Abstract
Malignant cells exhibit major metabolic alterations. The regulatory gene networks that regulate metabolism and the impact of these alterations on overall cellular fitness deserve further exploration. The let-7 microRNAs and their antagonists, the Lin28 RNA-binding proteins, are well-known for controlling the timing of embryonic development. This pathway has recently been shown to regulate glucose metabolism in adult mice and to reprogram metabolism during tissue injury and repair. In addition, many lines of evidence have established that Lin28 is an oncogene that drives tumorigenesis in part by suppressing let-7. The metabolic underpinnings of this oncogenic program are just beginning to be uncovered. Here, we will review the current understanding of how Lin28 exerts regenerative and oncogenic effects through metabolic mechanisms.
Keywords: Lin28, let-7; cancer; regeneration; metabolism
Introduction
Metabolism governs cellular homeostasis, growth, and survival. Alterations in cell metabolism are a common feature of cancer and have been increasingly viewed as one of the hallmarks of malignant transformation (1). In order to support their high proliferative rates, cancer cells adapt to their environment in part by reprogramming the metabolism of all major classes of macromolecules: proteins, carbohydrates, nucleic acids, and lipids (1). This link between metabolism and cancer is not a novel observation. Sixty years ago, Otto Warburg first noted that under normoxic conditions, normal cells metabolize glucose by using mitochondrial oxidative phosphorylation (OxPhos) instead of glycolysis to maximize the production of adenosine triphosphate (ATP), while some cancer cells rely more on aerobic glycolysis, a phenomenon later known as the Warburg effect (2). Since then, interest in this topic has increased and major areas of knowledge have been gained, but fundamentally important questions remain unresolved. The upstream signals that trigger metabolic alterations in cancer cells and what impact these changes have on overall tumor development and progression are still under intense investigation.
Many of the signaling pathways altered in oncogenesis, such as the PI3K-Akt-mTOR pathway (3), can reprogram cell metabolism in a way that promotes malignant growth (1,4). Recently, Lin28a and its homolog Lin28b (collectively referred to as Lin28) and the let-7 microRNA family, have been found to play a direct role in regulating glucose metabolism in adult tissues (5-8). In addition, mouse genetic studies have shown that the reactivation of Lin28 can drive tumor initiation and progression through let-7 dependent and independent mechanisms (9-13). Although all the connections have not been made, it is possible that reprogramming cell metabolism could be a major mechanism by which Lin28 exerts its oncogenic effects. Determining how Lin28 regulates metabolic reprogramming may provide an additional way to understand the interplay between oncogenic signaling pathways and cellular metabolism. We will first summarize roles for Lin28 and let-7 in regulating self-renewal and differentiation in stem cells and cancer. Then, we will discuss their roles in regulating metabolism.
The Lin28/let-7 axis temporally regulates self-renewal and differentiation
Lin28 and let-7 were first identified through mutagenesis screens as heterochronic genes that govern developmental timing in C. elegans (14-17). Lin28 homologs are RNA-binding proteins that consist of zinc fingers (consisting of cysteine and histidine residues in the order CCHC) zinc fingers and cold shock RNA binding domains (16). Subsequent genetic loss and gain-of-function studies in worms revealed that Lin28 promotes self-renewal and delays differentiation of the hypodermal and vulval progenitor cells (16). As expected from these functions, Lin28 expression is high during embryogenesis and early larval stage of development, and gradually declines to an undetectable level in adult tissues (18). In contrast, let-7 expression increases as Lin28 expression wanes from late larval stage and remains high thereafter (14,19). Loss-of-function let-7 mutations promote the division of seam cells and prevent them from cell cycle exit (20), phenocopying Lin28 gain-of-function mutants (19). Later, it was shown that let-7 promotes differentiation and inhibits self-renewal during the transition from larva to adulthood by repressing Lin28 expression through its 3’UTR (16,18).
