Abstract
γ-Catenin, an important component of desmosomes, may also participate in Wnt signaling. Herein, we dissect the role of γ-catenin in liver by generating conditional γ-catenin knockout (KO) mice and assessing their phenotype after bile duct ligation (BDL) and diethylnitrosamine-induced chemical carcinogenesis. At baseline, KO and wild-type littermates showed comparable serum biochemistry, liver histology, and global gene expression. β-Catenin protein was modestly increased without any change in Wnt signaling. Desmosomes were maintained in KO, and despite no noticeable changes in gene expression, differential detergent fractionation revealed quantitative and qualitative changes in desmosomal cadherins, plaque proteins, and β-catenin. Enhanced association of β-catenin to desmoglein-2 and plakophilin-3 was observed in KO. When subjected to BDL, wild-type littermates showed specific changes in desmosomal protein expression. In KO, BDL deteriorated baseline compensatory changes, which manifested as enhanced injury and fibrosis. KO also showed enhanced tumorigenesis to diethylnitrosamine treatment because of Wnt activation, as also verified in vitro. γ-Catenin overexpression in hepatoma cells increased its binding to T-cell factor 4 at the expense of β-catenin–T-cell factor 4 association, induced unique target genes, affected Wnt targets, and reduced cell proliferation and viability. Thus, γ-catenin loss in liver is basally well tolerated. However, after insults like BDL, these compensations at desmosomes fail, and KO show enhanced injury. Also, γ-catenin negatively regulates tumor growth by affecting Wnt signaling.
Plakoglobin or γ-catenin belongs to the catenin family of proteins and is a component of desmosomes. Desmosomes are submembranous plaques that provide resistance to mechanical and shear stresses within a tissue and contribute to intercellular adhesion.1 Analogous to β-catenin at adherens junctions (AJs), γ-catenin, like other desmosomal plaque proteins, such as desmoplakin (DP) and plakophilin (Pkp), adheres to neighboring cells by linking intracytoplasmic domains of transmembrane desmosomal cadherins like desmoglein (Dsg) and desmocollin (Dsc) to intermediate filaments.1 γ-Catenin can also be a structural component of AJ, where it may be exchangeable with its close homolog, β-catenin. Both catenins also contain an armadillo domain and can bind the T-cell factor (TCF) family of transcription factors to participate in Wnt signaling.2 We have previously identified an increase in γ-catenin in the liver-specific β-catenin knockout (KO) mice,3, 4 where it was able to compensate for β-catenin loss at AJs without compromising desmosomes. However, γ-catenin did not compensate for β-catenin loss in Wnt signaling, and β-catenin KO continued to lack liver-specific Wnt targets, such as glutamine synthetase (GS) and P450 2e1 and 1a2.5
The primary role of γ-catenin in liver pathobiology remains unknown. We addressed the function of γ-catenin in liver homeostasis through generation of liver-specific KO using the γ-catenin–floxed mice6, 7 and albumin-cre transgenic mice, as reported earlier for β-catenin.5 We show that γ-catenin loss in liver is well tolerated, and desmosomes remain intact because of the changes in protein expression of various desmosomal proteins along with their enhanced interactions with β-catenin. However, on challenging KO with bile duct ligation (BDL) that induces intrahepatic biliary pressure, these mice show enhanced hepatobiliary injury compared with wild-type (WT) littermates.8 This was because of loss of desmosomal integrity secondary to desmosomal protein and β-catenin alterations. We also address the role of γ-catenin in hepatocellular cancer (HCC), which remains ambiguous. A notable increase in tumorigenesis in response to diethylnitrosamine (DEN)9 was evident in KO compared with WT, which was because of enhanced β-catenin signaling. We discovered that γ-catenin suppression in HCC cells increases β-catenin–TCF complex, its activity, and expression of target genes. Intriguingly, γ-catenin overexpression led to enhanced γ-catenin–TCF complex as well as reporter activity and decreased β-catenin target gene expression, affecting cell viability and proliferation. Thus, we identify an important therapeutic benefit in HCC cells of enhancing γ-catenin levels to divert TCF from β-catenin and curb Wnt signaling.
Materials and Methods
Generation of Liver-Specific γ-Catenin Conditional KO Mice
All animal experiments and procedures were performed under the guidelines of the NIH and under an animal protocol approved by the Institutional Animal Use and Care Committees at the University of Pittsburgh (Pittsburgh, PA). Homozygous γ-catenin–floxed mice (C57/BL6 strain)6, 7 and albumin-cre transgenic mice were bred to generate KO mice using a similar breeding strategy, as previously described.5 Mice with genotype γ-cateninloxp/loxp;Alb-Cre+/− are KOs. Littermates with floxed γ-catenin genotypes and without cre allele are WT controls. Livers from age- and sex-matched 90- to 180-day-old KO and WT (n = 5 to approximately 7) were used, unless otherwise noted.
Chemical Carcinogenesis
Male KO (n = 5) and WT (n = 7) mice were injected i.p. with DEN (Sigma-Aldrich, Inc., St. Louis, MO) at a dose of 5 μg/g body weight at postnatal day 15 to study chemical carcinogenesis, as used previously.9 Mice were sacrificed at 9 months of age. Liver tissues were collected for gross, microscopic, and protein analysis.
BDL Data
BDL is commonly used as a model for extrahepatic biliary obstruction that causes acute cholestasis and periportal fibrosis.10 Briefly, under isoflurane anesthesia, the common bile duct was doubly ligated using 5.0 silk after a midline abdominal incision. Animals were sacrificed at 2 weeks after BDL (n = 5 to approximately 7). At the time of sacrifice, liver tissues were collected for histology and immunohistochemical (IHC) studies and protein analyses.
Histological and IHC Analysis
Parts of livers were fixed in 4% buffered formalin and paraffin embedded. Tissue sections (4 μm thick) were stained with hematoxylin and eosin (H&E). Liver sections were analyzed by indirect IHC for β-catenin, γ-catenin, cyclin-D1, GS (Santa Cruz Biotechnology, Dallas, TX), Ki-67, and CK19 (Thermo Scientific, Freemont, CA), as described elsewhere.3, 5, 9 Briefly, formalin-fixed sections were deparaffinized. Endogenous peroxide was inactivated using 3% hydrogen peroxide (Sigma-Aldrich, Inc.). Slides were microwaved in citrate buffer for 20 minutes, followed by blocking in the blue blocker (Shandon Lipshaw, Pittsburgh, PA). Sections were then incubated with horseradish peroxidase–conjugated secondary antibody (Chemicon, Temecula, CA) for 30 minutes, and the signal was detected using the Avidin-Biotin Complex Elite kit (Vector Laboratories, Burlingame, CA), according to the manufacturer's instructions.
Special Stains
For measurement of fibrosis, Sirius Red staining was performed. Briefly, sections were rehydrated and placed in Sirius Red staining solution for 1 hour at room temperature. Sections were washed with acidified water, dehydrated, and covered.
The ApopTag Peroxidase kit (Intergen Company, Billerica, MA) was used to detect apoptosis by the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) method. TUNEL-positive apoptotic nuclei were detected by the presence of brown staining.
Protein Extraction
Whole-cell lysates were obtained from the WT and KO livers using radioimmunoprecipitation assay buffer containing fresh protease and phosphatase inhibitor cocktails (Sigma-Aldrich, Inc.), as described previously.3, 5, 9 Detergent fractionation was used to isolate junctional proteins in a cytoskeleton-enriched fraction using a previously published protocol.4 Traditionally, the Triton X-100-insoluble fraction is enriched in desmosomes.11 Protein assays were performed using bicinchoninic acid protein assay.
Western Blot Analysis
A total of 20 to 30 μg of protein was resolved on Tris-HCl precast gels (Bio-Rad Laboratories, Hercules, CA) by SDS-PAGE analysis using the Mini-PROTEAN 3 Electrophoresis Module Assembly (Bio-Rad Laboratories). The resolved proteins were transferred to polyvinylidene difluoride membranes, and the signal was detected by SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Representative Western blots were scanned and assessed for quantitative changes by densitometry using ImageJ software version 1.49 (NIH, Bethesda, MD; http://imagej.nih.gov/ij). Mean integrated optical densitometry between various groups was compared for statistical analysis by t-test. Horseradish peroxidase–conjugated secondary antibodies were purchased from Millipore (Billerica, MA). The primary antibodies used were mouse antibody to β-catenin (BD610154; BD Biosciences, San Jose, CA), mouse antibody to γ-catenin (BD610153; BD Biosciences), mouse antibody to E-cadherin (BD610182; BD Biosciences), mouse antibody to Pkp3 (sc-166655; Santa Cruz Biotechnology), mouse antibody to Myc-Tag (2276; Cell Signaling Technology, Danvers, MA), rabbit antibody to β-catenin (sc-7199; Santa Cruz Biotechnology), rabbit antibody to E-cadherin (sc-7870; Santa Cruz Biotechnology), mouse antibody to TCF4 (T5817; Sigma-Aldrich, Inc.), rabbit antibody to Pkp3 (ab109441; Abcam, Cambridge, MA), rabbit antibody to Dsg1 (sc-20114; Santa Cruz Biotechnology), rabbit antibody to Dsg2 (sc-20115; Santa Cruz Biotechnology), rabbit antibody to DP I/II (sc-33555; Santa Cruz Biotechnology), rabbit antibody to γ-catenin (2309; Cell Signaling Technology), rabbit antibody to TCF4 (2569; Cell Signaling Technology), rabbit antibody to glyceraldehyde-3-phosphate dehydrogenase (sc-2577; Santa Cruz Biotechnology), goat antibody to β-catenin (sc-1496; Santa Cruz Biotechnology), goat antibody to Dsc2 (sc-34311; Santa Cruz Biotechnology), goat antibody to Dsg3 (sc-14867; Santa Cruz Biotechnology), goat antibody to Dsg4 (sc-28069; Santa Cruz Biotechnology), goat antibody to γ-catenin (sc-30996; Santa Cruz Biotechnology), and goat antibody to Pkp2 (sc-18977; Santa Cruz Biotechnology).
Immunoprecipitation
Coprecipitation studies were performed with 200 to 500 μg protein. Antibodies used for immunoprecipitation were rabbit anti–γ-catenin and TCF4 (Cell Signaling Technology), mouse anti–β-catenin (BD610154; BD Biosciences), mouse anti–E-cadherin (BD610182; BD Biosciences), rabbit anti-Pkp3 (Santa Cruz Biotechnology), rabbit anti-Dsg2 (Santa Cruz Biotechnology), and goat β-catenin–conjugated A/G agarose beads (Santa Cruz Biotechnology).
