M.W., Weigend S., Hillel J. Female-specific DNA sequences in the chicken genome. J. Hered. 2007;98:238–242. doi: 10.1093/jhered/esm010. [DOI] [PubMed] [Google Scholar]
- 90.Kalina J., Mucksová J., Yan H., Trefil P. Rapid sexing of selected galliformes by polymerase chain reaction. Czech J. Anim. Sci. 2012;57:187–192. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.