The expression and regulation of Lin28 and let-7 are highly conserved throughout evolution (18). In mice, Lin28 is expressed at high levels throughout the embryo at early developmental stages (~E6.5) (21). Then, its expression declines through development and remains present in only some adult tissues (21). Whether Lin28 also functions to promote stemness in mammals became more intriguing when overexpressing Lin28a, along with Sox2, Oct4, and Nanog, proved to be sufficient to reprogram human somatic fibroblasts into inducible Pluripotent Stem Cells (iPSCs) (22). Nevertheless, the mechanism by which Lin28 exerted this effect remained a mystery until a flurry of studies showed that in both mouse embryonic stem cells (ESCs) and C. elegans epithelial stem cells, Lin28 inhibits the post-transcriptional maturation of let-7. Lin28 binds to the primary immature form of let-7 and sequesters it from being processed by Drosha/DGCR8 (the small RNA generating machinery in the nucleus) (23,24). In the cytoplasm, Lin28 blocks the loading of let-7 into Dicer by binding the premature form of let-7 and recruiting Tutase4/7, which polyuridylates let-7’s tail, marking it for degradation by a exonuclease called Dis3l2 (24-35). Thus, when antagonized by Lin28, let-7 is rendered inactive.
Studies in mouse ESCs also demonstrated that let-7 antagonizes self-renewal and promotes differentiation. Melton et al. showed that Drosha/DGCR8 knockout ESCs, which are unable to produce most mature miRNAs, fail to silence the stem cell self-renewal program when placed under differentiation-inducing conditions. Introduction of mature let-7 into these Drosha/DGCR8 knockout ESCs is capable of rescuing differentiation and even inhibits ESC self-renewal in stem cell culture conditions (36). These phenotypes were in part due to the let-7 mediated suppression of pluripotency factors such as Lin28, Sal4, and N-Myc (36). More recently, Worringer et al. in Yamanaka’s group showed that let-7 acts as a barrier to counteract iPSC reprogramming by promoting the expression of differentiation genes (37). Thus, together Lin28 and let-7 form a highly conserved and highly regulated axis that temporally regulates the self-renewal and differentiation of stem cells.
Lin28a and Lin28b are oncogenes
In adult mammalian tissues, let-7 is one of the most abundant miRNAs (38). Although the exact roles of let-7 in adult tissues have not been fully characterized, let-7 is known to have tumor suppressor functions. There is copious evidence that let-7 expression is downregulated in a large number of cancers (39-43) and that let-7 overexpression inhibits growth and transformation of cancer cell lines and tumor xenografts (44-52). These anti-cancer effects are partly due to the suppression of let-7 target genes that are critical for cell cycle progression and proliferation, such as K-Ras, Cyclin D1, c-Myc, Cdc34, Hmga2, E2f2, and Lin28 (45-49,51,53). Most of these findings were discovered in cell lines, and thus our knowledge of let-7 functions would benefit from more definitive investigation in animal models.
In contrast to let-7, Lin28 expression is upregulated in multiple tumor types such as neuroblastoma, hepatocellular carcinoma (HCC), Wilms’ tumor, and melanoma (13). Several studies have demonstrated that activation of Lin28 is able to promote tumor development in various mouse tissues in part by suppressing let-7 (9-12). Furthermore, we recently showed that genetic deletion of Lin28a and Lin28b abrogated c-MYC-driven hepatocarcinogenesis and improved overall survival in mice (12). Similar results were achieved using in vivo siRNA to knockdown Lin28b, which resulted in greater levels of cell death in tumor tissues (12). In another study, He et al. isolated pre-malignant liver progenitor cells from Diethylnitrosamine mutagenized mice and showed that these cells have high expression of Lin28a and Lin28b, suggesting that Lin28 plays a role in malignant transformation within the chronically injured liver (54). These studies have functionally established the role of Lin28 in tumor initiation and progression and suggest that Lin28 could be a relevant target for either cancer prevention or therapy.
