Abstract
Upon encountering pathogens, T cells mount immune responses by proliferating, increasing cellular mass and differentiating. These cellular changes impose significant energetic challenges on T cells. It was believed that TCR and cytokine-mediated signaling are dominant dictators of T cell-mediated immune responses. Recently, it was recognized that T cells utilize metabolic transporters and metabolic sensors that allow them to rapidly respond to nutrient-limiting inflammatory environments. Metabolic sensors allow T cells to find a balance between energy consumption (anabolic metabolism) and production (catabolic metabolism) in order to mount effective immune responses. Also, metabolic regulators interact with cytokine-dependent transcriptional regulators, suggesting a more integrative and advanced model of T cell activation and differentiation. In this review, we will discuss recent discoveries regarding the roles of metabolic regulators in effector and memory T cell development and their interaction with canonical transcription factors.
Keywords: Treg, Effector and memory T cells, Proliferation, Differentiation, Catabolic metabolism, Anabolic metabolism, Aerobic glycolysis, Fatty acid oxidation, TCA cycle
1. Introduction to cellular metabolism and T cells
Cells constantly sense environmental changes and adapt to stress signals. Nutrients are essential for controlled cellular proliferation, growth and survival in organisms (Yuan et al., 2013). Macromolecular nutrients in the cell can be classified into polysaccharides, lipids, proteins and nucleic acids that are synthesized from fundamental building blocks: carbohydrates, fatty acids, amino acids and nucleotides. Cells can either break down (catabolic processes) or synthesize (anabolic processes) these macromolecules into, and from, their individual components. Catabolism provides cells adenosine triphosphate (ATP) that is critical for cellular proliferation and functions. On the other hand, the anabolic processes important for cellular growth require energy and are therefore dependent on high intracellular ATP levels.
Glucose is one of the building blocks that cells heavily rely on to maintain homeostasis. The simple sugar is utilized during the process called glycolysis, which generates two molecules of ATP leading to the final product of pyruvate. Under oxygen-rich conditions or in resting cells, pyruvate can be further processed by the tricarboxylic acid cycle (TCA cycle; also known as the Krebs cycle and the citric acid cycle) and oxidative phosphorylation in the mitochondria in order to generate maximal levels of ATP (Fig. 1). Catabolism of other molecules also provides substrates for the TCA cycle. Fatty acids are converted to acetyl-CoA (Ac-CoA) through a process called fatty acid oxidation (FAO) in the mitochondria. At the same time, amino acids are metabolized into 3-, 4-, and 5-carbon substrates that feed into the TCA cycle (Owen et al., 2002).
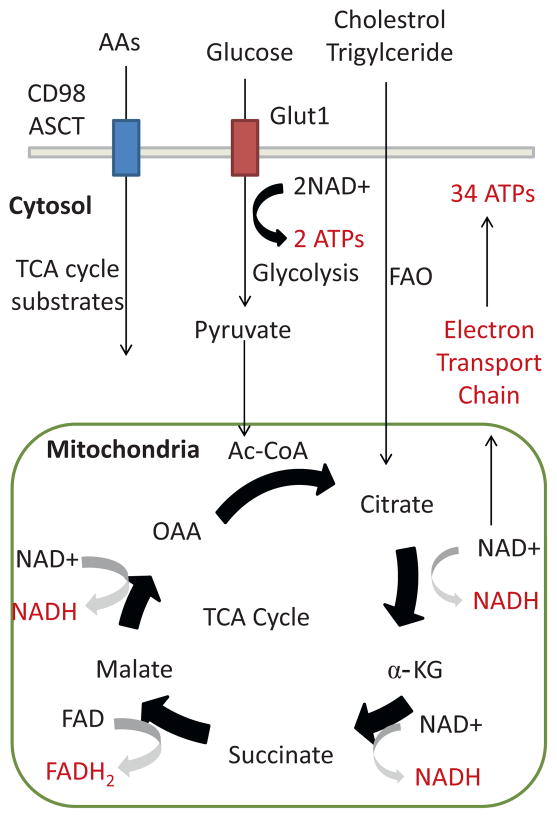
Fig. 1.
Catabolic metabolism in memory/regulatory T cells. Memory and regulatory T cells prefer catabolic metabolism that maximizes ATP production of the cells. Glucose will be converted to pyruvate through glycolysis resulting in 2 ATPs, and pyruvate is further converted to Ac-CoA that is used as a substrate to generate citrate in the TCA cycle. The TCA cycle is a series of circular enzymatic reaction that converts citrate to OAA generating NADH and FADH2. Substrates for the TCA cycle are provided by both fatty acid oxidation and glutaminolysis. Ac-CoA is generated by fatty acid oxidation of cholestrol and triglycerides. Glutaminolysis generates various 3-, 4-, 5-carbon substrates that replenish the levels of α-KG, malate and OAA. NADH and FADH2 pass through the electron transport chain and generate total of 34 ATPs. Abbreviations: amino acids (AAs), fatty acid oxidation (FAO), acetyl coenzyme A (Ac-CoA), oxaloacetate (OAA), α-ketogluterate (α-KG), flavin adenine dinucleo-tide (FAD), nicotinamide adenine dinucleotide (NAD), adenosine triphosphate (ATP), tricarboxylic acid cycle (TCA cycle).
On the other hand, rapidly proliferating cells utilize glycolysis in spite of oxygen availability (Warburg, 1956a). In cancer cells, the majority of ATP is derived from glycolysis without full-oxidation of pyruvate to CO2 and H2O through mitochondrial respiratory reaction through a process known as aerobic glycolysis (Warburg, 1956b). Rather, pyruvate is converted to lactate by lactate dehydrogenase (LDH) in order for cells to regenerate nicotinamide adenine dinucleotide (NAD) and continue aerobic glycolysis (Fig. 2). This is also called the Warburg effects and has provided significant insight into cellular metabolism and function. Meanwhile, proliferating cells need to undergo cellular growth that requires de novo synthesis of lipids and DNA. Lipids are synthesized from citrates by ATP citrate lyase and this process is critical for the accumulation of the plasma membrane (Hatzivassiliou et al., 2005). Glutamine, an amino acid and the most abundant nutrient in the blood, is also key for cell growth processes (Karinch et al., 2001; Newsholme, 2001). Intracellular glutamine can be converted to α-ketoglutarate (α-KG) during glutaminolysis in order to maintain homeostasis of the TCA cycle (DeBerardinis et al., 2008). Its carbon backbone can be also converted to lactate during the glutaminolysis process that generates NAD and NAD phosphate (NADPH). At the same time, glutamine can be utilized to replenish pyruvates in the face of robust aerobic glycolysis rates like those seen in activated T cells (Blagih et al., 2015).
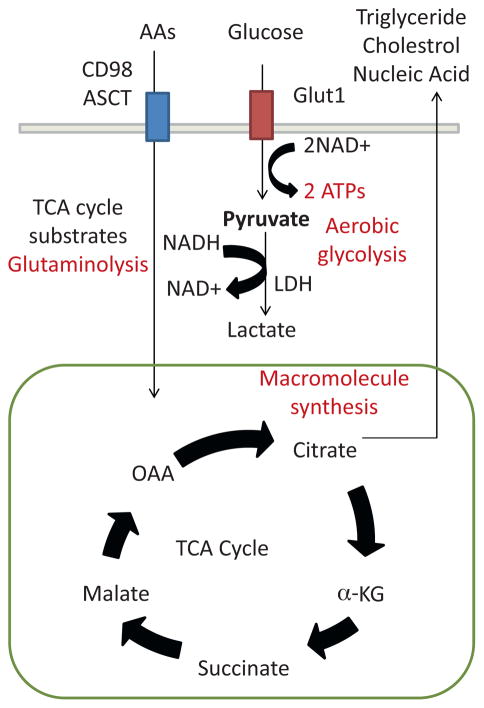
Fig. 2.