TEM Data
Liver tissue was fixed with 2.5% glutaraldehyde in phosphate-buffered saline by perfusion through the inferior vena cava, and transmission electron microscopy (TEM) was performed, as described previously.4, 12 Briefly, after fixation and dividing into sections, grids were hand stained with 2% uranyl acetate in 50% methanol for 10 minutes and 1% lead citrate for 7 minutes. Digital TEM images were taken with JEOL JEM-1011 Transmission Electron Microscope (Peabody, MA) at 80.0 kV. Quantitative analysis of junctions was performed with MetaMorph Software (Molecular Devices, Sunnyvale, CA). For quantification, at least six to seven desmosomes were counted in one KO and one WT liver at baseline and after BDL, and average of intercellular distance at desmosomes was compared for statistical significance by t-test.
Cell Culture and Transfection of siRNA and Expression Plasmids
Hep3B and SNU398 human HCC cell lines were obtained from ATCC (Manassas, VA). Cells were cultured in Eagle's minimal essential medium supplemented with 10% v/v fetal bovine serum at 37°C in a humidified 5% carbon dioxide atmosphere. The cells were transfected in 6-well plates in triplicate using Lipofectamine 2000 Reagent (Life Technologies, Grand Island, NY) and a total siRNA concentration of 50 nmol/L in Opti-MEMI Media (Life Technologies) or γ-catenin or Jup plasmid (500 to 1000 ng) for 24 to 72 hours, as per the manufacturer's instructions. Human γ-catenin siRNA (JUP, s7666) and Silencer Select Negative Control (4390847) siRNA were purchased from Ambion (Grand Island, NY). Plasmid for γ-catenin (plasmid 32228) was purchased from Addgene (Cambridge, MA).
MTT Assay
Cells were washed with 1× phosphate-buffered saline. One milliliter of 0.5 mg/mL MTT solution (M-2128; Sigma-Aldrich, Inc.) was added into each well of the 6-well plates. The cells were incubated at 37°C, 5% carbon dioxide for 30 minutes. The MTT solution was removed, and 0.5 mL of 0.04 N HCl/isopropanol was added. The plate was incubated at room temperature for 3 minutes with occasional shaking. The samples were transferred to 96-well plates in a 1:1 dilution with 0.04 N HCl/isopropanol. OD at 570 nm was measured using a Synergy HT plate reader (BioTek, Winooski, VT). An unpaired t-test was used for comparison. P < 0.05 was considered significant.
β-Catenin/TCF Transcription Reporter Assay
Hep3B cells grown on 6-well plates were transfected in triplicate with 0.8 μg of TOPflash construct (21-170; Millipore, Billerica, MA) along with 0.1 μg of internal control reporter Renilla reniformis luciferase (pRL-TK; Promega, Madison, WI) and JUP siRNA and plasmid, as described previously.4 The TOPflash construct contains three TCF binding sites and firefly luciferase reporter gene. The cells were harvested 48 hours after transfection using a Dual Luciferase Assay System kit, according to the manufacturer's protocol (E1910; Promega). TOPflash luminescent signals were normalized to Renilla luminescent signal, and an unpaired t-test was used to determine the P value for statistical significance. P < 0.05 was considered significant.
Thymidine Incorporation Assay
The cells were transfected with siRNA, according to the protocol described above. H3-thymidine (2.5 μCi/mL) was added 4 hours after transfection, and the pulse period was performed for 72 hours. The cells were then fixed with cold 5% trichloroacetic acid for at least 15 minutes, washed with cold water, and dried. The fixed cells were dissolved in 0.33 mol/L NaOH and incubated at room temperature for 10 minutes, followed by incubation at 4°C for 10 minutes. The sample solution was mixed with cold 1.2 mol/L HCl/40% trichloroacetic acid and centrifuged. The cell pellet was dissolved in 0.33 mol/L NaOH and mixed with scintillation solution at 1:10 dilution. The samples were incubated at room temperature in the dark for at least 1 hour and read in LS 6000 IC Beckman Scintillation Counter (Beckman Coulter, Pasadena, CA). A least significant differences multiple comparison test was used for comparison of multiple groups. P < 0.05 was considered significant.
Real-Time PCR
RNA was extracted from treated Hep3B cells using TRIzol (Life Technologies) and was DNAse treated using the TURBO DNase Kit (Ambion), as per the manufacturer's instructions. The comparative ΔΔCt method was used for analysis of the data. Real-time PCR was performed using Power SYBR Green (4367659; Life Technologies), 100 ng of cDNA, and 0.2 μmol/L of forward and reverse primers. The primers used were as follows: β-catenin, 5′-GAAACGGCTTTCAGTTGAGC-3′ (forward) and 5′-CTGGCCATATCCACCAGAGT-3′ (reverse); c-Myc, 5′-ATGGCCCATTACAAAGCCG-3′ (forward) and 5′-TTTCTGGAGTAGCAGCTCCTAA-3′ (reverse); cyclin-D1, 5′-GCTGCGAAGTGGAAACCATC-3′ (forward) and 5′-CCTCCTTCTGCACACATTTGAA-3′ (reverse); regucalcin, 5′-GATTGAGTGTGTTTTGCCAGA-3′ (forward) and 5′-GTCTACAAAGAGCAGAGAGTTGG-3′ (reverse); glyceraldehyde-3-phosphate dehydrogenase 1, 5′-TGCACCACCAACTGCTTAGC-GAPDH1-3′ (forward) and 5′-GGCATGGACTGTGGTCATGAG-3′ (reverse); 14-3-3, 5′-GTGTGTCCCCAGAGCCATGG-3′ (forward) and 5′-ACCTTCTCCCGGTACTCACG-3′ (reverse); Nm23-H1, 5′-CGCAGTTCAAACCTAAGCAGCAGCTGG-3′ (forward) and 5′-AGATCCAGTTCTGAGCACAGCTCG-3′ (reverse); and glyceraldehyde-3-phosphate dehydrogenase 2, 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′ (reverse).
Statistical Analysis
Data are presented as the means ± SEM. Data were analyzed by t-test or analysis of variance, and P < 0.05 was considered statistically significant.
Results
Generation of Liver-Specific γ-Catenin KO Mice
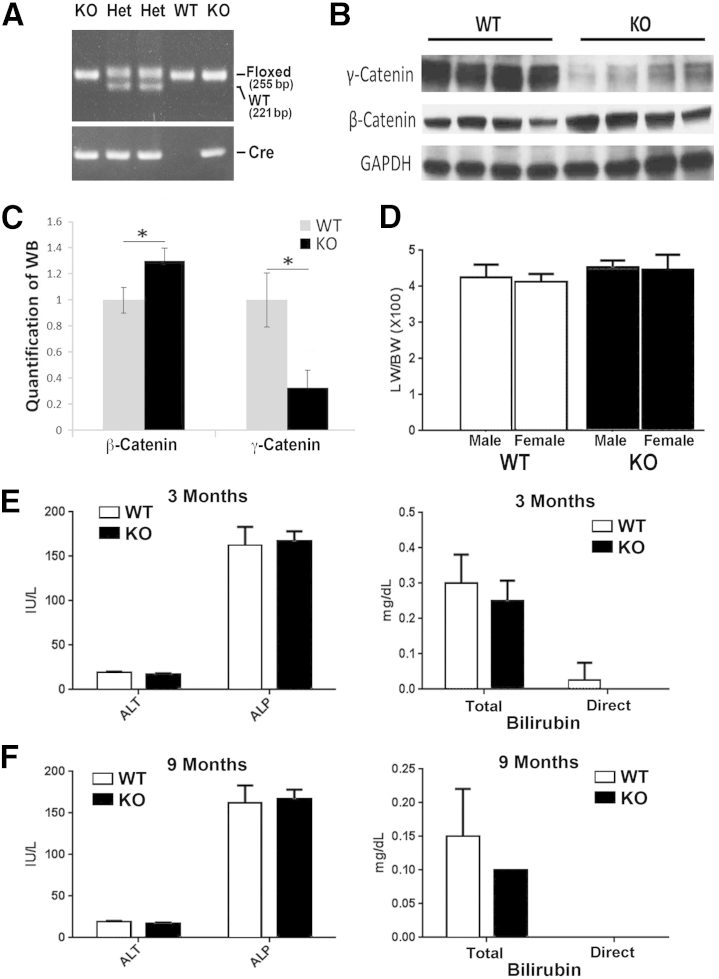
To determine the role of γ-catenin in the liver, floxed γ-catenin (exons 3 to 5) mice were bred to the Alb-Cre transgenic mice to eventually generate the γ-catenin conditional KOs. These mice were identified by PCR analysis on genomic DNA for simultaneous presence of floxed alleles (255 bp) and cre-recombinase (Figure 1A).6, 7 Littermate mice homozygous for floxed γ-catenin alleles without any cre, referred to as WT, were used as controls for all studies. Western blot analysis using whole cell lysates from livers of WT and KO showed a notable decrease in γ-catenin protein in the KO (Figure 1B).
Figure 1.
Lack of an overt phenotype in liver-specific γ-catenin conditional knockout (KO). A: Representative genomic PCR for genotyping mice used in the study. Mice with floxed γ-catenin allele and cre allele were KO, whereas those with floxed and wild-type (WT) alleles and cre allele are heterozygous (Het) mice used for breeding. Mice with floxed γ-catenin only without cre were labeled as WT and used as controls for all studies. B: Representative Western blot (WB) shows a dramatic decrease of γ-catenin in the tissue lysates from the KO livers. There is a modest increase in β-catenin in the same lysates. C: Mean integrated OD of a representative WB shown in B for β-catenin and γ-catenin in WT and KO livers. D: Insignificant difference in liver weight/body weight (LW/BW) ratio in both male and female KO and WT mice. E: No significant differences in serum alanine transaminase (ALT), alkaline phosphatase (ALP), and total and direct bilirubin in 3-month-old KO and WT. F: No significant differences in serum ALT, ALP, and total and direct bilirubin in 9-month-old KO and WT. *P < 0.05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Because β-catenin loss in the liver was compensated by γ-catenin increase at AJ only,3, 4 we wondered if γ-catenin loss could lead any changes in β-catenin protein and/or signaling. Furthermore, a β-catenin increase was reported in γ-catenin–null mice in skin.2 Intriguingly, in the liver lysates from γ-catenin KO, the β-catenin levels were modestly increased (Figure 1, B and C). To determine if increase in β-catenin protein was because of a change in β-catenin gene expression, we examined microarray performed on isolated mRNA from KO (n = 3) and WT (n = 3) livers. No notable difference in β-catenin was evident between the two groups (Table 1). We next queried the microarray data for changes in known β-catenin targets to address the state of Wnt/β-catenin signaling in the absence of γ-catenin. None of the known liver-specific β-catenin targets in the KO livers showed any significant differences from the WT, suggesting lack of any hyperactive β-catenin signaling in the γ-catenin KO livers (Table 1).