Although many major effects of Lin28 are mediated through let-7, Lin28 can also directly bind to and influence the translation of many mRNAs enriched with GGAGA (with G = guanosine and A = adenosine) sequences in their loop structures (55). Many of these mRNAs are oncogenic or growth-promoting genes, such as Igf2, Igf2-mRNA binding proteins, Hmga1, or those encoding ribosomal proteins, cell-cycle regulators, and metabolic enzymes (6, 12,55-59). Whether or not and to what extent these mRNA targets of Lin28 contribute to its oncogenic effects are important areas for future investigation.
The Lin28/let-7 axis regulates metabolism in mammalian ESCs
Studies in mammalian ESCs provided initial insights into the role of the Lin28/let-7 axis in metabolism. Genome-wide studies in human ESCs revealed that Lin28 binds to many mitochondrial enzyme mRNAs and interacts with RNA helicase A to enhance the translation of these mRNAs (56). In mouse ESCs, Wang et al. recently showed that threonine (Thr) oxidation into glycine (Gly) and acetyl-CoA catalyzed by threonine dehydrogenase is critical for cell growth (60). A follow-up study by Shyh-Chang et al. revealed that the catabolism of Thr also fuels the synthesis of S-adenosyl-methionine (SAM), which is important for methylation reactions and critical for pluripotency (61). Decreased SAM ultimately led to slowed growth and increased differentiation (61). Metabolic profiling also demonstrated that inducing Lin28 and let-7 had dramatic effects on the Thr-Gly-SAM pathway in mouse ESCs (61). Specifically, overexpressing Lin28 in mouse ESCs led to increased amount of many Thr-Gly-SAM metabolites, while overexpressing let-7 led to reduced amount of these metabolites (61). Together, these studies provided the first evidence that the Lin28/let-7 axis regulates metabolic networks in ESCs.
Lin28 regulates body size, metabolism, and tissue regeneration in adult mice
Lin28 and let-7 also modulate the expression of pathways that directly regulate metabolism in adult mammalian tissues. For gain of function studies, we previously engineered a tetracycline-inducible Lin28a transgenic mouse model [Lin28a transgenic (Tg)]. Due to leakiness of the transgene, Lin28a expression levels are modestly increased in the muscle, skin and connective tissues in the absence of doxycycline induction. We reported that Lin28a Tg mice, compared to control mice without the transgene, exhibited increased body size and delayed puberty onset (62). These phenotypes functionally validated a number of genome-wide association studies that identified connections between human height, puberty timing, and the LIN28B locus (63,64).
Most interestingly, Lin28a Tg mice exhibit enhanced glucose uptake in peripheral tissues (62). Enhanced glucose uptake in Lin28a Tg mice also led to higher levels of the glycolytic metabolite lactate (62). Similarly, whole body inducible human LIN28B overexpressing mice also exhibit superior glucose tolerance, indicating conserved functions between the Lin28 paralogs from two species (6). While gain-of-function Lin28a results in increased body size, loss-of-function Lin28a [Lin28a knockout (KO)] caused dwarfism from E13.5 to adulthood (65). Conditional deletion of Lin28a in skeletal muscles led to insulin resistance and impaired glucose uptake, indicating that Lin28 is physiologically required for normal glucose homeostasis (6,65). In contrast to the phenotypes seen in Lin28a Tg mice, inducible let-7 Tg mice not only have reduced body size and growth retardation, but also have hyperglycemia and glucose intolerance (6). Simultaneous Lin28a and let-7 whole body overexpression cancels out the glucose phenotype of each factor. Thus, there are likely to be mutually antagonistic effects including the possibility that Lin28a increases glucose uptake by suppressing let-7 (6) (Figure 1A).
