Abstract
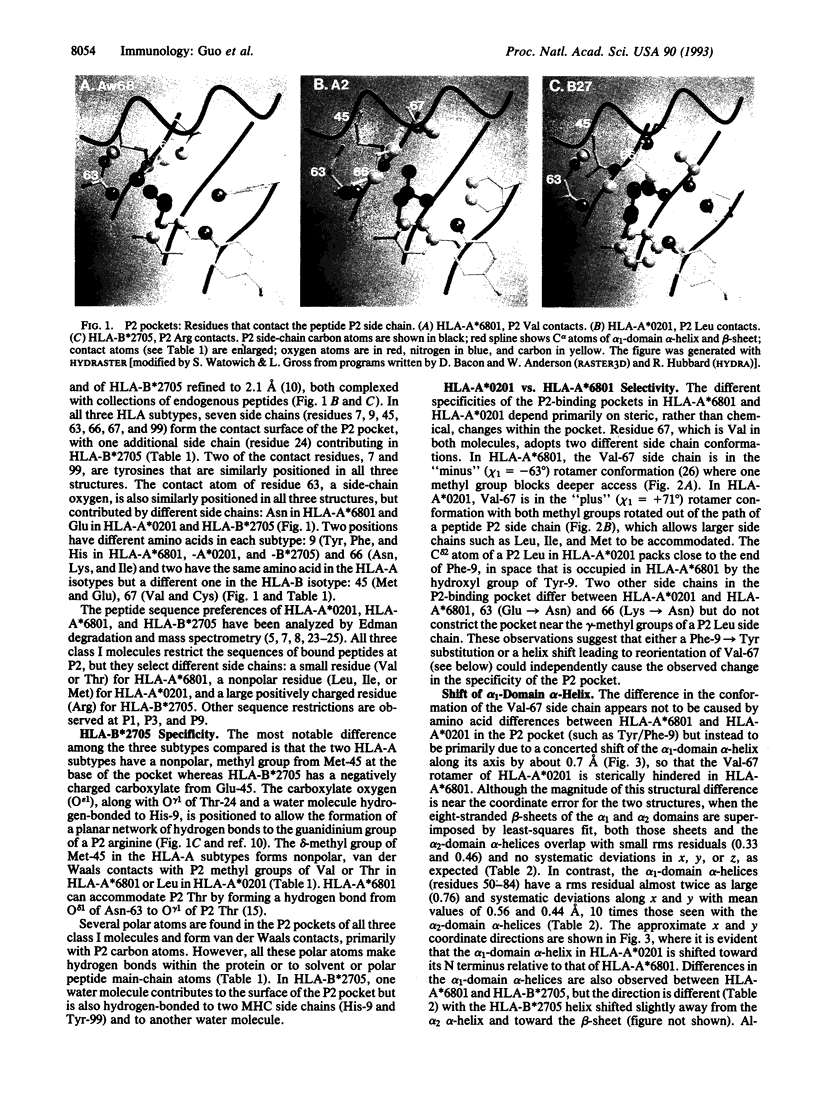
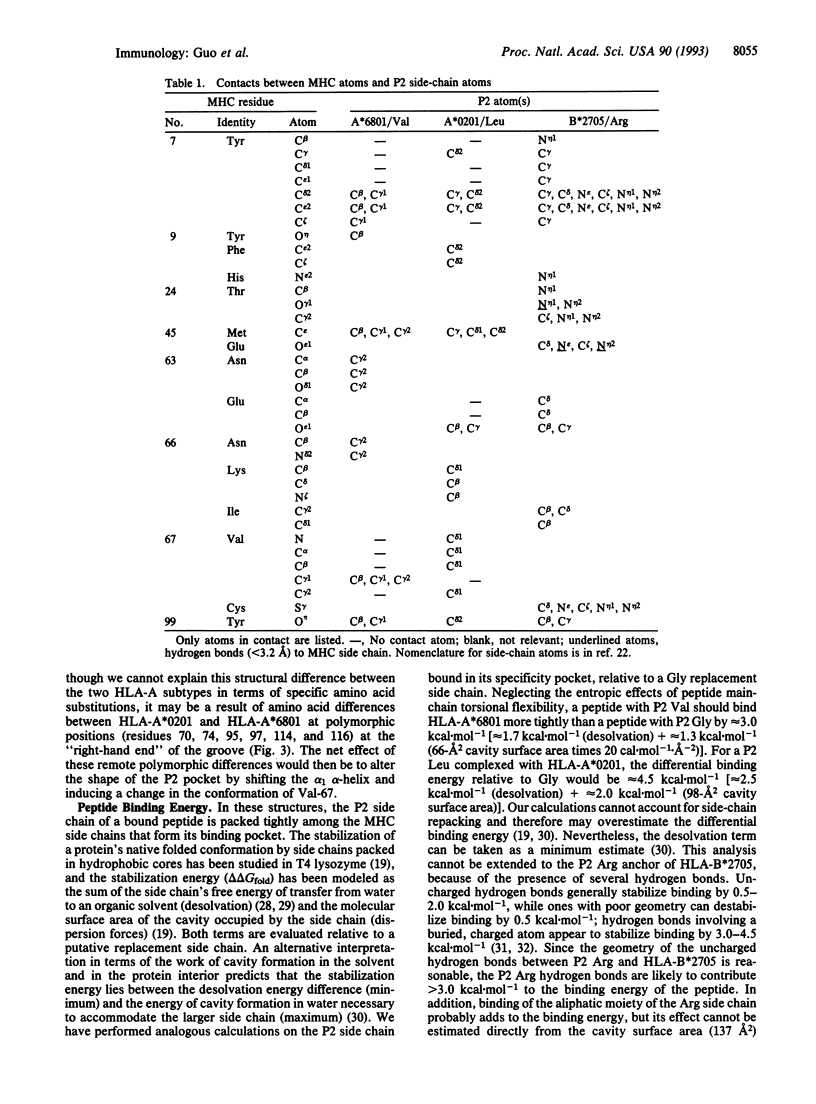
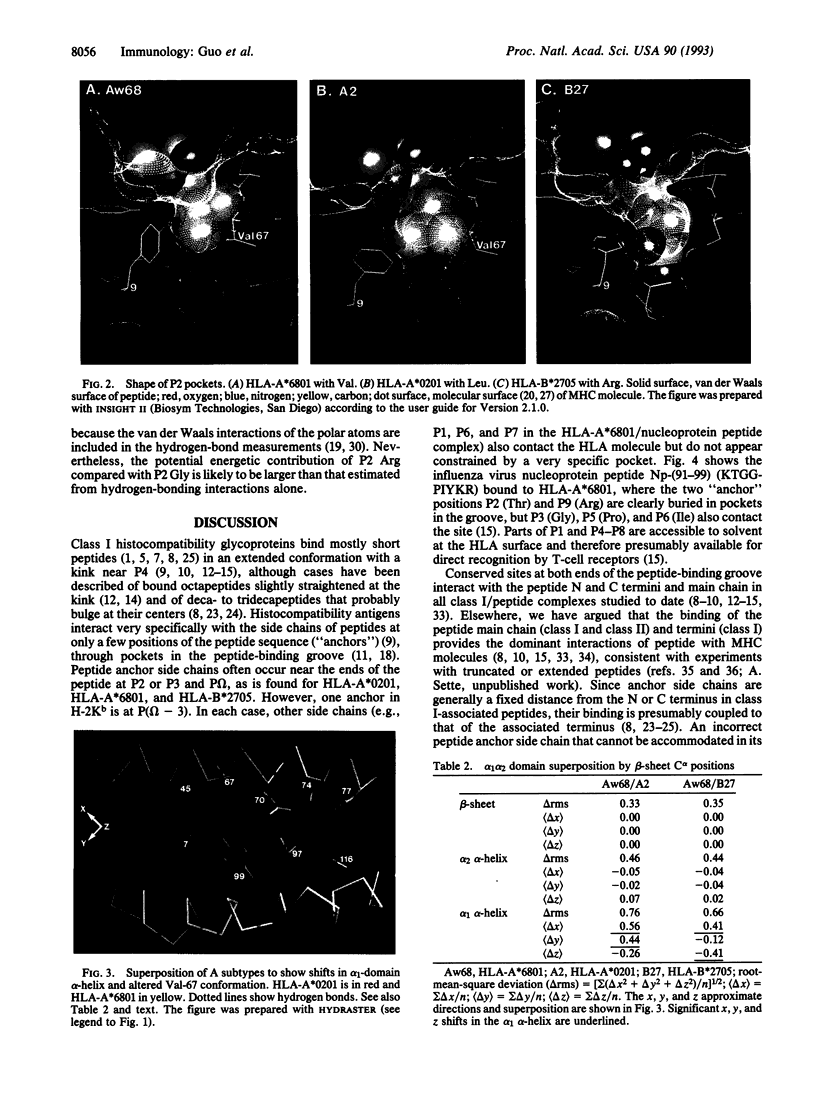
Coordinates from x-ray structures of HLA-A*6801, HLA-A*0201, and HLA-B*2705 were analyzed to examine the basis for their selectivity in peptide binding. The pocket that binds the side chain of the peptide's second amino acid residue (P2 residue) shows a preference for Val, Leu, and Arg in these three HLA subtypes, respectively. The Arg-specific pocket of HLA-B*2705 differs markedly from those of HLA-A*0201 and HLA-A*6801, as a result of numerous differences in the side chains that form the pocket's surface. The cause of the specificity differences between HLA-A*0201 and HLA-A*6801 is more subtle and depends both on a change in conformation of pocket residue Val-67 and on a sequence difference at residue 9. The Val-67 conformational change appears to be caused by a shift in the position of the alpha 1-domain alpha-helix relative to the beta-sheet in the cleft and may, in fact, depend on amino acid differences remote from the P2 pocket. Analysis of the stereochemistry of the P2 side chain interacting with its binding pocket permits an estimate to be made of its contribution to the free-energy change of peptide binding.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Cerundolo V., Elliott T., Elvin J., Bastin J., Rammensee H. G., Townsend A. The binding affinity and dissociation rates of peptides for class I major histocompatibility complex molecules. Eur J Immunol. 1991 Sep;21(9):2069–2075. doi: 10.1002/eji.1830210915. [DOI] [PubMed] [Google Scholar]
- Damodaran S., Song K. B. The role of solvent polarity in the free energy of transfer of amino acid side chains from water to organic solvents. J Biol Chem. 1986 Jun 5;261(16):7220–7222. [PubMed] [Google Scholar]