Table 1.
Changes in Gene Expression of Wnt Target Genes
| Gene name | WT | KO | Fold change (WT/KO) |
|---|---|---|---|
| Ornithine aminotransferase | 16,488 | 13,000 | 1.3 |
| β-Catenin gene | 8313 | 6887 | 1.3 |
| Axin 2 | 618 | 507 | 1.2 |
| Glutamate transporter | 639 | 514 | 1.2 |
| Cyclin D1 | 1472 | 1287 | 1.1 |
| Regucalcin | 27,415 | 27,857 | 1 |
| Leukocyte cell–derived chemotaxin 2 | 3048 | 3415 | 0.9 |
| Epidermal growth factor receptor | 1774 | 1946 | 0.9 |
| Claudin 2 | 941 | 1239 | 0.8 |
| Glutamine synthetase | 7233 | 9358 | 0.8 |
KO, knockout; WT, wild type.
Loss of γ-catenin in the liver also did not affect liver growth, as reflected by comparable liver weight/body weight ratios for both male and female KO and WT (Figure 1D). H&E staining of liver sections revealed no differences in histology between the KO and WT (data not shown). Likewise, no evidence of spontaneous hepatobiliary injury was observed at any age in KO (shown ages, 3 and 9 months), as also reflected by normal serum levels of alanine aminotransferase or direct and indirect bilirubin, similar to those observed in WT (Figure 1, E and F).
To identify any changes in global gene expression in the absence of γ-catenin in the liver, we next examined microarray analysis of three KO and three WT livers gene. Intriguingly, only a handful of genes showed greater than twofold change in expression in the KO livers (Table 2). The most down-regulated genes, other than the γ-catenin gene (Jup), were sphingomyelin phosphodiesterase 2 and early growth response 1. Among the genes most up-regulated in the absence of γ-catenin were secreted frizzled related protein 1, transforming growth factor-β–induced early growth response gene, heat shock protein 68, and inhibin A (Table 2). Thus, γ-catenin loss from liver had no significant impact on global gene expression. Thus, loss of γ-catenin in the liver is well tolerated at least at the baseline in mice.
Table 2.
Most Altered Genes in γ-Catenin Deleted Liver
| Gene name (symbol) | WT | KO | Fold change (WT/KO) |
|---|---|---|---|
| γ-Catenin (Jup) | 1060 | 282 | 3.8 |
| Sphingomyelin phosphodiesterase 2, neutral (Smpd2) | 1444 | 345.8 | 4.2 |
| Early growth response 1 (Egr1) | 4643 | 1937 | 2.4 |
| Secreted frizzled-related sequence protein 1 (SFRP1) | 364 | 752 | 0.5 |
| TGFB-inducible early growth response gene (Tieg/Klf10) | 727 | 1638 | 0.4 |
| Sphingosine kinase 2 (Sphk2) | 873 | 2011 | 0.4 |
| Regulator of G-protein signaling 16 (Rgs16) | 605 | 1915 | 0.3 |
| Heat shock protein (hsp68) | 1716.9/1093/1247 | 6696.1/3695/4177 | 0.25/0.3/0.3 |
| Inhibin β-A (Inhba) | 175 | 698 | 0.25 |
KO, knockout; TGFB, transforming growth factor-β; WT, wild type.
Qualitative and Quantitative Changes in Desmosomal Proteins in the Liver-Specific γ-Catenin KO Mice
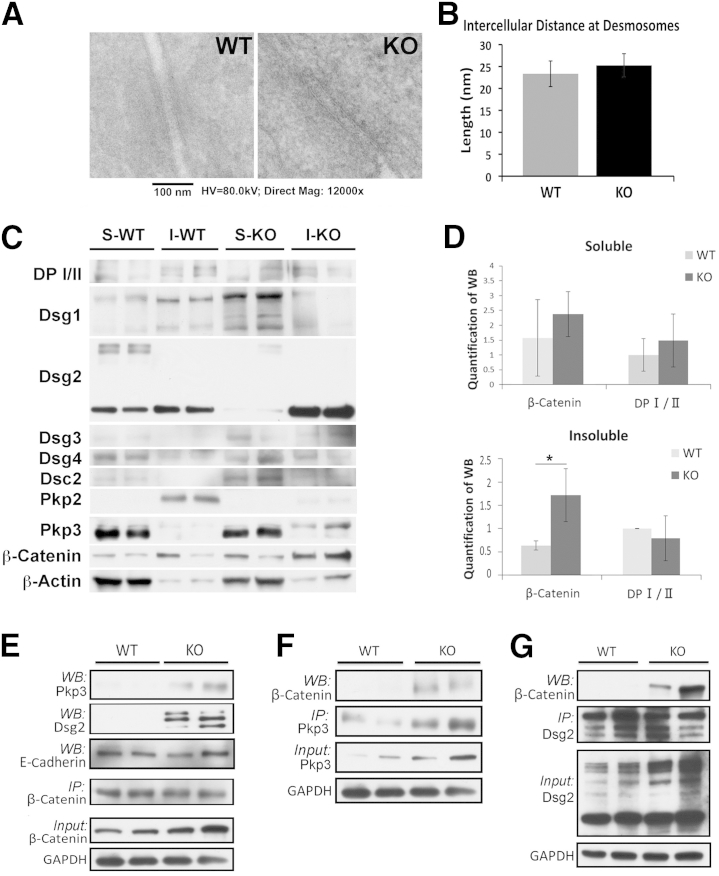
γ-Catenin is predominantly present at desmosomes in epithelial cells, including hepatocytes. Because γ-catenin KO mice lacked any overt phenotype, we next assessed the structure of desmosomes in KO. TEM showed presence of normal desmosome ultrastructure in KO and WT (Figure 2A). Furthermore, intercellular distance at the desmosomes was comparable in KO and WT, suggesting maintenance of adhesion at desmosomes in the absence of γ-catenin (Figure 2B).
Figure 2.
Maintenance of desmosomes in the absence of γ-catenin because of quantitative and qualitative changes in other junctional proteins. A: Transmission electron microscopy of γ-catenin knockout (KO) and wild-type (WT) livers shows the presence of normal desmosomes in both. B: Comparable intercellular distance between hepatocytes at desmosomes in KO and WT livers (mean distance: KO = 25.2 nm; WT = 23.3 nm; P > 0.3). C: Representative Western blot (WB) shows changes in various desmosomal proteins and β-catenin in the soluble (S) and insoluble (I) fractions in WT and KO. Desmoplakin (DP) I (250 kDa), DP II (210 kDa), desmoglein (Dsg) 1 (150 kDa), Dsg2 (59 to 150 kDa), Dsg3 (55 to 130 kDa), Dsg4 (100 to 150 kDa), desmocollin (Dsc) 2 (100 kDa), plakophilin (Pkp) 2 (100 kDa), Pkp3 (87 kDa), and β-catenin (92 kDa). D: Mean integrated OD of a representative WB shown in C for β-catenin and DP I/II in soluble and insoluble fractions. E: Representative immunoprecipitation (IP) studies using β-catenin pull down in whole cell lysates show notably increased association with Dsg2 and Pkp3 in KO compared with WT livers. F: Representative IP study shows Pkp3 pull down and its enhanced association with β-catenin in the KO versus WT liver lysates. G: Representative IP study shows Dsg2 pull down and its enhanced association with β-catenin in the KO versus WT liver lysates. *P < 0.05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HV, high voltage; Mag, magnification.
Microarray data were next queried for expression of genes encoding for various desmosomal proteins. No significant differences were observed in the expression of any desmosomal genes in KO when compared with WT (Table 3). Next, any changes in desmosomal proteins were assessed. Differential detergent fractionation was performed to isolate soluble and insoluble proteins from WT and KO livers and examined by Western blot analyses. Proteins within the desmosomes are mostly noticeable in the insoluble fraction, whereas their presence in the soluble fraction has traditionally been attributed to newly synthesized molecules not yet assembled into plaques, or represent cycling desmosomes.1, 11 In WT liver lysates, Dsg2 and Dsg1 were the two most abundant desmosomal cadherins, and although Dsg1 was mostly present in the insoluble fraction, appreciable Dsg2 was present in both insoluble and soluble compartments (Figure 2C). A notable loss of Dsg2 in soluble fraction and a gain in insoluble fraction were evident in the KO (Figure 2D). Another desmosomal cadherin, Dsc2, was evident at low levels, albeit in only the soluble fraction, which too was elevated in the same fraction in the absence of γ-catenin (Figure 2, C and D).
Table 3.
Changes in Expression of Genes Encoding for Desmosomal Proteins
| Gene name (symbol) | WT | KO | Fold change (WT/KO) |
|---|---|---|---|
| Desmoglein 2 (Dsg2) | 1283/1549 | 1261/1615 | 1.0/0.9 |
| Desmocollin 1 (Dsc1) | 91 | 115 | 0.8 |
| Desmocollin 2 (Dsc2) | 888 | 995 | 0.9 |
| Desmocollin 3 (Dsc3) | 238 | 242 | 1.0 |
| Plakophilin 1 (Pkp1) | 68 | 64 | 1.0 |
| Plakophilin 3 (Pkp3) | 379 | 354 | 1.1 |
KO, knockout; WT, wild type.
Because γ-catenin is a desmosomal plaque protein, we next assessed the expression of the other two major components of the plaques DP in both cellular fractions. No notable changes were evident in DP I/II between KO and WT in any of the two fractions (Figure 2, C and D). Pkp2 was expressed abundantly in the insoluble fraction in the WT livers, and a substantial decrease in the same fraction was observed in KO (Figure 2C). Intriguingly, Pkp3 was expressed at high levels in WT liver, albeit in the soluble fraction. However, in the absence of γ-catenin, Pkp3 was clearly evident in the insoluble fraction as well (Figure 2C).