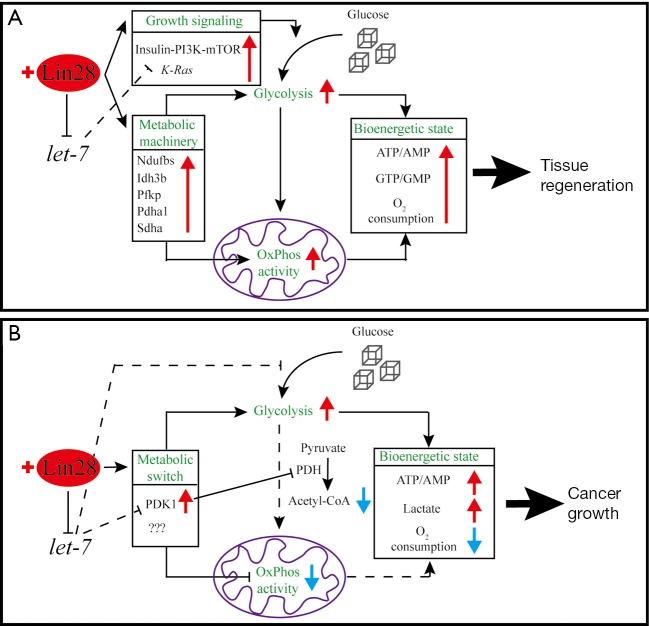
Figure 1.
Reactivation of Lin28 reprograms cell metabolism to enhance tissue regeneration and promote cancer growth. (A) Reactivation of Lin28 in physiological condition in mouse tissues enhances regeneration by suppressing let-7, upregulating K-Ras expression and the insulin-PI3K-mTOR signaling, and more importantly upregulating the expression of metabolic machinery. These changes lead to enhanced glucose uptake, increased activity of both glycolysis and OxPhos, resulting in Lin28-positive cells having a much higher bioenergetic state; (B) Reactivation of Lin28 promotes growth of multiple cancer cell lines in part by suppressing let-7 and upregulating PDK1 protein expression. When let-7 expression is reduced, glucose uptake is enhanced. PDK1 is a negative regulator of OxPhos activity since it inhibits the conversion of pyruvate to acetyl-CoA. When PDK1 expression is enhanced by Lin28 overexpression and let-7 suppression, it promotes glycolysis and blocks OxPhos activity. Consequently, Lin28-positive cancer cells switch to aerobic glycolysis for glucose metabolism.
Mechanistically, the overgrowth of Lin28a Tg mice could partly be due to the reduction of let-7 expression levels in organs where endogenous Lin28a is not normally present (62) and global increases in let-7-target protein production, such as Hmga1, Igf2, and Oct4—all of which are known to regulate body size (6,66-68). To further illustrate that the enhanced glucose uptake seen in Lin28a Tg mice was due to cell autonomous mechanisms, Lin28a was overexpressed in C2C12 myoblasts. Compared to control myoblasts, Lin28a overexpressing myoblasts take up glucose much faster (Figure 1A). This was the result of Lin28a suppressing let-7, which in turn suppresses the Insulin-PI3K-mTOR pathway at multiple nodes (namely, Igf1r, Insulin receptor, and Irs2) (Figure 1A). In terms of signaling output, Akt and S6 phosphorylation is increased in a let-7-dependent manner, which increases the insulin-sensitivity and glucose uptake of myoblasts (6). Furthermore, when muscle specific Tsc1 deficient mice, whose mTOR signaling is increased, were crossed with Lin28b deficient mice, Lin28b’s dwarfism phenotype was rescued. This further confirmed that mTOR signaling genetically interacts with the Lin28 program (65). Another line of evidence that further corroborates this concept is a recent report showing that under nutrient deprivation, let-7 prevents mTORC1 activation to induce autophagy in primary cortical neurons, muscle, and white fat (69). These studies show that the Lin28/let-7 axis strongly influences a known controller of organismal and cancer metabolism, but the following studies demonstrated more interesting mechanisms.
