Abstract
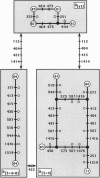
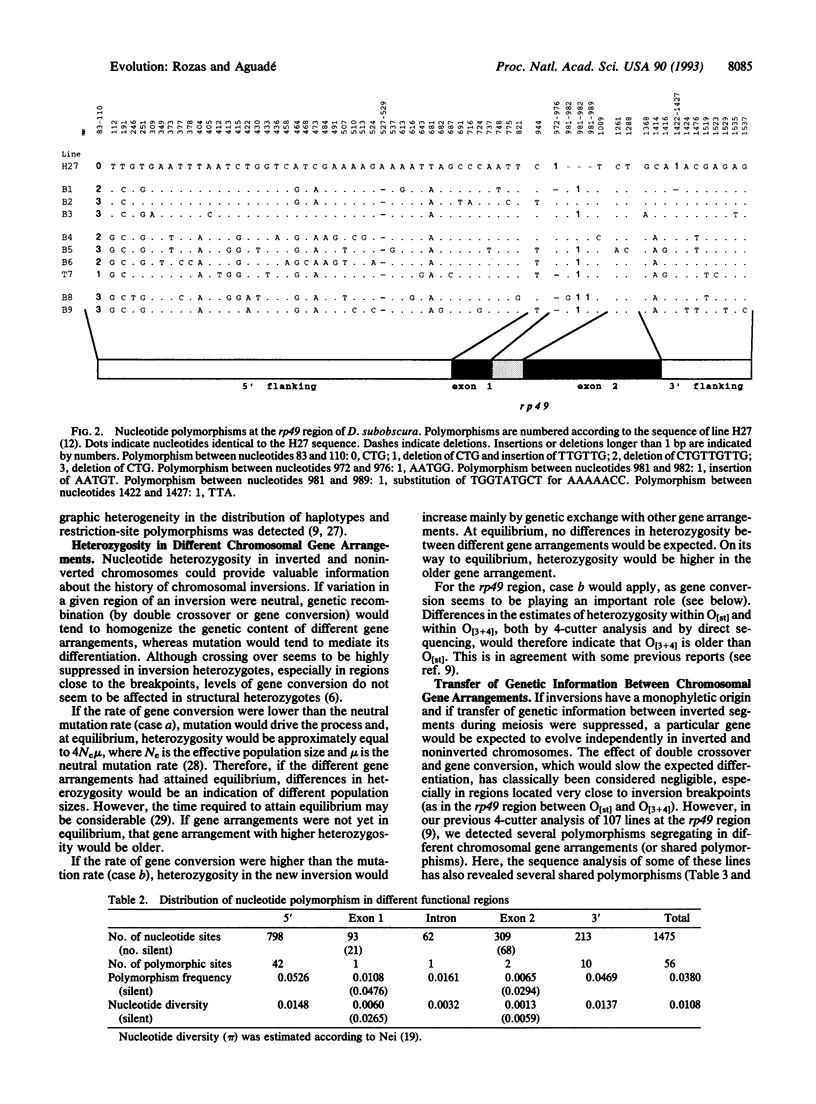
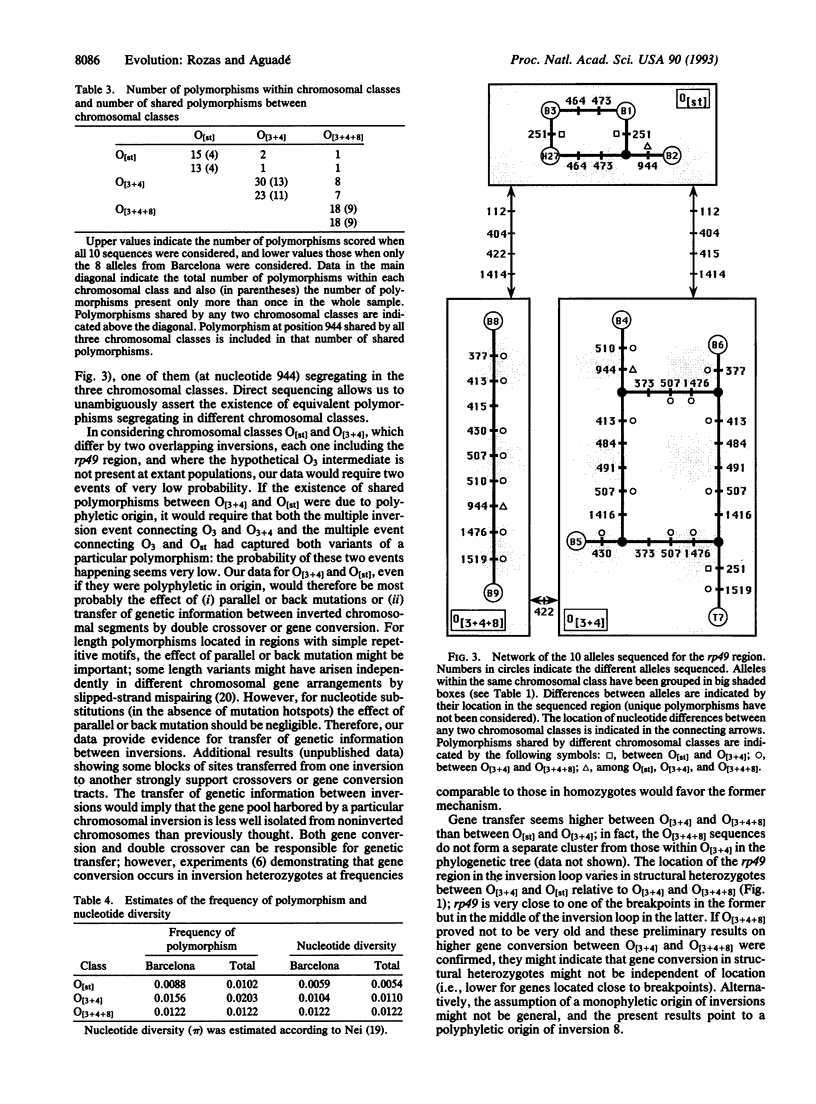
Nucleotide variation in the region including the ribosomal protein 49 (rp49) gene was investigated by direct sequencing of 10 alleles of Drosophila subobscura from chromosomes differing in gene arrangements. Fifty-six nucleotide and seven length polymorphisms were detected over a 1.5-kb region. Of the 20 nucleotide polymorphisms present more than once in the sample, 13 were segregating in O[3 + 4], 9 in O[3 + 4 + 8], and 4 in O[st] chromosomal classes. Several of these polymorphisms were segregating in more than one chromosomal class, a strong indication of genetic transfer between different chromosomal gene arrangements either by double crossover or gene conversion. Given the probable role played by gene conversion in the history of the rp49 region in D. subobscura, estimates of nucleotide diversity within chromosomal class indicate that the O[3 + 4] chromosomal gene arrangement is older than the O[st] arrangement.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguadé M. Nucleotide sequence comparison of the rp49 gene region between Drosophila subobscura and D. melanogaster. Mol Biol Evol. 1988 Jul;5(4):433–441. doi: 10.1093/oxfordjournals.molbev.a040502. [DOI] [PubMed] [Google Scholar]
- Aquadro C. F., Desse S. F., Bland M. M., Langley C. H., Laurie-Ahlberg C. C. Molecular population genetics of the alcohol dehydrogenase gene region of Drosophila melanogaster. Genetics. 1986 Dec;114(4):1165–1190. doi: 10.1093/genetics/114.