We also examined β-catenin expression in the two fractions in KO and WT. Although β-catenin was present in both fractions in WT lysates, a clear increase in its levels was observed in the insoluble fraction in the KO (Figure 2, C and D). This is in agreement with previously published studies in γ-catenin–null mice.2
Because we saw an increase in two major desmosomal proteins, Dsg2 and Pkp3, and β-catenin in the insoluble fraction in the KO, we next assessed any interaction among these proteins via immunoprecipitation studies. When pulling down for β-catenin, enhanced association with both Pkp3 and Dsg2 was evident with β-catenin in the γ-catenin KO liver lysates only as compared with WT (Figure 2E). Longer exposure revealed association of Pkp3 and β-catenin in a WT liver (data not shown). No change in β-catenin–E-cadherin association was evident in the KO (Figure 2E). When Pkp3 was immunoprecipitated, enhanced association with β-catenin was verified in KO (Figure 2F). When Dsg2 was pulled down, it also associated with β-catenin in the KO (Figure 2G).
Thus, these observations suggest specific reorganization of desmosome proteins in the absence of γ-catenin, which sufficiently maintained desmosomal integrity and function in the liver at baseline. Furthermore, β-catenin compensates for γ-catenin loss by binding to key desmosomal proteins, without disrupting its function at AJs.
Loss of γ-Catenin in the Liver Results in a More Pronounced Injury after BDL
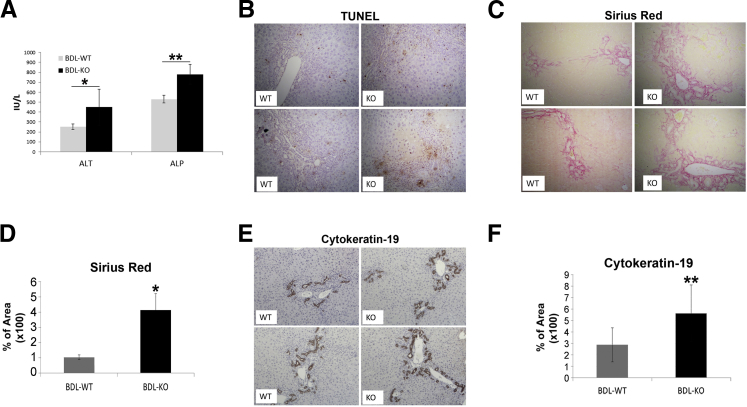
To determine whether γ-catenin KO mice, which show normal desmosomes, albeit with some altered molecular composition, comparably respond to hepatic injury, WT and KO mice were subjected to BDL. Fourteen days after the surgery, mice were sacrificed and examined for serum biochemistry and histology to determine liver functions. There was a significant increase in serum alanine transaminase and alkaline phosphatase in the KO after BDL, indicating greater hepatic injury (Figure 3A), which was further validated by IHC for TUNEL, which was notably higher in KO after BDL (Figure 3B). Furthermore, Sirius red staining for collagen deposition as an indicator of hepatic fibrosis was more pronounced in the KO at 14 days after BDL (Figure 3C). Quantification of Sirius red staining verified a significant increase in fibrosis in KO (Figure 3D). Because ductular reaction is a hallmark of cholestasis, we next determined CK19-positive ductules after BDL. KO showed notably higher numbers of CK19-positive ductules after BDL than WT (Figure 3E), and the difference was statistically significant (Figure 3F). Thus, γ-catenin KO mice are more susceptible to injury after BDL than WT.
Figure 3.
Increased injury in γ-catenin knockout (KO) mice after bile duct ligation (BDL). A: Serum alanine transaminase (ALT) and alkaline phosphatase (ALP) show a significant increase in KO mice after BDL. [Mean ALT for KO = 448.8 IU/L, wild type (WT) = 251.3 IU/L; mean ALP for KO mice = 780.6 IU/L, WT = 530.2 IU/L.] B: Immunohistochemistry (IHC) for terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) shows more cell death in KO liver after BDL. C: Sirius red staining shows greater collagen deposition in KO livers after BDL. D: Quantification of Sirius red staining shows greater area stained in KO livers after BDL (WT = 1.0%, KO = 4.1%). E: IHC for CK19 shows a greater biliary ductular reaction in KO livers after BDL. F: Quantification of CK19 staining shows a significant increase in CK19-positive ducts in KO livers compared with WT after BDL (KO = 5.6%, WT = 2.9%). *P < 0.05, **P < 0.01.
Decompensation at Desmosomes after BDL in the Absence of γ-Catenin after BDL
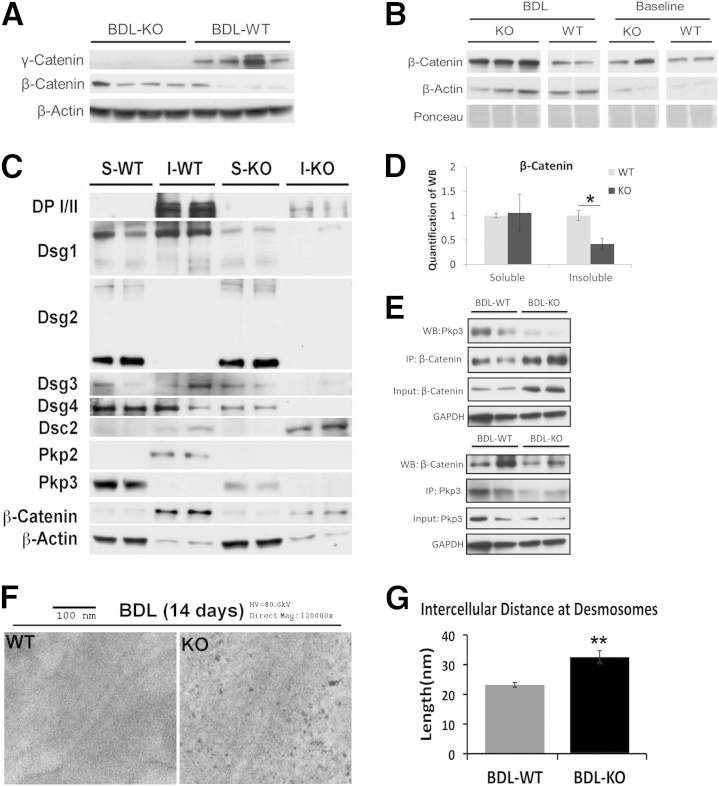
To address the mechanism of enhanced hepatobiliary injury in the KO after BDL, we examined various molecules that were originally altered in the KO to compensate for desmosomal structure and function. KO continued to lack γ-catenin after BDL, while expressing β-catenin (Figure 4A). In fact, an increase in β-catenin in the KO was more pronounced than WT after BDL (Figure 4B).
Figure 4.
Disparate changes in desmosomal proteins in knockout (KO) compared with wild type (WT) after bile duct ligation (BDL). A: Representative Western blot (WB) shows a notable increase in total levels of β-catenin in KO livers after BDL, whereas γ-catenin continues to be absent in the KO. B: Another representative WB shows a basal and BDL-induced increase in total β-catenin in KO compared with WT at baseline and after BDL. The samples were run in triplicate for KO BDL and in duplicate for all other groups. The samples in WT BDL have been spliced together. C: Representative WB shows changes in desmosome proteins and β-catenin in both soluble (S) and insoluble (I) fractions of WT and KO livers after BDL. D: Mean integrated OD of a representative WB shown in C for β-catenin in soluble and insoluble fractions. E: Representative immunoprecipitation (IP) shows a decrease in β-catenin– plakophilin (Pkp) 3 association in KO livers after BDL. Top panel: Pull down of β-catenin and WB for Pkp3. Bottom panel: IP of Pkp3 and WB for β-catenin using whole cell lysates. F: Representative transmission electron micrographs of KO and WT livers after BDL show a normal desmosome in WT compared with a loose desmosome in KO. G: Quantification of the intercellular distance at desmosomes after BDL in WT versus KO shows a significantly greater average distance in KO (average distance KO = 32.6 nm, WT = 23.2 nm). *P < 0.05, **P < 0.01. DP, desmoplakin; Dsc, desmocollin; Dsg, desmoglein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HV, high voltage; Mag, magnification.
Detergent fractionation was next performed on BDL samples. The analysis presented herein is dual. First, we assessed any changes in desmosomal proteins in WT after BDL when compared with baseline and contrasted them from changes occurring in KO before and after BDL. We found, in the insoluble fractions in WT after BDL, notable increases in DP I/II and Dsg1 and a decrease in Pkp2 (Figures 2C and 4C). In KO, there was a lack of increase in all these proteins in the same fractions. Second, we assessed after BDL the fate of compensatory changes that were evident in desmosomal proteins in KO at baseline. There was a notable loss of both Dsg2 and Pkp3 in the insoluble fractions after BDL in the KO (Figures 2C and 4C). Intriguingly, Dsc2 levels were increased in the insoluble fraction after BDL in both WT and KO, but more so in KO (Figure 4C). Finally, β-catenin levels in insoluble fractions that were increased normally in WT livers after BDL were not apparent in the KO (Figures 2C and 4, C and D).
To further clarify the relevance of these alterations, we next determined the fate of interactions that were apparent in KO at baseline. β-Catenin interacted with the desmosomal plaque protein Pkp3 in γ-catenin KO (Figure 2E). These proteins were predominantly present in the insoluble fractions of lysates in KO (Figure 2C). Intriguingly, a notable diminution in β-catenin–Pkp3 association was evident in KO after BDL, which coincided especially with the lack of Pkp3 in the insoluble fraction of KO after BDL (Figure 4E). Interestingly, an increase in β-catenin–Pkp3 association was evident in WT after BDL.
To directly assess if alterations in desmosomal proteins in KO after BDL affected its structure, TEM was performed. Although maintenance of desmosomal ultrastructure was evident in WT after BDL, loosening of desmosomal plaque was evident in KO (Figure 4F). This was associated with a significant increase in the intercellular distance at desmosomes (Figure 4G), validating loss of structural integrity in KO after injury because of enhanced intrahepatic biliary pressure.