Overexpressing just a modest amount of Lin28a led to increased body size and delayed mouse puberty (62). Even more striking is the superior tissue repair observed in Lin28a Tg mice (7). We and the Daley Lab showed that Lin28a Tg mice exhibited enhanced hair regeneration after shaving, digit repair after amputation, and ear wound healing after hole punch (7). We found that this superior regenerative phenotype in Lin28a Tg mice was not caused by suppression of let-7 alone, since let-7 antimiR delivered to wild-type (WT) mice failed to phenocopy the enhanced regeneration caused by Lin28a overexpression (7). By profiling metabolism during tissue repair, we demonstrated that Lin28a enhances both glycolysis and mitochondrial OxPhos activity through direct binding and translational enhancement of mRNAs that encode several major metabolic enzymes such as phosphofructokinase, pyruvate dehydrogenase (PDH), and isocitrate dehydrogenase (7) (Figure 1A). However, enhancement of OxPhos activity by Lin28a turned out to be required for better regeneration in all examined tissues, whereas enhancement of glycolysis was only required in some contexts (Figure 1A). To further understand how this metabolic enhancement influences tissue regeneration, Shyh-Chang et al. treated Lin28a Tg mouse embryonic fibroblasts (MEFs), which migrate significantly faster than WT MEFs, with an OxPhos inhibitor and found that the pro-migration phenotype was preferentially suppressed in Lin28a overexpressing cells (7). This suggested that Lin28a-mediated metabolic enhancements are sufficient to promote cell migration (7). Consistent with the fact that suppression of let-7 alone was not sufficient to recapitulate the regenerative phenotype, suppression of let-7 in MEFs had no effect on cell migration (7). Taken together, these lines of evidence demonstrated that Lin28 regulates metabolism through direct impact on the translation of core metabolic enzymes.
Metabolic reprogramming by the Lin28/let-7 axis in cancer
Recent evidence shows that these oncogenic effects have a metabolic basis. Ma et al. showed that overexpression of either LIN28A or LIN28B in Hep3B, a human liver cancer cell line, promotes the Warburg effect in the form of enhanced glucose uptake, lactate production, and O2 consumption rate (70) (Figure 1B). Treating these cells with let-7 mimics, however, resulted in the opposite effects (70) (Figure 1B). Unexpectedly, they found that LIN28-overexpressing cell lines under normoxic condition showed only marginally activated AKT-mTOR signaling when LIN28A or LIN28B is expressed (70). However, when they examined protein expression of metabolic enzymes, they found that PDH kinase 1 (PDK1) was highly upregulated in multiple LIN28-overexpressing cancer cell lines (70) (Figure 1B). PDK1 is a well-known metabolic regulator whose role is to inhibit the conversion of pyruvate to acetyl-CoA, which is used as a starting material for the Krebs cycle. PDK1 does so by phosphorylating PDH and inhibiting its activity (71) (Figure 1B).
The regulation of PDK1 by LIN28 most likely occurs post-transcriptionally since the PDK1 mRNA level was unchanged (70). Consistent with the fact that let-7 mimics block glucose uptake in the examined cancer cell lines, let-7 was shown to specifically suppress PDK1 expression but no other OxPhos enzymes (70). Luciferase experiments with PDK1 3’ UTR demonstrated that PDK1 is a direct target of let-7 (70) (Figure 1B). More importantly, Ma et al. showed that knocking down PDK1 in cell lines and xenografts impaired the growth-promoting effects of LIN28 overexpression (70). Together, this study made two important findings: first, when expressed in multiple cancer cell lines, Lin28 actively promotes aerobic glycolysis while inhibiting mitochondrial OxPhos, distinct from what we reported in the context of tissue repair. Second, blocking a metabolic effector of the Lin28 program in this context can disrupt cancer cell growth. Recently, we also showed that conditionally overexpressing human LIN28B in the liver resulted in the development of liver cancer, with histological features of both hepatoblastoma and HCC (12). Based on 2-deoxy-2-(18F) fluoro-D-glucose positron emission tomography imaging, human LIN28B-driven liver tumors are more glucose-avid than surrounding normal tissues, a feature that is seen only in a subset of aggressive human HCC (12). In light of the Lin28 and PDK1 connection, it would be interesting to determine if LIN28B preferentially promotes aerobic glycolysis in this endogenous cancer setting, and what role this metabolic mechanism has on tumor initiation and progression. If inhibition of aerobic glycolysis or glycolytic enzymes such as PDK1 can abrogate the oncogenic effects of LIN28B, it would not only support the idea that Lin28 promotes aerobic glycolysis, but also identifies a downstream effector of the Lin28 program that is potentially druggable. As molecules that effectively block Lin28 activity have not yet been developed, identifying a more readily actionable target could be beneficial in treating Lin28-expressing cancers.