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro C. F., Weaver A. L., Schaeffer S. W., Anderson W. W. Molecular evolution of inversions in Drosophila pseudoobscura: the amylase gene region. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):305–309. doi: 10.1073/pnas.88.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Chovnick A. Gene conversion and transfer of genetic information within the inverted region of inversion heterozygotes. Genetics. 1973 Sep;75(1):123–131. doi: 10.1093/genetics/75.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W. R., Preston C. R. Formation of chromosome rearrangements by P factors in Drosophila. Genetics. 1984 Aug;107(4):657–678. doi: 10.1093/genetics/107.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Montgomery E., Charlesworth B., Langley C. H. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 1987 Feb;49(1):31–41. doi: 10.1017/s0016672300026707. [DOI] [PubMed] [Google Scholar]
- Muki T., Watanabe T. K., Yamaguchi O. The genetic structure of natural populations of Drosophila melanogaster. XII. Linkage disequilibrium in a large local population. Genetics. 1974 Aug;77(4):771–793. doi: 10.1093/genetics/77.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Non-random association between electromorphs and inversion chromosomes in finite populations. Genet Res. 1980 Feb;35(1):65–83. doi: 10.1017/s001667230001394x. [DOI] [PubMed] [Google Scholar]
- Prakash S., Lewontin R. C. A molecular approach to the study of genic heterozygosity in natural populations. 3. Direct evidence of coadaptation in gene arrangements of Drosophila. Proc Natl Acad Sci U S A. 1968 Feb;59(2):398–405. doi: 10.1073/pnas.59.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]