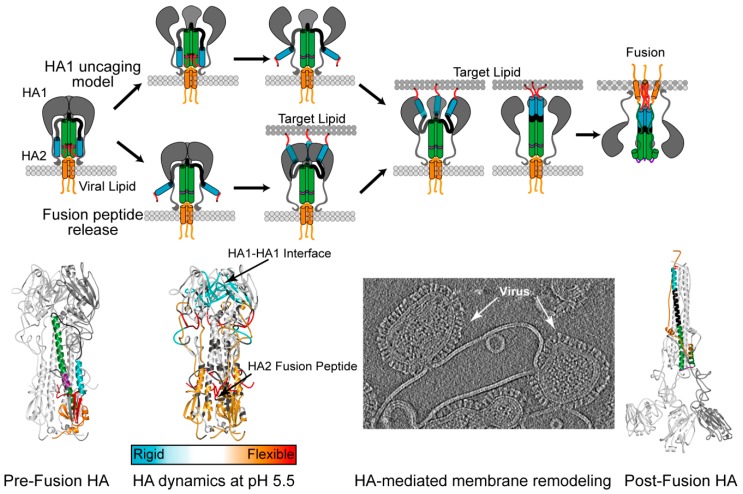
Figure 3.
Influenza Hemagglutinin pH-Activation Models. Pre-fusion HA is composed of the two subunits, HA1 (grey) and HA2 fusion domain with the fusion peptide and proximal regions (red), the short helix (blue), the B-loop (black), the long helix (green and purple), and the C-terminal regions (orange), colored identically on 3HMG crystal structure below. The pH-dependent membrane fusion is hypothesized to commence either by the uncaging of HA1 (top pathway) from the HA2 fusion machinery, or by the release of the fusion peptide from the core of the trimer (bottom pathway). Once the fusion peptide is released, it binds the target membrane and the subsequent refolding of the B-loop and long helix (purple), drives the fusion of the viral membrane with the target membrane. Membrane fusion ends in the post-fusion conformation depicted in the bottom panel where HA1 lobes are modeled onto the 1QU1 crystal structure. Recent solution-based techniques such as HDX-MS and cryo-electron tomography have provided evidence for the fusion peptide release model for H3 HA isolate fusion activation. The HDX dynamic changes are illustrated as regions more flexible at low pH (red) colors and more ordered regions at low pH (blue). In the cryo-electron tomogram, acid-activated influenza virus is shown at early stages of membrane fusion with a synthetic liposome.