Conclusions
Since the initial discovery of Lin28 and let-7 by Ruvkun and Ambros 30 years ago, we are closer to understanding the full spectrum of mammalian functions for this heterochronic pathway. Furthermore, these studies on Lin28 and let-7 have provided insights into the possible phenotypic outputs of post-transcriptional regulation. We have only recently identified their novel roles in metabolic regulation and great strides have been made in understanding how this translates to organismal homeostasis, regeneration, and disease. There are still many open questions. Do Lin28-expressing tumors have a distinctive metabolic signature when compared to those that are Lin28-negative? If Lin28 does promote the Warburg effect, what impact do these effects have on tumor initiation versus progression? If let-7 is to be used as an anti-cancer agent to target Lin28-positive tumors, will it alone be able to reverse the metabolic reprogramming events caused by Lin28? Since Lin28 interacts and enhances translation of thousands of genes, would targeting a subset of these genes be sufficient to abrogate Lin28’s oncogenic effects? We hope that future studies on Lin28 and let-7 can shed light on some of these questions.
Acknowledgements
Funding: LH Nguyen is supported by the International Pre-doctoral Fellowship from the Howard Hughes Medical Institute, H Zhu is supported by the Pollack Foundation, a NIH K08 grant (1K08CA157727), a Burroughs Welcome Career Medical Award, and a Cancer Prevention and Research Institute of Texas New Investigator grant.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011;11:85-95. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. On the origin of cancer cells. Science 1956;123:309-14. [DOI] [PubMed] [Google Scholar]
- 3.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev 2010;20:87-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7:11-20. [DOI] [PubMed] [Google Scholar]
- 5.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013;12:395-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H, Shyh-Chang N, Segrè AV, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011;147:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shyh-Chang N, Zhu H, Yvanka de Soysa T, et al. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013;155:778-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A 2011;108:21075-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molenaar JJ, Domingo-Fernández R, Ebus ME, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet 2012;44:1199-206. [DOI] [PubMed] [Google Scholar]
- 10.Urbach A, Yermalovich A, Zhang J, et al. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev 2014;28:971-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King CE, Cuatrecasas M, Castells A, et al. LIN28B promotes colon cancer progression and metastasis. Cancer Res 2011;71:4260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen LH, Robinton DA, Seligson MT, et al. Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 2014;26:248-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viswanathan SR, Powers JT, Einhorn W, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 2009;41:843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 1984;226:409-16. [DOI] [PubMed] [Google Scholar]
- 15.Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 1980;96:435-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 1997;88:637-46. [DOI] [PubMed] [Google Scholar]
- 17.Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell 2001;1:453-65. [DOI] [PubMed] [Google Scholar]
- 18.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol 2003;258:432-42. [DOI] [PubMed] [Google Scholar]
- 19.Abbott AL, Alvarez-Saavedra E, Miska EA, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 2005;9:403-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901-6. [DOI] [PubMed] [Google Scholar]
- 21.Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns 2003;3:719-26. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917-20. [DOI] [PubMed] [Google Scholar]
- 23.Kim SK, Lee H, Han K, et al. SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs. Cell Stem Cell 2014;15:735-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Wynsberghe PM, Kai ZS, Massirer KB, et al. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol 2011;18:302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science 2008;320:97-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 2008;10:987-93. [DOI] [PubMed] [Google Scholar]
- 27.Heo I, Joo C, Cho J, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 2008;32:276-84. [DOI] [PubMed] [Google Scholar]
- 28.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 2008;14:1539-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehrbach NJ, Armisen J, Lightfoot HL, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol 2009;16:1016-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam Y, Chen C, Gregory RI, et al. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011;147:1080-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 2009;16:1021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piskounova E, Polytarchou C, Thornton JE, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011;147:1066-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Copley MR, Babovic S, Benz C, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol 2013;15:916-25. [DOI] [PubMed] [Google Scholar]