Anabolic metabolism in effector T cells. Effector T cells maintain anabolic metabolism in order to prepare for proliferation and growth. Thus, effector T cells maintain their energy by aerobic glycolysis, and pyruvate is constantly converted to lactate by LDH in order to regenerate NAD and sustain aerobic glycolysis. At the same time, glucose must be replenished as a substrate for glycolysis and Glut1 expression, a glucose transporter, increases during T cell activation. Instead of utilizing the TCA cycle as an energy source, effector T cells utilized products of the TCA cycle as substrates for the synthesis of membrane lipids and nucleic acids. In order to maintain the levels of TCA cycle products (citrate, α-KG, malate and OAA), activated T cell utilize the glutaminolysis process. Increased demand for amino acid consumption is met by enhanced amino acid transporter expression such as CD98 and ASCT. Abbreviations: lactate dehydrogenase (LDH).
T-lymphocytes (T cells) have been an ideal system to study the disparate metabolic requirements that arise during an immune response because of their distinct developmental stages: (1) naïve or resting (2) effector or activated (3) memory T cells (Kaech et al., 2002). Naïve T cells are activated in response to antigens interacting with the T-cell receptor (TCR) and major-histocompatibility complex (MHC). During the effector phase of T cell development, T cells proliferate, grow and differentiate in response to antigens. Activated CD4+ helper T cells (Th) can be further divided into four different subsets that produce characteristic cytokines and posses highly specialized functions. These include the type-1 (Th1), type-2 (Th2) and type-17 (Th17) and regulatory T cells (Tregs) (Zhu et al., 2010). Activated CD8+ T cells differentiate into cytolytic T cells (CTLs) that secret granzyme B, perforin, interferon-γ (IFN-γ), tumor necrosis factor (TNF-α), which are critical for the clearance of pathogens (Pearce et al., 2003). After expansion during the primary immune responses, T cells undergo a contraction phase mediated by pathways of programmed death and only a fraction of the expanded T cell population survives to become memory T cells (Kaech and Cui, 2012).
Recently, it became apparent that the three developmental stages of T cells have differential metabolic requirements. Much of our current knowledge in cellular metabolism originated from studies utilizing tumor cells, and interestingly, some T cell subsets have metabolic regulation analogous to tumor cells. Proliferating and activated CD8+ and CD4+ T cells utilize aerobic glycolysis as their energy source (Rathmell et al., 2000). At the same time, activated T cells decrease the catabolic process and rather increase fatty acid, nucleic acid and amino acid synthesis in order to meet the demands of cellular division. Interestingly, naïve, Tregs and memory T cells show higher fatty acid oxidation rates suggesting distinct metabolic requirements from activated T cells (Michalek et al., 2011; MacIver et al., 2013; O’Sullivan et al., 2014).
2. Nutrient transporters and metabolic regulators
Many macromolecule-transporters and sensors allow T cells to rapidly adapt to extracellular environments (Table 1). Metabolic transporters (amino acid and glucose transporters) serve as a bridge between the extracellular and intracellular environment, and provide substrates for the TCA cycle. Glut1 (gene name: Slc2a1) is a member of the Glut family that transports glucose and its expression is induced upon T cell activation (Macintyre et al., 2014; Jacobs et al., 2008). Also, T cell activation induces amino acid transporter expression on the cellular membrane including a single System L transporter (also known as CD98; a heterodimer of Slc7a5/Slc3a2) that preferentially transports large and branched amino acids and ASCT2 (also known as Slc1a5) that transports glutamine (Nakaya et al., 2014; Sinclair et al., 2013).
Table 1.
Metabolic regulators of T cell development and quiescence.
| Effector T cells (Th1, Th2, Th17 and CTL) | Naïve T cells | Tregs | Memory CD8+ T cells | |
|---|---|---|---|---|
| Preferred metabolic mode | Anabolic metabolism | Catabolic metabolism | ||
| Primary source of energy | Aerobic glycolysis glutaminolysis | Fatty acid oxidation | ||
| Metabolic transporter | CD98 (amino acid transporter) ASCT (amino acid transporter) Glut1 (glucose transporter) |
Low nutrient uptake | ||
| Metabolic kinases | AMPK↓ mTORC1/2↑ |
N/A | AMPK↑ | mTORC1/2↓ |
| Metabolic transcription factors | Myc HIF-1α IRF4 AP4 SREBP |
TSC KLF FoxO Foxp Tob |
HIF-1α | ACC1 |
Distinct stages of T cell development correlate with different metabolic states. Naïve, Tregs and memory T cells can be grouped for their reliance on catabolic metabolism and fatty acid oxidation. These cells also have low rates of nutrient uptake, and naïve T cells express high levels of Tob, KLF and Fox that maintain quiescence and survival. Tregs and memory T cells have high AMPK and low mTOR activity, and HIF-1α can post-transcriptionally regulate Treg development. Effector CD4+ and CD8+ T cells utilize aerobic glycolysis and glutaminolysis for their energy supply, and prefer anabolic metabolism in order to synthesize macromolecules. They express high levels of glucose and amino acid transporters, and high rates of nutrient uptake lead to high mTOR and low AMPK activity. At the same time, effector T cells express high levels of Myc and HIF-1α that initiate metabolic and transcriptional changes.
Abbreviations: helper CD4+ T cell type-1 (Th1), type-2 (Th2), type-17 (Th17), regulatory T cells (Tregs), cytolytic T cells (CTLs).
In addition, many intracellular regulators serve as sensors for the energy and metabolic status of T cells (Table 1). Intra-cellular regulators modulate the expression of aforementioned nutrient transporters and many enzymes that are essential for catabolic and anabolic metabolism. Expression of intracellular regulators is induced upon T cell activation by both TCR signaling and CD28-mediated costimulation, and can be classified into metabolic kinases and transcriptional regulators (Fig. 3). Kinases include phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt), AMP-activated protein kinase (AMPK), and mammalian target of rapamycin (mTOR). Together with TCR and co-stimulation, these metabolic kinases regulate the expression and activity of transcriptional regulators including myelocytomatosis oncogene (Myc), and hypoxia-inducible factor-1α (HIF-1α). At the same time, the expression of transcriptional regulators that maintain T cell quiescence decreases upon T cell activation (Fig. 3). In this review, we will describe how these different transporters and intracellular regulators interact and regulate quiescence, activation and immunological memory in T lymphocytes.
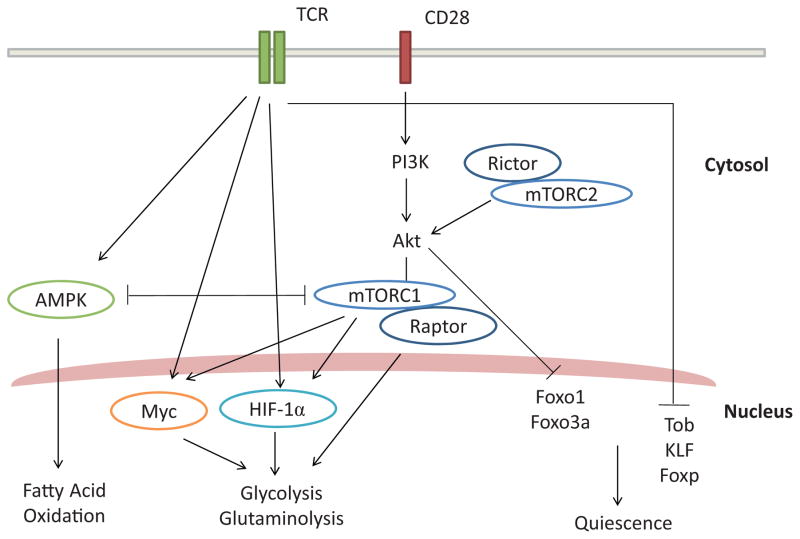
Fig. 3.