Enhanced Hepatocarcinogenesis in γ-Catenin KO Mice Exposed to Chemical Carcinogen
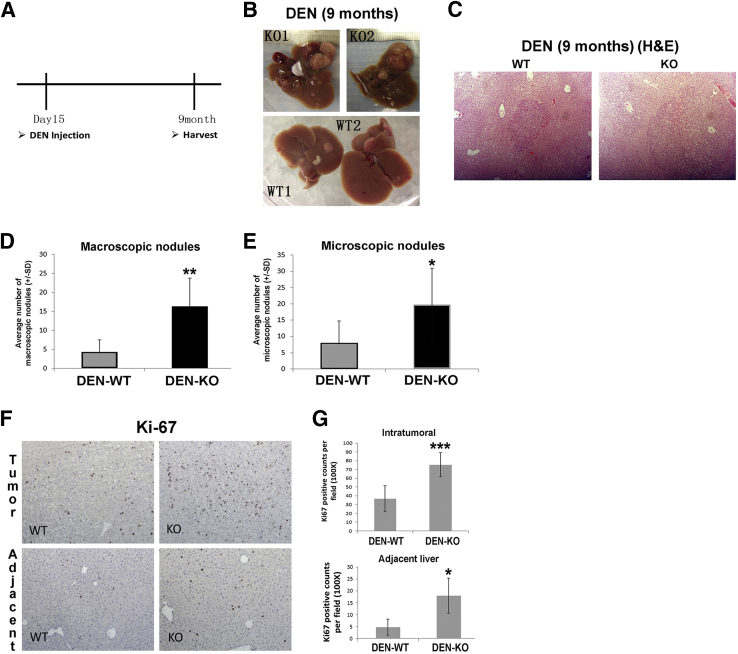
Because of its ambiguous role in carcinogenesis, we next assessed the role of γ-catenin in hepatic tumorigenesis. We subjected KO (n = 5) and WT (n = 7) males to a single i.p. injection of 5 μg/g body weight DEN at 15 days after birth and sacrificed at 9 months for assessing tumor burden (Figure 5A). None of the non–DEN-injected KO (n = 5) or WT (n = 4) male mice showed any evidence of tumorigenesis at 9 months (data not shown). However, KO exposed to DEN showed a notable increase in gross hepatic tumorigenesis compared with WT (Figure 5B). H&E on liver sections showed several microscopic tumor foci in DEN-injected KO and WT (Figure 5C). Numbers of macroscopic tumor nodules counted showed a significantly higher gross tumor burden in the KO (Figure 5D). Because at least four lobes from each WT and KO animal were represented on each slide, H&E sections were scanned for all microscopic tumor nodules (magnification, ×50), which were counted, averaged, and compared between the two groups. This analysis also revealed that KO had significantly higher microscopic tumor burden than WT (Figure 5E). Finally, these sections were stained for Ki-67, an indicator of cells in the cell cycle. Indeed, a greater number of Ki-67–positive cells were observed in KO than WT (Figure 5F). In fact, significantly higher numbers of Ki-67–positive cells were seen in KO after DEN in both intratumoral and extratumoral areas (Figure 5G). Thus, lack of γ-catenin promoted chemical carcinogenesis in the livers of γ-catenin KO mice, although no spontaneous increase in tumorigenesis was observed.
Figure 5.
Enhanced liver tumors in γ-catenin knockout (KO) mice after diethylnitrosamine (DEN) exposure. A: Scheme depicting DEN-induced carcinogenesis protocol used in the study. B: Gross images of livers showing macroscopic tumor nodules in 9-month-old KO and wild-type (WT) livers that were given a single injection of DEN at postnatal day 15. C: Representative hematoxylin and eosin (H&E) images from DEN-treated KO and WT livers showing microscopic tumor nodules. D: Quantification of macroscopic nodules shows a significantly higher tumor burden in KO compared with WT after DEN (average: KO = 12.2, WT = 4.3). E: Quantification of microscopic nodules shows significantly more foci in KO livers after DEN injection compared with WT (average: KO = 19.8, WT = 8). F: Representative immunohistochemical staining for Ki-67 shows notably more positive cells in both tumor tissue and adjacent hepatic tissue in KO compared with WT. G: Quantification of Ki-67 staining shows significantly more positive cells in both tumor and adjacent tissue in KO compared with WT (average in tumor: KO = 76.4, WT = 36.5; average in adjacent tissue: KO = 18, WT = 4.8). *P < 0.05, **P < 0.01, and ***P < 0.001. Original magnification, ×50 (C).
Enhanced HCC in γ-Catenin KO Is Partially Because of β-Catenin Activation
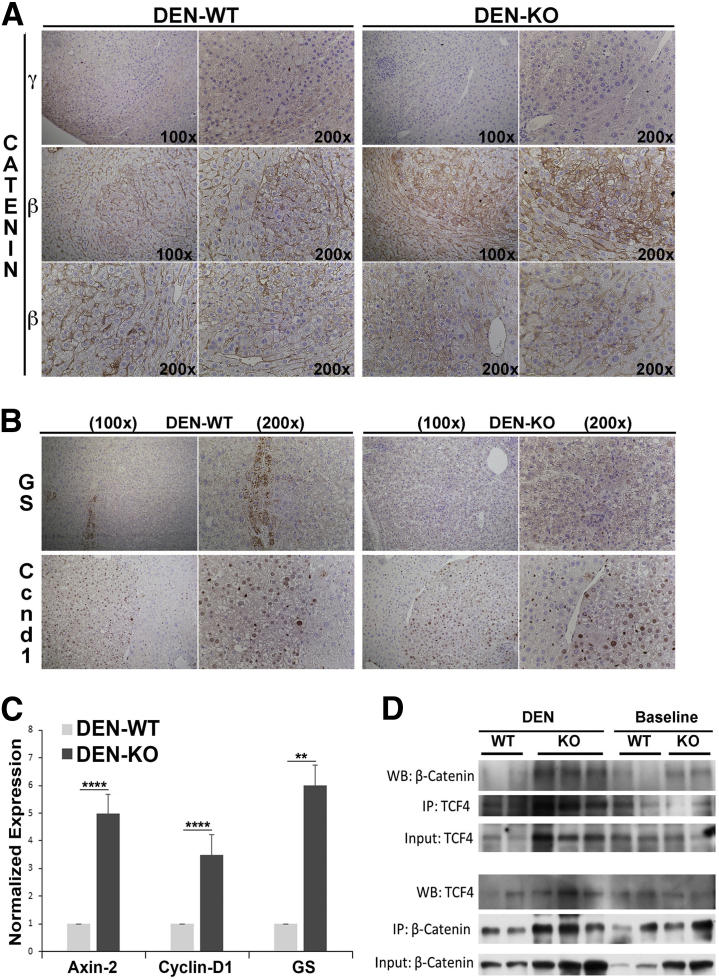
To try and address the mechanism of enhanced tumorigenesis in KO after DEN, we first examined γ-catenin expression in the tumors in WT and KO. Every tumor in WT lacked any staining for γ-catenin, whereas some expression was evident in the hepatocytes surrounding the tumor nodule by IHC (Figure 6A). As expected, neither the tumor nodules nor the adjacent area was γ-catenin positive in KO (Figure 6A). Next, we examined expression of β-catenin, a known oncogene, in the tumors. IHC showed that most of the tumors in WT retained only membranous β-catenin, although a subset showed mild cytoplasmic staining of occasional tumor cells (Figure 6A). In fact, approximately 35% of all tumor nodules in γ-catenin KO showed several cells within tumor nodules, with cells displaying clear cytoplasmic and/or nuclear β-catenin (Figure 6A). Thus, there was both intratumoral and intertumoral heterogeneity in tumors in KO in regards to β-catenin redistribution, as identified by IHC (Figure 6A).
Figure 6.
Enhanced tumor development in γ-catenin knockout (KO) exposed to diethylnitrosamine (DEN) is partially because of greater β-catenin activation. A: Immunohistochemistry (IHC) for γ-catenin shows reduced staining in tumor nodules in wild-type (WT) livers compared with adjacent liver, whereas tumors and adjacent liver are completely negative in KO. IHC for β-catenin shows tumor to be composed of cells with membranous β-catenin only. However, a notable heterogeneity is evident in tumors observed in KO, with a subset of tumors composed of cells with clear cytoplasmic and nuclear β-catenin and others with predominant membranous β-catenin. B: IHC for glutamine synthetase (GS) shows no GS-positive nodules in WT or KO livers. However, IHC for cyclin-D1 shows equally strong staining in nodules in both KO and WT livers. C: Real-time PCR shows a significant increase in mRNA expression of axin-2, cyclin-D1, and GS in DEN-treated, tumor-bearing KO compared with DEN-treated, tumor-bearing WT livers. D: Representative immunoprecipitation (IP) of β-catenin and T-cell factor (TCF) 4 in whole cell lysates of baseline WT and KO livers shows a notable increase in association with DEN-treated, tumor-bearing KO compared with DEN-treated, tumor-bearing WT livers. **P < 0.01, ****P < 0.0001. WB, Western blot.
To further determine whether β-catenin signaling is active in the tumors in KO livers, we looked at the expression of some known β-catenin targets. GS is a surrogate low-sensitivity, high-specificity β-catenin target that has been shown to be a biomarker for activating CTNNB1 mutations.13, 14, 15 None of the tumors in either WT or KO were GS positive (Figure 6B). This was also true for tumors in KO that showed clear nuclear/cytoplasmic β-catenin. Cyclin-D1 is another liver-specific β-catenin target,5, 9 although β-catenin–independent regulation of its expression is also known.16 Intriguingly, all tumors in both WT and KO were strongly cyclin-D1 positive (Figure 6B). To assess β-catenin signaling quantitatively, we isolated RNA from tumor-bearing WT and KO livers and assessed them for known Wnt targets by real-time PCR. A significant increase in the gene expression of Axin-2, Cyclin-D1, and GS was observed in the KO compared with WT (Figure 6C). To further determine whether β-catenin signaling was active in γ-catenin KO after DEN treatment, whole cell lysates from tumor-bearing KO and WT livers were examined for binding of β-catenin to TCF4, a known transcription factor that regulates, binds, and activates Wnt target genes. A notably higher β-catenin–TCF4 association was observed in DEN-treated, tumor-bearing KO compared with DEN-treated, tumor-bearing WT mice (Figure 6D).
Taken together, the data demonstrate enhanced β-catenin signaling in a subset of tumors occurring in the livers of DEN-treated γ-catenin KO mice.