- 34.Chang HM, Triboulet R, Thornton JE, et al. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013;497:244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thornton JE, Chang HM, Piskounova E, et al. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012;18:1875-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010;463:621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worringer KA, Rand TA, Hayashi Y, et al. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 2014;14:40-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002;12:735-9. [DOI] [PubMed] [Google Scholar]
- 39.Inamura K, Togashi Y, Nomura K, et al. let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer 2007;58:392-6. [DOI] [PubMed] [Google Scholar]
- 40.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004;64:3753-6. [DOI] [PubMed] [Google Scholar]
- 41.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007;67:6092-9. [DOI] [PubMed] [Google Scholar]
- 42.Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A 2007;104:11400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [DOI] [PubMed] [Google Scholar]
- 44.Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673-7. [DOI] [PubMed] [Google Scholar]
- 45.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007;67:7713-22. [DOI] [PubMed] [Google Scholar]
- 46.Schultz J, Lorenz P, Gross G, et al. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 2008;18:549-57. [DOI] [PubMed] [Google Scholar]
- 47.Dong Q, Meng P, Wang T, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One 2010;5:e10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007;21:1025-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007;315:1576-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasinski AL, Kelnar K, Stahlhut C, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene 2014. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635-47. [DOI] [PubMed] [Google Scholar]
- 52.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008;7:759-64. [DOI] [PubMed] [Google Scholar]
- 53.Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007;67:9762-70. [DOI] [PubMed] [Google Scholar]
- 54.He G, Dhar D, Nakagawa H, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell 2013;155:384-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilbert ML, Huelga SC, Kapeli K, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell 2012;48:195-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng S, Chen LL, Lei XX, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells 2011;29:496-504. [DOI] [PubMed] [Google Scholar]
- 57.Polesskaya A, Cuvellier S, Naguibneva I, et al. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev 2007;21:1125-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janiszewska M, Suvà ML, Riggi N, et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev 2012;26:1926-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hafner M, Max KE, Bandaru P, et al. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 2013;19:613-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Alexander P, Wu L, et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science 2009;325:435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shyh-Chang N, Locasale JW, Lyssiotis CA, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 2013;339:222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu H, Shah S, Shyh-Chang N, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet 2010;42:626-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lettre G, Jackson AU, Gieger C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 2008;40:584-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Widén E, Ripatti S, Cousminer DL, et al. Distinct variants at LIN28B influence growth in height from birth to adulthood. Am J Hum Genet 2010;86:773-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shinoda G, Shyh-Chang N, Soysa TY, et al. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 2013;31:1563-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trumpp A, Refaeli Y, Oskarsson T, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 2001;414:768-73. [DOI] [PubMed] [Google Scholar]
- 67.Weedon MN, Lettre G, Freathy RM, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet 2007;39:1245-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christiansen J, Kolte AM, Hansen Tv, et al. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol 2009;43:187-95. [DOI] [PubMed] [Google Scholar]
- 69.Dubinsky AN, Dastidar SG, Hsu CL, et al. Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell Metab 2014;20:626-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma X, Li C, Sun L, et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat Commun 2014;5:5212. [DOI] [PubMed] [Google Scholar]
- 71.Kolobova E, Tuganova A, Boulatnikov I, et al. Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem J 2001;358:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]