TCR signaling and metabolic regulators. TCR and CD28-mediated co-stimulation in naïve T cells inhibit the expression of Tob and LKLF that inhibit proliferation and growth (quiescence). At the same time, TCR/CD28 signaling induces the expression and activity of metabolic kinases and transcriptional regulators. TCR induces c-Myc and HIF-1α that are critical for glycolysis, glutaminolysis and activation of T cells. Also, CD28-dependent activation of PI3K and Akt leads to mTORC1 activation and subsequent modulation of c-Myc and HIF-1α activity. At the same time, mTORC1 itself can promote cellular growth and protein synthesis. Little is known about the upstream activators of mTORC2, but mTORC2-mediated activation of Akt can prevent nuclear localization of FoxO1/3a that promotes T cell quiescence. AMPK acts as a negative regulator of mTOR and glycolysis/glutaminolysis. At the same time, AMPK promotes fatty acid oxidation and memory T cell development. Abbreviations: T-cell receptor (TCR), phosphatidylinositol 3-kinase (PI3K), protein kinase B (Akt), AMP-activated protein kinase (AMPK), mammalian target of rapamycin (mTOR), regulatory associated protein of mTOR (Raptor), rapamycin-insensitive companion of mTOR (Rictor), myelocytomatosis oncogene (MYC), hypoxia-inducible factor (HIF), forkhead box O (FoxO), Krupple-like factor (KLF), transducer of ErbB-2 (Tob).
3. Naïve T cells – quiescence and homeostasis
Naïve T cells are those that have yet to encounter their cognate antigens, and their quiescence is maintained by many environmental signals (Hamilton and Jameson, 2012). Quiescence is defined in terms of cellular growth, proliferation and death. Naïve T cells are relatively small in size, and they are arrested at the G0 stage of the cell cycle. They maintain their survival through TCR-and interleukin-7 (IL-7)-triggered signaling (Marrack and Kappler, 2004). It appears that quiescence is an active process regulated by transcriptional regulators in T cells because TCR signaling inhibits expression of many genes (Yusuf and Fruman, 2003).
Members of the forkhead box (FOX) family of transcription factors are found to be critical to prevent T cell activation and differentiation (Ouyang et al., 2009; Freitas and Rocha, 2009). In naïve T cells, FoxO transcription factors are active and localized in the nucleus, but their function can be inhibited by activation-induced metabolic regulators. Upon activation, PI3K-dependent Akt signaling inhibits and retains FoxO in the cytoplasm, negating their activity as transcriptional regulators. Among different members of FoxO transcription factors, FoxO1 is critical to maintain homeostasis of naïve T cells. Foxo1 knock-out (KO) mice display enhanced T cell activation (as evidenced by high CD44 and low CD62L expression). Impaired homeostatic control in Foxo1 KO mice was due to decreased IL-7 receptor (IL-7R) expression on both CD4+ and CD8+ T cells (Ouyang et al., 2009; Kerdiles et al., 2009). Similarly, T cells deficient in FoxO3a, another member of the FoxO transcript factor family, are prone to spontaneous activation due to increased activation of nuclear factor of κB (NF-κB), and Foxo3a KO mice have autoimmune phenotypes (Lin et al., 2004).
Foxp1 deficiency in mice also leads to activated phenotype of thymocytes and decreased accumulation of peripheral CD4+ and CD8+ T cells (Feng et al., 2010). In order to exclude developmental effects of Foxp1 on T cell quiescence, another study utilized inducible Foxp1 deletion in mature T cells and found that Foxp1 KO T cells are hyperproliferative in response to IL-7 in vitro (Feng et al., 2011). Interestingly, Foxp1 deficiency in T cells leads to enhanced IL-7R expression in contrast to Foxo1 deficiency. Further analysis in the same study suggests that Foxp1 antagonizes Foxo1 binding to IL-7R regulatory regions.
The Krupple-like factor (KLF) transcription factor family includes 15 mammalian members that contain zinc-finger domains. Particularly, KLF2 (also known as LKLF) is expressed in the lungs and the spleen (Anderson et al., 1995). In T cells, KLF2 expression is upregulated after positive selection in the thymus and its expression is maintained in naïve T cells, but downregulated after T cell activation (Kuo et al., 1997; Schober et al., 1999). Correlative to their expression patterns, KLF2 is known to maintain quiescence and activation of naïve T cells. Exogenous expression of KLF2 is sufficient to arrest T cell proliferation and growth by inhibiting c-myc, a gene that promotes glycolysis and activation in T cells (Buckley et al., 2001). Nevertheless, KLF2 may not be required for maintenance of quiescence per se as KLF2 deficiency in T cells does not lead to spontaneous activation (Carlson et al., 2006). Although its exact mechanism is still not clear, phenotypic analysis of KLF2 deficiency in T cells is consistent across multiple studies. KLF2 KO mice have higher accumulation of naive CD4+ and CD8+ T cells in the thymus, but their peripheral migration is significantly impaired (Carlson et al., 2006; Sebzda et al., 2008). KLF2 regulates various surface receptors in naïve T cells, particularly chemokine receptors that affect T cell migration (Haaland et al., 2005). Naive T cells from KLF2 deficient mice express lower levels of CD62L, CCR7 and β7-integrin that are critical for migration to secondary lymphoid organs (Carlson et al., 2006). At the same time, these naïve T cells also express higher levels of inflammatory chemokine receptors including CCR3 and CCR5 that divert naïve T cells to non-lymphoid peripheral organs (Sebzda et al., 2008).
Tuberous sclerosis complex (TSC) also regulates quiescence in naïve T cells but its mechanism is quite distinct from KLF and Fox proteins. Tsc1 deficient mice have normal accumulation of CD4+ and CD8+ thymocytes, but their accumulation in the peripheral lymphoid organs is significantly lower than WT littermates (Yang et al., 2011). This defect was attributed to increased cell-intrinsic apoptotic pathways dependent on Caspase activity and Bcl-2 expression. This survival defect in Tsc1 KO T cells is also due to higher levels of reactive oxygen species (ROS) that are toxic to T cells in spite of lower mitochondria contents in Tsc1 KO T cells in comparison to WT T cells (O’Brien et al., 2011). Interestingly, enhanced cellular apoptosis resulting from Tsc1 deficiency was insensitive to rapamycin treatment suggesting that it is independent of mTOR activity (O’Brien et al., 2011; Wu et al., 2011).
Transducer of ErbB-2 (Tob) is a member of the Tob and BTG anti-proliferative protein family that has been isolated from various cell lines (Rouault et al., 1992; Matsuda et al., 1996). Members of the Tob family transcription factors include Tob1 and Tob2 that share amino acid sequence homology (Ikematsu et al., 1999). Only Tob1 is highly expressed on resting human T cells in circulation, and it is rapidly degraded by activation (Tzachanis et al., 2001). The same study also found that introduction of ectopic Tob expression in CD4+ T cells decreases their proliferation and their effector T cell function upon activation. Collectively, these data support the idea that the down-regulation of quiescence mediators is critical for optimal T cell activation. Nevertheless, genetic deletion of a single mediator is often not sufficient to trigger autoimmune phenotypes and these mediators may play redundant roles in order to prevent uncontrolled immune responses.
4. Effector T cells – aerobic glycolysis and fatty acid synthesis
4.1. Nutrient transporters
Activated T cells undergo proliferation, growth and differentiation into effector T cells. This cellular change is accompanied by altered global gene expression profiles (Best et al., 2013). TCR and CD28-dependent co-stimulation suppress aforementioned “quiescent” genes, and at the same time, induce “activation” genes to meet the increased metabolic demands of activated T cells (Teague et al., 1999). In order to maintain aerobic glycolysis, T cells must replenish their intracellular glucose by up-regulating glucose transporter expression. The GLUT transporter family includes 14 different members (GLUT1-14) with various substrate specificities (Mueckler and Thorens, 2013). Resting human peripheral blood T cells express GLUT2 and GLUT3, while mitogenic stimulation predominantly induces GLUT1 expression (Chakrabarti et al., 1994). Subsequently, it was found that CD28-dependentt PI3K/Akt signaling induces GLUT1 expression and increases glucose influx into the cytoplasm of T cells (Frauwirth et al., 2002). Further supporting the importance of glucose influx during activation, murine T cells deficient in GLUT1 failed to proliferate, grow and survive during activation (Macintyre et al., 2014).