γ-Catenin Suppression in Vitro Enhances β-Catenin Signaling in Liver Tumor Cells
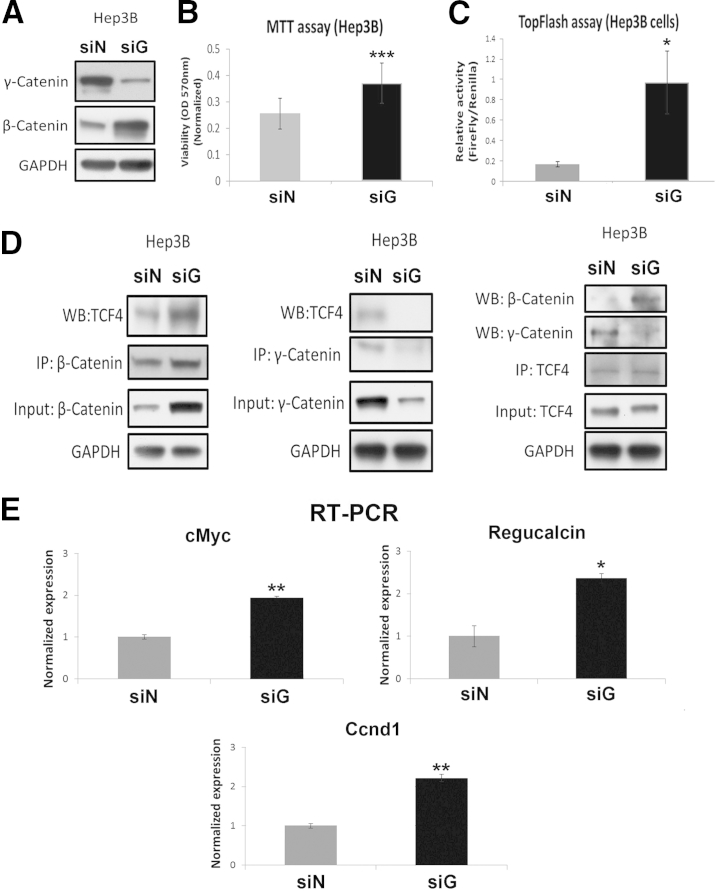
To further ascertain the γ-catenin–β-catenin relationship in β-catenin–TCF4 signaling and HCC, we performed in vitro analysis in Hep3B cells. siRNA mediated knockdown of γ-catenin in Hep3B cells. As also shown previously, this led to a notable increase in β-catenin protein (Figure 7A). Simultaneously, an increase in Hep3B cell viability was evident, as assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide or MTT assay (Figure 7B).
Figure 7.
In vitro suppression of γ-catenin in hepatocellular cancer cells leads to enhanced β-catenin activity. A: Representative Western blot (WB) shows successful knockdown of γ-catenin at 48 hours after γ-catenin siRNA (siG) compared with negative control siRNA (siN) transfection of Hep3B cells. A notable increase in β-catenin levels was evident in siG-transfected cells. B: MTT assay shows increased Hep3B cell viability at 48 hours after siG transfection compared with siN. C: TopFlash assay shows a significant increase in T-cell factor (TCF) activity in Hep3B cells at 48 hours after siG transfection (ratio of firefly/Renilla: siG = 0.97, siN = 0.17). D: Representative immunoprecipitation (IP) shows increased association of β-catenin and TCF4 in Hep3B cells after 48 hours of siG transfection compared with siN. γ-Catenin–TCF4 association is evident basally in siN-transfected Hep3B cells, which is abrogated after 48 hours’ transfection with siG. E: Real-time PCR shows a significant increase in mRNA expression of c-Myc, regucalcin, and cyclin-D1 (Cnnd1) in Hep3B cells in the siG-transfected group. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To establish if β-catenin increase after γ-catenin knockdown translates to increased activation, we next assessed γ-catenin–TCF4 activity by TopFlash reporter. Indeed, γ-catenin suppression in Hep3B cells led to a significant increase in TopFlash reporter assay (Figure 7C). Indeed, enhanced β-catenin–TCF4 complex was visible in Hep3B cells after γ-catenin siRNA transfection compared with control siRNA transfection (Figure 7D). There was also a concomitant decrease in γ-catenin–TCF4 association after γ-catenin knockdown. Last, when TCF4 was pulled down, there was a basal γ-catenin–TCF4 association in Hep3B cells, which, on γ-catenin knockdown, led to an enhanced β-catenin–TCF4 association at the expense of the γ-catenin–TCF4 complex. Finally, we assessed β-catenin target gene expression after γ-catenin silencing in Hep3B cells. There was a significant increase in the mRNA expression of c-Myc, cyclin-D1, and regucalcin after γ-catenin suppression (Figure 7E).
Thus, there is a γ-catenin, TCF4, and β-catenin dynamic in HCC cells, which regulates β-catenin transactivation and has implications in tumor biology.
γ-Catenin Overexpression Suppresses β-Catenin Activity and Target Genes
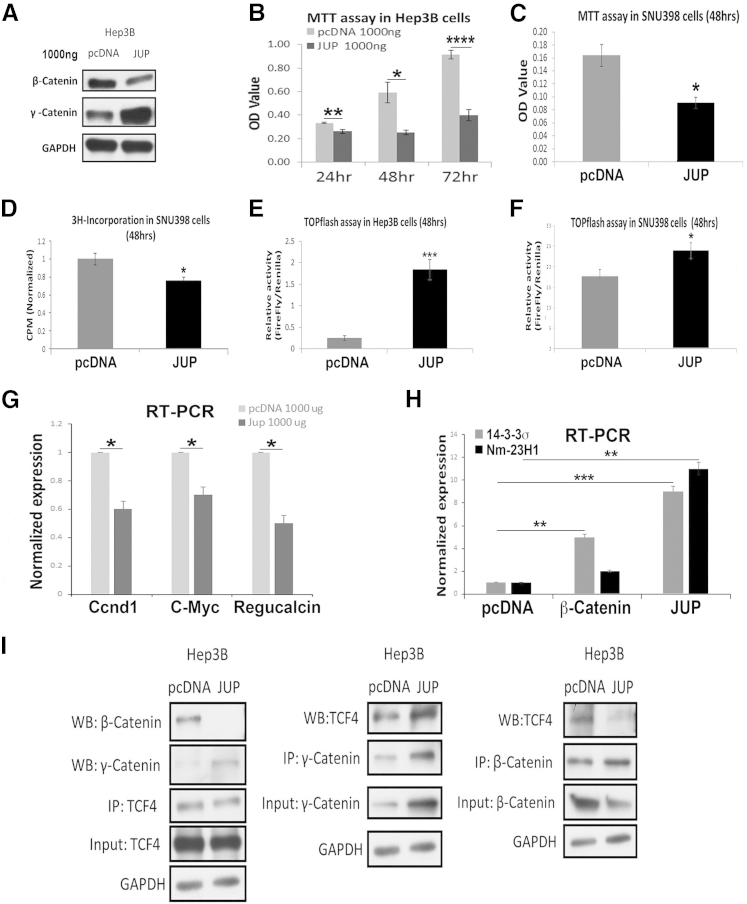
To further ascertain the dynamic interactions between β-catenin, γ-catenin, and TCF4, we next overexpressed γ-catenin in HCC cell lines. γ-Catenin gene or Jup was cloned in pcDNA3 plasmid (pcDNA) and transiently transfected in Hep3B cells. Jup overexpression led to a notable decrease in β-catenin protein compared with pcDNA-only transfected Hep3B cells (Figure 8A). Simultaneously, a significant decrease in cell viability by MTT assay was observed on Jup overexpression (Figure 8B). To determine whether γ-catenin overexpression can similarly affect β-catenin activation and downstream biological activity in more than one cell line, we overexpressed Jup in HCC cells that harbor active β-catenin because of a point mutation in exon 3 of the CTNNB1 gene. As in Hep3B cells, overexpression of γ-catenin in Snu-398 HCC cells decreased cell viability, as seen by MTT assay (Figure 8C), and proliferation, as seen by thymidine incorporation (Figure 8D).
Figure 8.
Overexpression of γ-catenin in hepatocellular cancer cells suppresses β-catenin activity to reduce cell viability and proliferation. A: Representative Western blot (WB) shows the efficiency of γ-catenin plasmid (Jup) transfection (1000 ng, 48 hours) compared with pcDNA plasmid only. A notable decrease in β-catenin protein is evident in Jup-transfected cells. B: MTT assay shows a significant decrease in viability of Hep3B cells at 24, 48, and 72 hours after Jup overexpression. C: MTT assay shows decreased viability of Snu-398 cells after γ-catenin plasmid transfection at 48 hours. D: Thymidine incorporation assay shows decreased cell proliferation in γ-catenin plasmid transfection group compared with pcDNA group in Snu-398 cells. E: TopFlash assay shows increased T-cell factor (TCF) activity in Hep3B cells after Jup overexpression at 48 hours (ratio of firefly/Renilla: pcDNA = 0.25, JUP = 1.84). F: TopFlash assay shows increased TCF4 activity in Snu-398 cells after γ-catenin plasmid transfection at 48 hours (ratio of firefly/Renilla: pcDNA 1 μg = 17.8, JUP 1 μg = 23.9). G: Real-time PCR shows a significant decrease in mRNA expression of cyclin-D1 (Cnnd1), c-Myc, and regucalcin in Hep3B cells at 48 hours after Jup overexpression. H: Real-time PCR shows a significant increase in mRNA expression of 14-3-3σ and nonmetastatic protein 23 H1 (Nm-23H1) in Hep3B cells at 48 hours after γ-catenin plasmid transfection. However, Hep3B cells transfected with β-catenin plasmid cause a modest increase in 14-3-3σ but no change in Nm-23H1 mRNA levels. I: Representative immunoprecipitation (IP) shows loss of β-catenin–TCF4 association after Jup overexpression at 48 hours in Hep3B, whereas there is a concomitant increase in γ-catenin–TCF4 association in these cells at the same time. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To directly assess if β-catenin transactivational activity was impeded by γ-catenin overexpression, we performed TopFlash reporter assay on lysates from Hep3B and Snu-398 cells transiently transfected with either Jup or pcDNA plasmids. This is because of the fact that γ-catenin also binds TCF4 to stimulate its transcriptional activity and can be measured by TopFlash reporter.17 Unexpectedly, a significant increase in TopFlash reporter activity was evident after γ-catenin overexpression in both cell types (Figure 8, E and F).
To determine the basis of this paradoxical observation, we next focused on elucidating the mechanism in Hep3B cells. First, we directly determined the impact of γ-catenin–TCF4 activation on β-catenin target gene expression after Jup overexpression. Not only did β-catenin targets not increase after γ-catenin overexpression, as expected by the observation of increased TopFlash activity, there was actually a significant decrease in the mRNA expression of cyclin-D1, c-Myc, and regucalcin, when compared with pcDNA transfection (Figure 8G). This suggested that, despite increased TopFlash reporter activity with γ-catenin overexpression, β-catenin transactivational activity is actually decreased. To begin and address the disconnect between enhanced TopFlash activity and decrease in β-catenin target gene expression, we next assessed for expression of genes that have been shown to be more closely regulated by γ-catenin.18 Indeed, a notable increase in the expression of both 14-3-3σ and nonmetastatic protein 23 H1 (Nm-23H1) was evident after Jup overexpression compared with either control plasmid or β-catenin overexpression in Hep3B cells (Figure 8H).