- Eriksson A. E., Baase W. A., Zhang X. J., Heinz D. W., Blaber M., Baldwin E. P., Matthews B. W. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992 Jan 10;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H. G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991 May 23;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Knill-Jones J., Lowe D. M., Wilkinson A. J., Blow D. M., Brick P., Carter P., Waye M. M., Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985 Mar 21;314(6008):235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Hunt D. F., Henderson R. A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A. L., Appella E., Engelhard V. H. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992 Mar 6;255(5049):1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- Jardetzky T. S., Gorga J. C., Busch R., Rothbard J., Strominger J. L., Wiley D. C. Peptide binding to HLA-DR1: a peptide with most residues substituted to alanine retains MHC binding. EMBO J. 1990 Jun;9(6):1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor D. A., Zemmour J., Ennis P. D., Parham P. Evolution of class-I MHC genes and proteins: from natural selection to thymic selection. Annu Rev Immunol. 1990;8:23–63. doi: 10.1146/annurev.iy.08.040190.000323. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. The structure of HLA-B27 reveals nonamer self-peptides bound in an extended conformation. Nature. 1991 Sep 26;353(6342):321–325. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC. Cell. 1992 Sep 18;70(6):1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Fremont D. H., Peterson P. A., Wilson I. A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992 Aug 14;257(5072):927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- Rötzschke O., Falk K., Deres K., Schild H., Norda M., Metzger J., Jung G., Rammensee H. G. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990 Nov 15;348(6298):252–254. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- Saper M. A., Bjorkman P. J., Wiley D. C. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol. 1991 May 20;219(2):277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- Serrano L., Kellis J. T., Jr, Cann P., Matouschek A., Fersht A. R. The folding of an enzyme. II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol. 1992 Apr 5;224(3):783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- Silver M. L., Guo H. C., Strominger J. L., Wiley D. C. Atomic structure of a human MHC molecule presenting an influenza virus peptide. Nature. 1992 Nov 26;360(6402):367–369. doi: 10.1038/360367a0. [DOI] [PubMed] [Google Scholar]
- Townsend A. R., Gotch F. M., Davey J. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell. 1985 Sep;42(2):457–467. doi: 10.1016/0092-8674(85)90103-5. [DOI] [PubMed] [Google Scholar]
- Townsend A., Bodmer H. Antigen recognition by class I-restricted T lymphocytes. Annu Rev Immunol. 1989;7:601–624. doi: 10.1146/annurev.iy.07.040189.003125. [DOI] [PubMed] [Google Scholar]
- Tsomides T. J., Walker B. D., Eisen H. N. An optimal viral peptide recognized by CD8+ T cells binds very tightly to the restricting class I major histocompatibility complex protein on intact cells but not to the purified class I protein. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11276–11280. doi: 10.1073/pnas.88.24.11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M. L., Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature. 1992 Apr 2;356(6368):443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- Zhang W., Young A. C., Imarai M., Nathenson S. G., Sacchettini J. C. Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bleek G. M., Nathenson S. G. The structure of the antigen-binding groove of major histocompatibility complex class I molecules determines specific selection of self-peptides. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11032–11036. doi: 10.1073/pnas.88.24.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]