At the same time, amino acids are important components of T cell activation (Carr et al., 2010), and TCR signaling induces amino acid transporter expression and localization on to the membrane (Nakaya et al., 2014; Sinclair et al., 2013). Slc7a5 is a subunit of heterodimeric System L amino-acid transporters that regulate transport of large and branched neutral amino acids such as leucine and phenylalanine. Slc7a5 is highly upregulated on antigen-specific murine CD8+ T cells during in vitro activation and in vivo Listeria monocytogenes (L. monocytogenes) infection (Sinclair et al., 2013). Slc7a5 deficient murine CD4+ and CD8+ T cells failed to proliferate, grow and differentiate into effector T cells. In contrast to Slc7a5, ASCT2 (Slc1a5) that preferentially transports glutamine is dispensable for T cell proliferation, but is critical for T cell differentiation and effector function (Nakaya et al., 2014). Slc1a5 KO CD4+ T cells fail to differentiate into Th1 and Th17 cells in vitro and Slc1a5 KO mice develop less severe paralysis in the experimental autoimmune encephalomyelitis (EAE) model These data suggest that different amino acid transporters have distinct regulatory effects on T cell activation.
4.2. Metabolic sensors
TCR signaling and CD28-dependent co-stimulation also lead to the activation of the mTOR pathway through PI3K/Akt (Chi, 2012). mTOR is a serine/threonine kinase that promotes aerobic glycolysis and anabolic metabolism for T cell activation. mTOR senses glucose and amino acid availability in the cells (Nicklin et al., 2009), increases the surface expression of the appropriate transporters (Edinger and Thompson, 2002), and enhances protein translation by phosphorylating the eukaryotic initiation factor 4E (eIF4E)-binding protein-1 (4E-BP1) and the p70 ribosomal S6 kinase 1 (S6K1) (Ma and Blenis, 2009). mTOR can form two complexes, mTOR-complex1 (mTORC1) and mTOR-complex2 (mTORC2) (Laplante and Sabatini, 2009). mTORC1 contains regulatory associated protein of mTOR (Raptor) and its activity is inhibited by rapamycin. mTORC2 contains rapamycin-insensitive companion of mTOR (Rictor), and, not surprisingly, its activity is insensitive to rapamycin except when exposure to drug is prolonged (Powell and Delgoffe, 2010). mTORC1 activity is regulated by a heterodimer of TSC1 and TSC2, a GTPase-activating protein, and PI3K/Akt mediates its phosphorylation releases RAS homolog enriched in brain (Rheb) that activates mTORC1 activity (Zoncu et al., 2011). In contrast, relatively little is known about the upstream mediator of mTORC2 activity.
In order to study the role of the mTOR pathway on T cell development, various genetic mouse models have been utilized. Rheb KO CD4+ T cells have decreased mTORC1 activity and fail to differentiate into Th1/Th17 cells in vitro and in vivo (Delgoffe et al., 2011; Kurebayashi et al., 2012). On the other hand, Rictor KO CD4+ T cells that are deficient of mTORC2 activity fail to differentiate into Th2 cells in vitro and in vivo (Delgoffe et al., 2011). Interestingly, T cells deficient in mTOR (impaired mTORC1 and mTORC2 activity) failed to differentiate into all effector CD4+ T cells. Nevertheless, the selective effects of mTORC1 or mTORC2 on effector CD4+ T cell development are not clear. Others have shown that Rptor KO CD4+ T cells have defective Th2 development in association with decreased glycolysis and lipid synthesis (Yang et al., 2013). Such discrepancy was due to differential requirements of Raptor and Rheb for mTORC1 activation. Raptor deficiency inhibits mTORC1 activity for longer duration than Rheb deficiency in CD4+ T cells. Also, another study found that Rictor regulates the development of Th1 cells through Akt in addition to Th2 through PKCθ, as Rictor KO CD4+ T cells fail to develop Th1 and Th2 cells (Lee et al., 2010). Alternatively, serum- and glucocorticoid-regulated kinase 1 (SGK1), another downstream molecule of mTORC2, regulates Th1 and Th2 development as well (Heikamp et al., 2014). Sgk1 KO CD4+ T cells preferentially develop into Th1 cells even under Th2 polarizing conditions representing a mechanism distinct from Akt or PKCθ.
mTOR activation also promotes effector CD8+ T cell development (Araki et al., 2009; Pollizzi et al., 2015; Rao et al., 2010). Utilizing Tsc2 KO mice that show enhanced mTORC1 activity, another study found that mTORC1 promotes effector CD8+ T cell development in association with high glycolysis during vaccinia infection (Pollizzi et al., 2015). The data are not necessarily consistent with impaired anti-bacterial effector T cell responses in Tsc1 KO mice (Yang et al., 2011). Although Tsc1 and Tsc2 are known to form heterodimers and deficiency of either factor leads to enhanced mTOR activity, Tsc1 and Tsc2 may have differential downstream effects. Alternatively, anti-bacterial and anti-viral immune responses may have differential requirements of Tsc1 and Tsc2. At a molecular level, mTOR promotes T-box transcription factor (T-bet) and Blimp1 that drive transcriptional profiles of effector CD8+ T cells (Rao et al., 2010). Collectively, these observations support the role of mTOR as a glycolytic switch critical for the development of both effector CD4+ and CD8+ T cells.
Another serine/threonine kinase that is induced by T cell activation is adenosine monophosphate (AMP) activated protein kinase (AMPK) (Andris and Leo, 2015). AMPK consists of three subunits: a catalytic α-subunit, a regulatory β-subunit and an AMP-binding γ-subunit (Mihaylova and Shaw, 2011). AMPK expression is induced by TCR signaling and intracellular Ca2+ influx in T cells (Tamas et al., 2006). AMPK is further activated by the high intracellular ratio of AMP and adenosine triphosphate (ATP), and in general, its role can be classified as a promoter of catabolic metabolism in order to restore ATP production in cells. Thus, AMPK can be considered as an antagonist of mTOR activity that promotes aerobic glycolysis and anabolic metabolism. AMPK induces the expression of carnitine palmitoyl transferase I (CPT I), a rate-limiting enzyme in FAO, and thus increases FAO (O’Neill et al., 2014; Faubert et al., 2013). In contrast, PI3K-dependent mTOR signaling inhibits CPT I expression and FAO, while enhancing aerobic glycolysis (Deberardinis et al., 2006). Furthermore, AMPK and mTOR can directly antagonize each other’s activity (Blagih et al., 2015; MacIver et al., 2011).
Reciprocal inhibition between AMPK and mTOR suggests that AMPK deficiency during T cell activation should enhance effector T cell development. Some studies have found that AMPKα1-deficient mice develop more severe paralysis compared to WT littermates in EAE with elevated Th1 responses in vivo (Nath et al., 2009a). Similarly, AMPKα1-deficient CD8+ T cells have enhanced secretion of IFN-γ in vitro without significant effects on proliferation (MacIver et al., 2011). Nevertheless, other studies are not consistent with this hypothesis. AMPK1α deficiency in CD8+ T cells did not affect proliferation and primary CTL responses against L. monocytogenes infection (Rolf et al., 2013). Rather, the most recent study found that AMPK1α-deficiency inhibits cellular proliferation and effector cytokine secretion of murine CD4+ and CD8+ T cells only under glucose-limiting conditions in vitro (Blagih et al., 2015). In the same study, in vivo analysis showed intact development of effector CD4+ and CD8+ T cells, but decreased total number of Th1/Th17 and INF-γ+ CD8+ T cells in response to influenza virus and L. monocytogenes infection. It is plausible that physiological glucose levels can affect differential phenotypes by AMPKα1-deficient mice. Alternatively, the effect of AMPKα1 is solely due to cellular survival given the in vivo change of absolute numbers of effector T cells (Blagih et al., 2015).
4.3. Metabolic transcription factors
The role of the MYC family transcription factor as a glucose metabolic regulator has been implicated in rapidly proliferating tumor cells (Dang and Semenza, 1999). The MYC family consists of L-MYC, N-MYC and c-MYC, and they form heterodimers with a helix–loop–helix leucine zipper domain containing protein, Max. Myc-Max heterodimers bind to DNA regulatory elements and regulate gene expression involved in cellular growth and proliferation including p27, a cyclin-dependent kinase (CDK) inhibitor (Dang et al., 1999). Myc is an immediate early gene induced by mitogenic stimulation in various cells including T cells, and c-Myc is predominantly expressed in T cells (Douglas et al., 2001; Kelly et al., 1983). C-myc KO mice are embryonic lethal, supporting this molecule’s critical role in cellular development, and conditional deletion of c-myc in T cells greatly enhanced our understanding of its role in T cells. C-myc KO CD4+ and CD8+ T cells manifest impaired cellular proliferation and growth both in vitro and in vivo (Wang et al., 2011). In addition to proliferation, the metabolic analysis of c-myc KO CD4+ and CD8+ T cells further revealed significantly impaired glycolysis and glucose influx (i.e. Glut1 expression) with relatively intact FAO. Also, MYC regulates amino acid influx (i.e. Slc7a5 and Slc1a5 expression) and glutaminolysis suggesting its critical roles in T cell activation-dependent metabolic regulation.