To more conclusively corroborate the differential interactions of TCF4 with γ-catenin at the expense of β-catenin after Jup overexpression, we directly assessed Jup or pcDNA plasmid transfected Hep3B cells for association of TCF4 to γ-catenin and β-catenin. A notably greater γ-catenin–TCF4 association, which came at the expense of the β-catenin–TCF4 association, was evident after Jup overexpression in Hep3B cells (Figure 8I).
Discussion
Although the role of β-catenin, an important component of the AJ and the canonical Wnt signaling pathway, is well understood in various aspects of liver pathobiology, there is little information on the role of γ-catenin. Similarly, the role of desmosomes, which are the primary location of γ-catenin in epithelial cells, is not well understood in the liver as well. γ-Catenin belongs to the catenin family of proteins with a notable homology to β-catenin,17 and can sometimes replace β-catenin for its function in the Wnt signaling pathway.19 We generated liver-specific γ-catenin KO mice to examine its role in hepatic pathophysiology. At baseline, we saw no notable differences in either global gene expression or specifically Wnt signaling, which regulates centrizonal gene expression in an adult liver.5 This suggests minimal participation of γ-catenin in any major signaling mechanism in the liver.
Although there is significant information on the structure and function of desmosomes,1 little is known about these in a normal liver. We show that normal desmosomal proteins in the insoluble fractions of liver include desmosomal cadherins Dsg1 and Dsg2 and plaque proteins γ-catenin, Pkp2, and DP I/II. Intriguingly, there were appreciable levels of desmosomal cadherin Dsg2 in the soluble fraction as well, whereas Dsc2 and plaque protein Pkp3 were exclusively in the soluble fractions in a normal liver. Although the significance of these observations is unclear, it is likely that the soluble compartment may represent a desmosome-independent pool or cycling desmosomes.11 Interestingly, Pkp3 was shown earlier to be not expressed in the liver.20 However, we not only identified Pkp3 in a normal liver, but it was also found to complex with β-catenin, albeit at low levels. This is not entirely surprising because Pkps, although primarily desmosomal in location, are known to also interact with AJ proteins. For example, Pkp2 associated with β-catenin when it was ectopically introduced into cultured keratinocytes.21 The role and regulation of Pkp3–β-catenin complex in a normal liver definitely deserves additional investigation.
However, in the absence of γ-catenin in the liver, there was a significant change in the composition of desmosome that maintained its structure and function at baseline. Not only did the total levels of β-catenin, Dsg2, and Pkp3 increase in the insoluble fraction in KO, but also there was a clear association among them. Indeed, the presence of β-catenin at desmosome in the absence of γ-catenin has been reported earlier in tissues like skin.2 Also, deletion of desmosome proteins has been shown to cause anomalous mixing of junctions.22 β-Catenin–E-cadherin association remained intact in the γ-catenin KO livers. Dsg2 is reported to maintain tight junction integrity and the epithelial barrier in the intestine.23 Pkp3 has been shown to promote plaque formation and cell adhesion24, 25 and can also inhibit tumor progression and metastasis.26 Thus, β-catenin–Pkp3/Dsg2 complexes in KO may be a result of junctional intermingling, suggesting an important role of γ-catenin in proper junctional sorting, as has been shown in global γ-catenin KO with cardiac defects.27 Although the compensations in the absence of γ-catenin appear sufficient to maintain desmosome structure and function at baseline, more in-depth studies will be essential to address the primary role of proteins, like Dsg2 and Pkp3, in liver pathophysiology and in the context of γ-catenin loss.
Ligation of the extrahepatic bile duct in BDL model causes cholestasis as bile accumulates in the intrahepatic bile ducts to induce backpressure and shear stress, leading to injury and fibrosis.10 This procedure is known to bring about changes in tight junctions and causes an increase in desmosomes, but has no impact on gap junctions.8 However, a detailed analysis on desmosomes after BDL has not been performed. We observed increased levels of DP I/II, Dsg1, Dsc2, and even β-catenin and decreased levels of Dsg2 and Pkp2 in the insoluble fractions after BDL in a control animal. These changes may represent extensive hepatic remodeling as liver undergoes injury and repair. Interestingly, we observed an increase in β-catenin–Pkp3 association in a WT liver after BDL. Because these two proteins were commonly present only in the insoluble fraction after BDL, this association may represent desmosomal turnover or formation of hybrid junctions. Recently, mixed junctions, also called area composita, have been observed in normal cardiac myocytes.28 Future studies will address if formation of such hybrid junctions in liver after injury is a more global response with biological implications. In the heart, Pkp2 has been shown to interact with α-catenin at these hybrid junctions to assist with sorting of various junctional proteins. It is possible that Pkp3, like Pkp2 and, in addition to γ-catenin, may play a role in junctional sorting, especially after BDL-induced hepatic remodeling.
In the absence of γ-catenin, KO mice showed a more extensive injury after BDL. In fact, there was a definite loss of desmosomal structure, as indicated by increased intercellular distance at desmosomes. This appears to be because of failure of baseline compensatory changes at desmosomes, as reflected by loss of Dsg2, Pkp3, and β-catenin in insoluble fractions and their interactions after BDL. This is also explained by failure in KO of increases in desmosomal proteins that normally occurred in the insoluble fractions in WT after BDL, including DP I/II, Dsg1, and even β-catenin. This highlights a nonredundant role of γ-catenin in junctional sorting, especially during acute liver injury, as was visible in γ-catenin global KO.27
The changes in desmosomal structural proteins are traditionally associated with cardiac and dermal diseases with both genetic and autoimmune bases. Specifically, γ-catenin truncation in its C-terminal is associated with Naxos disease.6, 7, 29 However, no hepatic pathology is associated with γ-catenin aberrations thus far, other than preliminary observations in liver cancer.30, 31 However, other junctional anomalies, especially in tight junction proteins, have recently been associated with cholestatic liver diseases, like progressive familial intrahepatic cholestasis.32 Because γ-catenin loss led to a more extensive hepatic injury during cholestasis, it will be relevant to characterize desmosomal proteins in cholestatic liver diseases as possible disease initiators or disease modifiers.
The role of γ-catenin and desmosomes in tumorigenesis is less well understood.33 In various tumors, loss of desmosomal proteins is associated with enhanced tumorigenesis or tumor metastasis. Loss of Pkps is associated with increased tumor cell invasion in colorectal cancers26 and prostate adenocarcinoma.34 Loss of Dsc-2 is associated with enhanced tumorigenicity of colorectal cancer through β-catenin activation.35 The role of γ-catenin in tumorigenesis is more perplexing. This stems from its role in desmosomes and Wnt signaling.17 γ-Catenin has been shown to promote rat kidney epithelial cell transformation.36 However, loss of γ-catenin promotes prostate cancer growth.37 In HCC, both increase and loss of γ-catenin expression has been reported.30, 31 We demonstrate a tumor suppressive role of γ-catenin in HCC such that its loss from the liver did not yield spontaneous tumors, but led to enhanced tumorigenesis in response to a chemical carcinogen. Furthermore, in vivo and in vitro analysis suggests that at least a partial mechanism of these observations is activation of Wnt signaling, as reflected by enhanced expression of various Wnt targets in the tumor-bearing livers from DEN-treated γ-catenin KO mice.
The role of γ-catenin in Wnt signaling, a known oncogenic pathway, is ambiguous. γ-Catenin can act as both a positive and a negative regulator of the Wnt signaling.33 In the current study, loss of γ-catenin in hepatocytes led to increased activation of β-catenin in vitro and in vivo. In fact, knockdown of γ-catenin in HCC cells led to enhanced TCF activity and increase in expression of several β-catenin targets. This occurred secondary to greater β-catenin–TCF4 association at the expense of γ-catenin–TCF4 complex, suggesting that the latter may be acting as a dominant-negative for the Wnt signaling.
Intriguingly, γ-catenin overexpression in HCC cells (including one that has activating β-catenin gene mutations) led to enhanced TopFlash reporter activity. This seemingly paradoxical observation, when characterized further, did in fact verify γ-catenin overexpression to reduce β-catenin–TCF4 transactivation, as reflected by down-regulation of β-catenin gene targets and associated biological activity. It has been suggested previously that γ-catenin binds TCF4 and stimulates gene expression, albeit to a lesser extent.38 Others have shown γ-catenin to bind TCF4 adjacent to β-catenin, thus preventing binding of TCF4 to DNA.39 However, increased TopFlash activity after γ-catenin overexpression does suggest that γ-catenin–TCF4 complex is transcriptionally active and may itself promote expression of certain target genes. What may be the downstream targets of γ-catenin in HCC cells that lead to decreased cell viability and decreased cell proliferation or if it is solely because of β-catenin suppression remains under investigation. Although c-Myc has been shown to be a more sensitive target of γ-catenin,40 we found it to be down-regulated after γ-catenin overexpression in HCC cells. Previous studies have also reported a few additional γ-catenin–specific targets, such as 14-3-3σ and Nm-23, in other cell types.41, 42 The expression levels of both 14-3-3σ and Nm-23H1 were significantly increased on γ-catenin overexpression compared with β-catenin. Although a modest increase in 14-3-3σ gene expression was also evident after β-catenin overexpression, Nm-23H1 was exclusively regulated by γ-catenin in Hep3B cells. Together, these observations suggest that, although both catenins are capable of binding to TCFs, γ-catenin–TCF4 is functionally weaker than β-catenin–TCF4 complex, but more important, regulates a repertoire of genes that are distinct from those regulated by β-catenin–TCF4 complex.
Thus, γ-catenin appears to have tumor suppressive function, which could also have therapeutic implications because its overexpression in HCC cells with β-catenin gene mutations, an event observed in approximately 30% of all HCCs in patients,14, 15 led to a considerable decrease in tumor cell proliferation and viability. Thus, agents that may enhance γ-catenin expression could be useful to mop TCF4, making it unavailable for binding to β-catenin and hence curb Wnt signaling for treatment of a subset of HCCs.
Footnotes
Supported by NIH grants 1R01DK62277 and 1R01DK100287 and Endowed Chair for Experimental Pathology (S.P.M.) and in part by 5T32DK63922-13 (T.P.-S.). L.Z. was also supported by China Scholarship Council.