Although Myc expression is required for initial metabolic switch to aerobic glycolysis during T cell activation, Myc expression is transient and decreases after activation. Additional transcription factors have been found to sustain aerobic glycolysis after Myc expression decreases, but these are not required for initial metabolic switch. AP4 is induced by Myc and sustains effector T cell development (Chou et al., 2014). AP4 expression is maintained highly after Myc expression diminishes and AP4 shares many target genes with Myc that regulate aerobic glycolysis in CD8+ T cells. Accordingly, AP4 deficiency in T cells leads to failed clearance of acute viral infection. Interferon regulatory factor-4 (IRF4) is another transcription factor induced by high-affinity TCR signaling and is required for T cell effector development (Man et al., 2013). Similar to AP4, IRF4 expression is not required during the early phase of T cell activation and proliferation, and rather IRF4 deficiency in T cells failed to sustain aerobic glycolysis in later phase. In addition to metabolic regulation, IRF4 also inhibits cell cycle inhibitors and Bim expression further promoting effector T cell proliferation and survival (Yao et al., 2013). IRF4 KO T cells have comparable expression of Myc to WT T cells. This suggests that IRF4 may regulate effector T cell development independently of Myc, but it is also plausible that IRF4 is a downstream target of Myc. Given the similar phenotype of AP4- and IRF4-deficient T cells, further analysis of IRF4 expression kinetics will be of interest in relation to AP4 during T cell activation.
Hypoxia-inducible factor-1 (HIF1) is another transcription factor that regulates glycolytic enzyme expression (Kaelin, 2005). HIF1 is a heterodimeric transcription factor composed of α- and β-subunits (HIF1α and HIF1β), and HIF-1α expression is induced upon TCR activation together with MYC. Under normoxic conditions, HIF-1 is rapidly degraded following hydroxylation and subsequent von Hippel-Lindau (VHL) tumor suppressor-mediated ubiquitin ligase action. In contrast, HIF1 is stabilized under hypoxic conditions (Semenza, 2000). Similar to IRF4 and AP4, HIF-1 appears to be dispensable for T cell proliferation as Hif-1α KO T cells show comparable proliferation to wild-type (WT) T cells (Wang et al., 2011; Finlay et al., 2012). Rather, HIF-1 regulates differentiation and effector function of CD4+ and CD8+ T cells (Doedens et al., 2013; Lukashev et al., 2006). In CD4+ T cells, HIF-1α reciprocally regulates Th17 and Treg development (Pan et al., 2012). HIF1α expression is highly sustained in the Th17 subset, even in the presence of oxygen, through IL-6-mediated signal transducer and activator of transcription 3 (STAT3) (Dang et al., 2011). Hif-1α KO CD4+ T cells are defective in Th17 differentiation but are more prone to differentiate into Treg cells in vitro (Dang et al., 2011; Shi et al., 2011). Downstream, HIF-1αdirectly regulates retinoic acid related orphan receptor (RORγt) expression, a transcription factor critical for Th17 development. At the same time, it inhibits forkhead box P3 (Foxp3) expression by promoting post-translational modification (ubiquitination) of the Treg master regulator (Dang et al., 2011). Accordingly, Hif-1α KO mice have ameliorated paralysis during EAE in vivo with lower central nervous system infiltration of Th17 cells and a more prominent population of Foxp3+ T cells. Besides developmental effects, another study found that HIF-1α may actually contribute to the suppressive capacity of Tregs in vivo as HIF-1α-deficient Tregs were less effective than their WT counterparts at preventing T-cell transfer mediated colitis in mice (Clambey et al., 2012).
HIF-1αalso regulates effector CD8+ T cell development although its effects are not completely clear. One study found that stabilized HIF1 activity enhances the maintenance of CTL responses (Doedens et al., 2013). Accordingly, Vhl KO mice that have enhanced HIF-1α expression can better control murine B16-melanoma growth and lymphocytic choriomeningitis virus (LCMV) infection. Consistent with this finding, Vhl KO CD8+ T cells have enhanced expression of granzyme B and effector cytokines including IFN-γ and TNF-α. In contrast, another study utilizing HIF-1α deficient mice found that Hif-1α KO CD8+ T cells show increased expression of proinflammatory cytokines, and Hif-1α KO mice are more resistant to bacterial sepsis than WT littermates (Thiel et al., 2007). Our group also observed that HIF-1α KO mice better control B16 tumor growth and mount more robust anti-tumor immune responses (unpublished data). Given that VHL is known to target various genes in addition to HIF-1, VHL deficiency may represent more than hypoxic conditions or enhanced HIF-1 activity alone and this may explain the discrepancies in the conclusions of these studies (Maina et al., 2005).
In addition to aerobic glycolysis, de novo lipid synthesis is critical for the development of effector T cells (Chen et al., 1975; Bensinger et al., 2008). Sterol regulatory element-binding proteins (SREBPs) regulate fatty acid and cholesterol synthesis in cells (Horton et al., 2002). Genetic deletion of SCAP, a molecule that is associated with SREBP and regulates its activity in CD8+ T cells, leads to significantly impaired growth and proliferation of T cells upon activation (Kidani et al., 2013). As a result, Scap KO mice could not develop effector CD8+ T cell responses during acute LCMV infection. One of the many enzymes that are under the control of SREBP includes acetyl-CoA carboxylase 1 (ACC1), a key enzyme in fatty acid synthesis. ACC1 deficiency in CD8+ T cells did not alter proliferation nor effector function of CD8+ and CD4+ T cells (Lee et al., 2014). Instead, the frequency of effector CD8+ T cells in Acc1 KO mice was significantly lower than that in WT mice, suggesting Acc1 maintains effector T cell survival. Thus, ACC1 appears to act as a metabolic checkpoint rather than direct regulator of proliferation and growth.
Each metabolic regulator appears to have distinct effects on T cell activation. HIF-1α and mTOR have more specific effects, while c-Myc and AMPK have more global effects on proliferation, survival and development. These differential effects attest to interaction among metabolic sensors and transcription factors. For example, SREBP, c-Myc and HIF-1α are under direct control of mTOR suggesting that mTOR regulates effector T cell development through these transcription factors (Wang et al., 2011; Finlay et al., 2012; Duvel et al., 2010). Also, HIF-1α expression is under direct control by IRF4 and in turn, IRF4 activity depends on mTOR (Man et al., 2013; Yao et al., 2013). Thus, direct hierarchical relationships among metabolic sensors and transcription factors remain to be established.
5. Tregs and memory T cells – fatty acid oxidation
Although Tregs and memory T cells are distinct subsets, they share common metabolic requirements: FAO (Michalek et al., 2011; Pearce et al., 2009). Tregs belong to the subset of effector CD4+ T cells that are critical to maintain homeostasis and tolerance in host organisms (Rudensky, 2011). Treg development also requires TCR and co-stimulation as other effector CD4+ T cells, yet Tregs predominantly utilize catabolic FAO rather than aerobic glycolysis in order to maintain their survival (Michalek et al., 2011). Thus, the promotion of FAO and inhibition of aerobic glycolysis often leads to Treg development. For example, inhibition of mTOR signaling by genetic deletion or small molecule inhibitor in CD4+ T cells during activation results in Treg development in association with increased FAO and decreased aerobic glycolysis (Delgoffe et al., 2011). Also, ACC1 deficient CD4+ T cells have enhanced activity of AMPK that correlates with increased FAO and strongly favors Treg development (Berod et al., 2014).
The development of memory T cells is closely related to effector T cells as T cells undergo activation, clonal expansion followed by contraction after exposure to pathogens (Kaech and Cui, 2012). Memory CD8+ T cells can rapidly mount effector responses upon secondary infection, and their metabolic requirements are distinct from effector CD8+ T cells. Memory T cells have higher spare respiratory capacity (SRC) than effector T cells (van der Windt et al., 2012, 2013) and SRC is critical for the survival of diverse cells under energetic stress (Nicholls, 2009). Higher SRC in memory T cells was due to greater mitochondrial mass and enhanced expression of regulatory key enzymes in FAO (van der Windt et al., 2012).
Many metabolic sensors and transcription factors that modulate glycolysis also regulate FAO and thus the development of memory T cells. The first line of evidence that FAO is required for memory T cell development comes from microarray analysis in Traf6 KO mice (Pearce et al., 2009). Although it is not clear how Traf6 directly modulates FAO, Traf6 deficiency in CD8+ T cells show impaired fatty acid metabolism in comparison to WT CD8+ T cells. One downstream effect of Traf6 deficiency was decreased AMPK activity, and impaired fatty acid metabolism in Traf6 KO CD8+ T cells could be restored by AMPK agonist treatment. Further supporting the role of AMPK in memory T cell development, AMPKα1-deficient CD8+ T cells have impaired memory responses during secondary infection by L. monocytogenes, suggesting that AMPKα1 is critical for CD8+ T cells to switch anabolic effector to catabolic memory responses (Rolf et al., 2013).
mTOR activation in T cells promotes aerobic glycolysis and inhibits FAO that are essential for effector T cell development. Thus, inhibition of mTOR activity promotes catabolic FAO and thus memory T cell development (Araki et al., 2009; Pollizzi et al., 2015; Rao et al., 2010). Accordingly, rapamycin treatment in T cells enhanced memory CD8+ T cell development (Araki et al., 2009; Rao et al., 2010). Because rapamycin can inhibit both mTORC1 and mTORC2, further genetic analysis characterizes the role of specific mTOR complex on memory T cell development. Thus far, the data suggest that either complex can non-redundantly inhibit memory T cell development. RNAi-mediated knock-down of Raptor in CD8+ T cells generated enhanced memory T cell development (Araki et al., 2009). Conversely, Tsc1 KO CD8+ T cells that have enhanced mTORC1 activity show decreased memory T cell responses during L. monocytogenes infection (Shrestha et al., 2014). mTORC2 also plays a role as Rictor KO mice have enhanced memory T cell development in association with increased FAO (Pollizzi et al., 2015). Collectively, these observations suggest that the Tsc-mTOR pathway inhibits catabolic metabolism during memory T cell development. In addition to its metabolic regulation, mTOR regulates the expression of eomesodermin (Eomes), a transcription factor that is critical for the development of memory CD8+ T cells (Rao et al., 2010). Whether ACC1 or AMPK directly affects the expression Eomes and other transcriptional regulators involved in memory T cells remains to be explored.
6. Therapeutic perspectives
Many studies provide compelling evidence that metabolic regulators can modulate effector and memory T cell development in response to various pathogens. Furthermore, it has been proposed that different stages of memory T cells provide better protective immunity than terminal effector T cells against viral infection and tumor (Gattinoni et al., 2005; Klebanoff et al., 2005). Thus, many recent studies have focused on identifying small-molecule inhibitors that target metabolic regulators and presented therapeutically promising results. Several synthetic analogs have been discovered to effectively inhibit aerobic glycolysis and amino acid transport. 2-Deoxyglucose (2-DG) is a glucose analog and competitive inhibitor of hexokinase-2. 2-DG can enhance memory T cell development and anti-tumor immunity (Sukumar et al., 2013). Also, 2-DG treatment can enhance murine Treg development while inhibiting Th17 development in vitro, and 2-DG-conditioned Th17 cells were less capable of inducing EAE in mice after adoptive transfer (Shi et al., 2011). Other transport inhibitors include 2-aminobicylo-(2,2,1)-heptane-2-carboxylic acid (BCH) and brasilicardin A that inhibit System L amino-acid transporters (Usui et al., 2006; Zheng et al., 2009). These compounds are known immunosuppressants and inhibitors of T cell proliferation, yet its efficacy in autoimmune diseases or organ transplants remains to be investigated.
Similarly, rapamycin appears to be effective in treatment of autoimmune diseases and increasing vaccine responses. Rapamycin was discovered as a potent immunosuppressant that was effective in organ transplants (Augustine et al., 2007), and mTORC1 is its major target of inhibition. Recently, it was found that long-term rapamycin exposure of cells can also inhibit mTORC2 activity that has been considered to be resistant (Sarbassov et al., 2006). Supporting the critical role of mTOR in effector CD4+ T cell development, oral administration of rapamycin ameliorates pathogenesis of murine systemic lupus erythematosus (SLE) in vivo (Warner et al., 1994). Rapamycin can also reduce disease severity in SLE patients (Fernandez et al., 2006). Alternatively, rapamycin and its derivatives, so-called “rapalogues”, have been tested for efficacy in vaccine development in many pre-clinical disease models. Sirolimus, one of the rapalogues, can enhance memory CD8+ T cell responses in vaccinia vaccination in non-human primates (Turner et al., 2011). Short-term, high-dose rapamycin inhibiting mTOR activity and glycolysis can enhance vaccine-induced anti-tumor immune responses as well (Li et al., 2012). In order to further overcome the non-specific effects of rapamycin, another study utilized an aptamer conjugated RNAi approach to knockdown mTORC1. This led to more effective control of tumor growth in vivo (Berezhnoy et al., 2014).
Pharmacological activators of AMPK include N,N-dimethylbiguanide (metformin) and 5-aminoimidazole-4-carboxamide-ribonucleoside (AICAR) that act as 5′ AMP analog (Sajan et al., 2010). Metformin belongs to the biguanide class of anti-diabetic drugs that also include phenformin and buformin and has been an effective regimen for type-2 diabetes (Foretz et al., 2014). Both metformin and AICAR can suppress T cell activation and IL-2 secretion (Zheng et al., 2009; Jhun et al., 2005), and decrease murine CD8+ T cell effector function in vitro and in vivo (Blagih et al., 2015). Consistent with such observation, administration of metformin and AICAR in mice during EAE reduced disease severity with decreased inflammatory cytokine expression of CD4+ T cells (Nath et al., 2005, 2009b). Instead, metformin treatment during L. monocytogenes infection strongly enhanced memory CD8+ T cell development in mice (Pearce et al., 2009). Although not consistent with the role of AMPK, others have found that metformin can enhance CD8+ T cell effector function and anti-tumor immunity (Eikawa et al., 2015).
Several synthetic compounds have been also discovered to inhibit Myc and HIF-1 activity. 10058-F4 is a small molecule inhibitor that inhibits heterodimerization between Myc and Max and their transcriptional activity in tumor cells (Yin et al., 2003). 10058-F4 can also inhibit Th1 and Th17 development in vitro and 10058-F4 treated mice were resistant to EAE development in vivo (Bandukwala et al., 2012). Digoxin and other cardiac glycosides can inhibit HIF-1α expression and its target genes (Zhang et al., 2008). Digoxin treatment can inhibit Th17 development in vitro and ameliorate pathogenesis during EAE in mice (Huh et al., 2011). Although digoxin also inhibits ROR-γt activity, digoxin-mediated inhibition of HIF-1α and glycolysis can contribute to its regulation of Th17 development as well. Overall, the effects of these synthetic analogs on effector and memory T cell development are similar to those observed in genetic mouse models. This fact greatly strengthens the idea of metabolic regulation in effector versus memory T cell fate decisions.
7. Concluding remarks
T cell-mediated immune responses are tightly coordinated and involve cellular proliferation, growth, and differentiation. The metabolic regulators that are described in this review control different aspects of T cell activation. Some of them appear to selectively affect T cell differentiation without significant effects on proliferation, while others affect proliferation and growth. It is intuitive that T cells incapable of proliferation and growth cannot further support their differentiation, and thus in this perspective, some are more potent targets for T cell activation and are ideal candidates in organ transplants or graft-versus host diseases. At the same time, their selective effects provide more targeted therapeutic approaches such as vaccine development. Furthermore, we have begun to understand cross-talk among metabolic regulators as well as their interaction with conventional transcription factors during T cell development in addition to metabolic changes. Thus, our increasing knowledge on metabolic requirements for T cell activation demands a more integrated and comprehensive model of T cell-mediated immune responses.
Acknowledgments
Funding support comes from grants from the Melanoma Research Alliance, the National Institutes of Health (RO1AI099300 and RO1AI089830), “Kelly’s Dream” Foundation, the Janey Fund, and the Seraph Foundation, and gifts from Bill and Betty Topecer and Dorothy Needle. FP is a Stewart Trust Scholar and a recipient of Johns Hopkins University Catalytic Award.
Footnotes
Conflict of interest
The authors declare no financial conflicts of interest.
References
- Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andris F, Leo O. AMPK in lymphocyte metabolism and function. Int Rev Immunol. 2015;34:67–81. doi: 10.3109/08830185.2014.969422. [DOI] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine JJ, Bodziak KA, Hricik DE. Use of sirolimus in solid organ transplantation. Drugs. 2007:369–391. doi: 10.2165/00003495-200767030-00004. [DOI] [PubMed] [Google Scholar]
- Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, Molesworth AM, Smithers N, Lee K, Witherington J, Tough DF, Prinjha RK, Peters B, Rao A. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc Natl Acad Sci U S A. 2012:14532–14537. doi: 10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, Tontonoz P. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezhnoy A, Castro I, Levay A, Malek TR, Gilboa E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J Clin Invest. 2014:188–197. doi: 10.1172/JCI69856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bahre H, Tschirner SK, Gorinski N, Gohmert M, Mayer CT, Huehn J, Ponimaskin E, Abraham WR, Muller R, Lochner M, Sparwasser T. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, Dustin ML, Goldrath AW. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vazquez G, Yurchenko E, Raissi TC, van der Windt GJ, Viollet B, Pearce EL, Pelletier J, Piccirillo CA, Krawczyk CM, Divangahi M, Jones RG. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity. 2015;42:41–54. doi: 10.1016/j.immuni.2014.12.030. [DOI] [PubMed] [Google Scholar]
- Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat Immunol. 2001:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Jung CY, Lee TP, Liu H, Mookerjee BK. Changes in glucose transport and transporter isoforms during the activation of human peripheral blood lymphocytes by phytohemagglutinin. J Immunol. 1994;152:2660–2668. [PubMed] [Google Scholar]
- Chen HW, Heiniger HJ, Kandutsch AA. Relationship between sterol synthesis and DNA synthesis in phytohemagglutinin-stimulated mouse lymphocytes. Proc Natl Acad Sci U S A. 1975;72:1950–1954. doi: 10.1073/pnas.72.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C, Pinto AK, Curtis JD, Persaud SP, Cella M, Lin CC, Edelson BT, Allen PM, Colonna M, Pearce EL, Diamond MS, Egawa T. c-Myc-induced transcription factor AP4 is required for host protection mediated by CD8+ T cells. Nat Immunol. 2014;15:884–893. doi: 10.1038/ni.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clambey ET, McNamee EN, Westrich JA, Glover LE, Campbell EL, Jedlicka P, de Zoeten EF, Cambier JC, Stenmark KR, Colgan SP, Eltzschig HK. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci U S A. 2012:E2784–E2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikawa S, Nishida M, Mizukami S, Yamazaki C, Nakayama E, Udono H. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci U S A. 2015:1809–1814. doi: 10.1073/pnas.1417636112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, Mamer OA, Avizonis D, DeBerardinis RJ, Siegel PM, Jones RG. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Ippolito GC, Tian L, Wiehagen K, Oh S, Sambandam A, Willen J, Bunte RM, Maika SD, Harriss JV, Caton AJ, Bhandoola A, Tucker PW, Hu H. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. 2010;115:510–518. doi: 10.1182/blood-2009-07-232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat Immunol. 2011;12:544–550. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D, Bonilla E, Mirza N, Niland B, Perl A. Rapamycin reduces disease activity and normalizes T cell activation-induced calcium fluxing in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2983–2988. doi: 10.1002/art.22085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M, Guigas B, Bertrand L, Pollak M, Viollet B. Metformin: from mechanisms of action to therapies. Cell Metab. 2014;20:953–966. doi: 10.1016/j.cmet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- Freitas AA, Rocha B. Homeostasis of naive T cells: the Foxo that fixes. Nat Immunol. 2009:133–134. doi: 10.1038/ni0209-133. [DOI] [PubMed] [Google Scholar]
- Gattinoni L, Klebanoff CA, Palmer DC, Wrzesinski C, Kerstann K, Yu Z, Finkelstein SE, Theoret MR, Rosenberg SA, Restifo NP. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J Clin Invest. 2005;115:1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaland RE, Yu W, Rice AP. Identification of LKLF-regulated genes in quiescent CD4+ T lymphocytes. Mol Immunol. 2005:627–641. doi: 10.1016/j.molimm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Hamilton SE, Jameson SC. CD8 T cell quiescence revisited. Trends Immunol. 2012:224–230. doi: 10.1016/j.it.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Heikamp EB, Patel CH, Collins S, Waickman A, Oh MH, Sun IH, Illei P, Sharma A, Naray-Fejes-Toth A, Fejes-Toth G, Misra-Sen J, Horton MR, Powell JD. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nat Immunol. 2014;15:457–464. doi: 10.1038/ni.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Leung MWL, Huang P, Ryan DA, Krout MR, Malapaka RRV, Chow J, Manel N, Ciofani M, Kim SV, Cuesta A, Santori FR, Lafaille JJ, Xu HE, Gin DY, Rastinejad F, Littman DR. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing ROR[ggr]t activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, Ohsugi M, Onda M, Hirai M, Fujimoto J, Yamamoto T. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 1999;18:7432–7441. doi: 10.1038/sj.onc.1203193. [DOI] [PubMed] [Google Scholar]
- Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhun BS, Oh YT, Lee JY, Kong Y, Yoon KS, Kim SS, Baik HH, Ha J, Kang I. AICAR suppresses IL-2 expression through inhibition of GSK-3 phosphorylation and NF-AT activation in Jurkat T cells. Biochem Biophys Res Commun. 2005:339–346. doi: 10.1016/j.bbrc.2005.04.126. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr The von Hippel-Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem Biophys Res Commun. 2005:627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- Karinch AM, Pan M, Lin CM, Strange R, Souba WW. Glutamine metabolism in sepsis and infection. J Nutr. 2001;131:2535S–2538S. doi: 10.1093/jn/131.9.2535S. discussion 2550S–2531S. [DOI] [PubMed] [Google Scholar]
- Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, Graeber TG, Reue K, Brooks DG, Bensinger SJ. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff CA, Gattinoni L, Torabi-Parizi P, Kerstann K, Cardones AR, Finkelstein SE, Palmer DC, Antony PA, Hwang ST, Rosenberg SA, Waldmann TA, Restifo NP. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc Natl Acad Sci U S A. 2005:9571–9576. doi: 10.1073/pnas.0503726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Leiden JM. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, Matsuda S, Koyasu S. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Rep. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Walsh MC, Hoehn KL, James DE, Wherry EJ, Choi Y. Regulator of fatty acid metabolism, acetyl coenzyme a carboxylase 1, controls T cell immunity. J Immunol. 2014;192:3190–3199. doi: 10.4049/jimmunol.1302985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Rao R, Vazzana J, Goedegebuure P, Odunsi K, Gillanders W, Shrikant PA. Regulating mammalian target of rapamycin to tune vaccination-induced CD8(+) T cell responses for tumor immunity. J Immunol. 2012:3080–3087. doi: 10.4049/jimmunol.1103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Lukashev D, Klebanov B, Kojima H, Grinberg A, Ohta A, Berenfeld L, Wenger RH, Sitkovsky M. Cutting edge: hypoxia-inducible factor 1alpha and its activation-inducible short isoform I.1 negatively regulate functions of CD4+ and CD8+ T lymphocytes. J Immunol. 2006:4962–4965. doi: 10.4049/jimmunol.177.8.4962. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, Rathmell JC. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver NJ, Blagih J, Saucillo DC, Tonelli L, Griss T, Rathmell JC, Jones RG. The liver kinase B1 is a central regulator of T cell development, activation, and metabolism. J Immunol. 2011:4187–4198. doi: 10.4049/jimmunol.1100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina EN, Morris MR, Zatyka M, Raval RR, Banks RE, Richards FM, Johnson CM, Maher ER. Identification of novel VHL target genes and relationship to hypoxic response pathways. Oncogene. 2005:4549–4558. doi: 10.1038/sj.onc.1208649. [DOI] [PubMed] [Google Scholar]
- Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, Kallies A. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- Marrack P, Kappler J. Control of T cell viability. Annu Rev Immunol. 2004;22:765–787. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Kawamura-Tsuzuku J, Ohsugi M, Yoshida M, Emi M, Nakamura Y, Onda M, Yoshida Y, Nishiyama A, Yamamoto T. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 1996;12:705–713. [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMP-activated protein kinase (AMPK) signaling pathway coordinates cell growth, autophagy, & metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34:121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, Blonska M, Lin X, Sun SC. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N, Giri S, Prasad R, Salem ML, Singh AK, Singh I. 5-Aminoimidazole-4-carboxamide ribonucleoside: a novel immunomodulator with therapeutic efficacy in experimental autoimmune encephalomyelitis. J Immunol. 2005:566–574. doi: 10.4049/jimmunol.175.1.566. [DOI] [PubMed] [Google Scholar]
- Nath N, Khan M, Rattan R, Mangalam A, Makkar RS, de Meester C, Bertrand L, Singh I, Chen Y, Viollet B, Giri S. Loss of AMPK exacerbates experimental autoimmune encephalomyelitis disease severity. Biochem Biophys Res Commun. 2009a:16–20. doi: 10.1016/j.bbrc.2009.05.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N, Khan M, Paintlia MK, Singh I, Hoda MN, Giri S. Metformin attenuated the autoimmune disease of the central nervous system in animal models of multiple sclerosis. J Immunol. 2009b:8005–8014. doi: 10.4049/jimmunol.0803563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme P. Why is L-glutamine metabolism important to cells of the immune system in health, postinjury, surgery or infection? J Nutr. 2001;131:2515S–2522S. doi: 10.1093/jn/131.9.2515S. discussion 2523S-2514S. [DOI] [PubMed] [Google Scholar]
- Nicholls DG. Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem Soc Trans. 2009;37:1385–1388. doi: 10.1042/BST0371385. [DOI] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TF, Gorentla BK, Xie D, Srivatsan S, McLeod IX, He YW, Zhong XP. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. Eur J Immunol. 2011;41:3361–3370. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill HM, Lally JS, Galic S, Thomas M, Azizi PD, Fullerton MD, Smith BK, Pulinilkunnil T, Chen Z, Samaan MC, Jorgensen SB, Dyck JR, Holloway GP, Hawke TJ, van Denderen BJ, Kemp BE, Steinberg GR. AMPK phosphorylation of ACC2 is required for skeletal muscle fatty acid oxidation and insulin sensitivity in mice. Diabetologia. 2014;57:1693–1702. doi: 10.1007/s00125-014-3273-1. [DOI] [PubMed] [Google Scholar]
- O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, Hsu FF, Birnbaum MJ, Pearce EJ, Pearce EL. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- Pan F, Barbi J, Pardoll DM. Hypoxia-inducible factor 1: a link between metabolism and T cell differentiation and a potential therapeutic target. Oncoimmunology. 2012;1:510–515. doi: 10.4161/onci.19457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, Powell JD. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J Clin Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JD, Delgoffe GM. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 2010;33:301–311. doi: 10.1016/j.immuni.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol Cell. 2000:683–692. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Rolf J, Zarrouk M, Finlay DK, Foretz M, Viollet B, Cantrell DA. AMPKalpha1: a glucose sensor that controls CD8 T-cell memory. Eur J Immunol. 2013;43:889–896. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault JP, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud JP. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky AY. Regulatory T cells and Foxp3. Immunol Rev. 2011;241:260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan MP, Bandyopadhyay G, Miura A, Standaert ML, Nimal S, Longnus SL, Van Obberghen E, Hainault I, Foufelle F, Kahn R, Braun U, Leitges M, Farese RV. AICAR and metformin, but not exercise, increase muscle glucose transport through AMPK-, ERK-, and PDK1-dependent activation of atypical PKC. Am J Physiol Endocrinol Metab. 2010:E179–E192. doi: 10.1152/ajpendo.00392.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Schober SL, Kuo CT, Schluns KS, Lefrancois L, Leiden JM, Jameson SC. Expression of the transcription factor lung Kruppel-like factor is regulated by cytokines and correlates with survival of memory T cells in vitro and in vivo. J Immunol. 1999;163:3662–3667. [PubMed] [Google Scholar]
- Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Yang K, Wei J, Karmaus PW, Neale G, Chi H. Tsc1 promotes the differentiation of memory CD8+ T cells via orchestrating the transcriptional and metabolic programs. Proc Natl Acad Sci U S A. 2014;111:14858–14863. doi: 10.1073/pnas.1404264111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, Roychoudhuri R, Palmer DC, Muranski P, Karoly ED, Mohney RP, Klebanoff CA, Lal A, Finkel T, Restifo NP, Gattinoni L. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas P, Hawley SA, Clarke RG, Mustard KJ, Green K, Hardie DG, Cantrell DA. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J Exp Med. 2006:1665–1670. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague TK, Hildeman D, Kedl RM, Mitchell T, Rees W, Schaefer BC, Bender J, Kappler J, Marrack P. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc Natl Acad Sci U S A. 1999;96:12691–12696. doi: 10.1073/pnas.96.22.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel M, Caldwell CC, Kreth S, Kuboki S, Chen P, Smith P, Ohta A, Lentsch AB, Lukashev D, Sitkovsky MV. Targeted deletion of HIF-1alpha gene in T cells prevents their inhibition in hypoxic inflamed tissues and improves septic mice survival. PLoS ONE. 2007;2:e853. doi: 10.1371/journal.pone.0000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AP, Shaffer VO, Araki K, Martens C, Turner PL, Gangappa S, Ford ML, Ahmed R, Kirk AD, Larsen CP. Sirolimus enhances the magnitude and quality of viral-specific CD8+ T-cell responses to vaccinia virus vaccination in rhesus macaques. Am J Transplant. 2011;11:613–618. doi: 10.1111/j.1600-6143.2010.03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzachanis D, Freeman GJ, Hirano N, van Puijenbroek AA, Delfs MW, Berezovskaya A, Nadler LM, Boussiotis VA. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol. 2001:1174–1182. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- Usui T, Nagumo Y, Watanabe A, Kubota T, Komatsu K, Kobayashi J, Osada H. Brasilicardin A, a natural immunosuppressant, targets amino acid transport system L. Chem Biol. 2006:1153–1160. doi: 10.1016/j.chembiol.2006.09.006. [DOI] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, O’Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci U S A. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956a;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956b;124:269–270. [PubMed] [Google Scholar]
- Warner LM, Adams LM, Sehgal SN. Rapamycin prolongs survival and arrests pathophysiologic changes in murine systemic lupus erythematosus. Arthritis Rheum. 1994;37:289–297. doi: 10.1002/art.1780370219. [DOI] [PubMed] [Google Scholar]
- Wu Q, Liu Y, Chen C, Ikenoue T, Qiao Y, Li CS, Li W, Guan KL, Zheng P. The tuberous sclerosis complex-mammalian target of rapamycin pathway maintains the quiescence and survival of naive T cells. J Immunol. 2011;187:1106–1112. doi: 10.4049/jimmunol.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, Guertin DA, Lamb RF, Chi H. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity. 2013;39:833–845. doi: 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Mol Cell. 2013;49:379–387. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I, Fruman DA. Regulation of quiescence in lymphocytes. Trends Immunol. 2003:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- Zhang H, Qian DZ, Tan YS, Lee K, Gao P, Ren YR, Rey S, Hammers H, Chang D, Pili R, Dang CV, Liu JO, Semenza GL. Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc Natl Acad Sci U S A. 2008:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. J Immunol. 2009:6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]