Disclosures: None declared.
References
- 1.Delva E., Tucker D.K., Kowalczyk A.P. The desmosome. Cold Spring Harb Perspect Biol. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierkamp C., Schwarz H., Huber O., Kemler R. Desmosomal localization of beta-catenin in the skin of plakoglobin null-mutant mice. Development. 1999;126:371–381. doi: 10.1242/dev.126.2.371. [DOI] [PubMed] [Google Scholar]
- 3.Wickline E.D., Awuah P.K., Behari J., Ross M., Stolz D.B., Monga S.P. Hepatocyte gamma-catenin compensates for conditionally deleted beta-catenin at adherens junctions. J Hepatol. 2011;55:1256–1262. doi: 10.1016/j.jhep.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickline E.D., Du Y., Stolz D.B., Kahn M., Monga S.P. gamma-Catenin at adherens junctions: mechanism and biologic implications in hepatocellular cancer after beta-catenin knockdown. Neoplasia. 2013;15:421–434. doi: 10.1593/neo.122098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan X., Behari J., Cieply B., Michalopoulos G.K., Monga S.P. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 6.Li D., Liu Y., Maruyama M., Zhu W., Chen H., Zhang W., Reuter S., Lin S.F., Haneline L.S., Field L.J., Chen P.S., Shou W. Restrictive loss of plakoglobin in cardiomyocytes leads to arrhythmogenic cardiomyopathy. Hum Mol Genet. 2011;20:4582–4596. doi: 10.1093/hmg/ddr392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D., Zhang W., Liu Y., Haneline L.S., Shou W. Lack of plakoglobin in epidermis leads to keratoderma. J Biol Chem. 2012;287:10435–10443. doi: 10.1074/jbc.M111.299669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robenek H., Herwig J., Themann H. The morphologic characteristics of intercellular junctions between normal human liver cells and cells from patients with extrahepatic cholestasis. Am J Pathol. 1980;100:93–114. [PMC free article] [PubMed] [Google Scholar]
- 9.Nejak-Bowen K.N., Thompson M.D., Singh S., Bowen W.C., Jr., Dar M.J., Khillan J., Dai C., Monga S.P. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant beta-catenin. Hepatology. 2010;51:1603–1613. doi: 10.1002/hep.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezure T., Sakamoto T., Tsuji H., Lunz J.G., 3rd, Murase N., Fung J.J., Demetris A.J. The development and compensation of biliary cirrhosis in interleukin-6-deficient mice. Am J Pathol. 2000;156:1627–1639. doi: 10.1016/s0002-9440(10)65034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer B., Tucker G.C., Valles A.M., Franke W.W., Thiery J.P. Rearrangements of desmosomal and cytoskeletal proteins during the transition from epithelial to fibroblastoid organization in cultured rat bladder carcinoma cells. J Cell Biol. 1989;109:1495–1509. doi: 10.1083/jcb.109.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wack K.E., Ross M.A., Zegarra V., Sysko L.R., Watkins S.C., Stolz D.B. Sinusoidal ultrastructure evaluated during the revascularization of regenerating rat liver. Hepatology. 2001;33:363–378. doi: 10.1053/jhep.2001.21998. [DOI] [PubMed] [Google Scholar]
- 13.Bioulac-Sage P., Rebouissou S., Thomas C., Blanc J.F., Saric J., Sa Cunha A., Rullier A., Cubel G., Couchy G., Imbeaud S., Balabaud C., Zucman-Rossi J. Hepatocellular adenoma subtype classification using molecular markers and immunohistochemistry. Hepatology. 2007;46:740–748. doi: 10.1002/hep.21743. [DOI] [PubMed] [Google Scholar]
- 14.Cieply B., Zeng G., Proverbs-Singh T., Geller D.A., Monga S.P. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology. 2009;49:821–831. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zucman-Rossi J., Benhamouche S., Godard C., Boyault S., Grimber G., Balabaud C., Cunha A.S., Bioulac-Sage P., Perret C. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 16.Klein E.A., Assoian R.K. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci. 2008;121:3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhurinsky J., Shtutman M., Ben-Ze'ev A. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J Cell Sci. 2000;113(Pt 18):3127–3139. doi: 10.1242/jcs.113.18.3127. [DOI] [PubMed] [Google Scholar]
- 18.Aktary Z., Pasdar M. Plakoglobin: role in tumorigenesis and metastasis. Int J Cell Biol. 2012;2012:189521. doi: 10.1155/2012/189521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhurinsky J., Shtutman M., Ben-Ze'ev A. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol Cell Biol. 2000;20:4238–4252. doi: 10.1128/mcb.20.12.4238-4252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt A., Langbein L., Pratzel S., Rode M., Rackwitz H.R., Franke W.W. Plakophilin 3: a novel cell-type-specific desmosomal plaque protein. Differentiation. 1999;64:291–306. doi: 10.1046/j.1432-0436.1999.6450291.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen X., Bonne S., Hatzfeld M., van Roy F., Green K.J. Protein binding and functional characterization of plakophilin 2: evidence for its diverse roles in desmosomes and beta-catenin signaling. J Biol Chem. 2002;277:10512–10522. doi: 10.1074/jbc.M108765200. [DOI] [PubMed] [Google Scholar]
- 22.Bass-Zubek A.E., Godsel L.M., Delmar M., Green K.J. Plakophilins: multifunctional scaffolds for adhesion and signaling. Curr Opin Cell Biol. 2009;21:708–716. doi: 10.1016/j.ceb.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlegel N., Meir M., Heupel W.M., Holthofer B., Leube R.E., Waschke J. Desmoglein 2-mediated adhesion is required for intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2010;298:G774–G783. doi: 10.1152/ajpgi.00239.2009. [DOI] [PubMed] [Google Scholar]
- 24.Gosavi P., Kundu S.T., Khapare N., Sehgal L., Karkhanis M.S., Dalal S.N. E-cadherin and plakoglobin recruit plakophilin3 to the cell border to initiate desmosome assembly. Cell Mol Life Sci. 2011;68:1439–1454. doi: 10.1007/s00018-010-0531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kundu S.T., Gosavi P., Khapare N., Patel R., Hosing A.S., Maru G.B., Ingle A., Decaprio J.A., Dalal S.N. Plakophilin3 downregulation leads to a decrease in cell adhesion and promotes metastasis. Int J Cancer. 2008;123:2303–2314. doi: 10.1002/ijc.23797. [DOI] [PubMed] [Google Scholar]
- 26.Khapare N., Kundu S.T., Sehgal L., Sawant M., Priya R., Gosavi P., Gupta N., Alam H., Karkhanis M., Naik N., Vaidya M.M., Dalal S.N. Plakophilin3 loss leads to an increase in PRL3 levels promoting K8 dephosphorylation, which is required for transformation and metastasis. PLoS One. 2012;7:e38561. doi: 10.1371/journal.pone.0038561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz P., Brinkmann V., Ledermann B., Behrend M., Grund C., Thalhammer C., Vogel F., Birchmeier C., Gunthert U., Franke W.W., Birchmeier W. Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J Cell Biol. 1996;135:215–225. doi: 10.1083/jcb.135.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pieperhoff S., Franke W.W. The area composita of adhering junctions connecting heart muscle cells of vertebrates, IV: coalescence and amalgamation of desmosomal and adhaerens junction components–late processes in mammalian heart development. Eur J Cell Biol. 2007;86:377–391. doi: 10.1016/j.ejcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 29.McKoy G., Protonotarios N., Crosby A., Tsatsopoulou A., Anastasakis A., Coonar A., Norman M., Baboonian C., Jeffery S., McKenna W.J. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 30.Endo K., Ueda T., Ueyama J., Ohta T., Terada T. Immunoreactive E-cadherin, alpha-catenin, beta-catenin, and gamma-catenin proteins in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, and patients' survival. Hum Pathol. 2000;31:558–565. doi: 10.1053/hp.2000.6683. [DOI] [PubMed] [Google Scholar]
- 31.Zhai B., Yan H.X., Liu S.Q., Chen L., Wu M.C., Wang H.Y. Reduced expression of E-cadherin/catenin complex in hepatocellular carcinomas. World J Gastroenterol. 2008;14:5665–5673. doi: 10.3748/wjg.14.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrotta M., Strautnieks S., Papouli E., Rushton P., Clark B.E., Parry D.A., Logan C.V., Newbury L.J., Kamath B.M., Ling S., Grammatikopoulos T., Wagner B.E., Magee J.C., Sokol R.J., Mieli-Vergani G., Smith J.D., Johnson C.A., McClean P., Simpson M.A., Knisely A.S., Bull L.N., Thompson R.J. Mutations in TJP2 cause progressive cholestatic liver disease. Nat Genet. 2014;46:326–328. doi: 10.1038/ng.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chidgey M., Dawson C. Desmosomes: a role in cancer? Br J Cancer. 2007;96:1783–1787. doi: 10.1038/sj.bjc.6603808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breuninger S., Reidenbach S., Sauer C.G., Strobel P., Pfitzenmaier J., Trojan L., Hofmann I. Desmosomal plakophilins in the prostate and prostatic adenocarcinomas: implications for diagnosis and tumor progression. Am J Pathol. 2010;176:2509–2519. doi: 10.2353/ajpath.2010.090737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolegraff K., Nava P., Helms M.N., Parkos C.A., Nusrat A. Loss of desmocollin-2 confers a tumorigenic phenotype to colonic epithelial cells through activation of Akt/beta-catenin signaling. Mol Biol Cell. 2011;22:1121–1134. doi: 10.1091/mbc.E10-10-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolligs F.T., Hu G., Dang C.V., Fearon E.R. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franzen C.A., Todorovic V., Desai B.V., Mirzoeva S., Yang X.J., Green K.J., Pelling J.C. The desmosomal armadillo protein plakoglobin regulates prostate cancer cell adhesion and motility through vitronectin-dependent Src signaling. PLoS One. 2012;7:e42132. doi: 10.1371/journal.pone.0042132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conacci-Sorrell M.E., Ben-Yedidia T., Shtutman M., Feinstein E., Einat P., Ben-Ze'ev A. Nr-CAM is a target gene of the beta-catenin/LEF-1 pathway in melanoma and colon cancer and its expression enhances motility and confers tumorigenesis. Genes Dev. 2002;16:2058–2072. doi: 10.1101/gad.227502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miravet S., Piedra J., Miro F., Itarte E., Garcia de Herreros A., Dunach M. The transcriptional factor Tcf-4 contains different binding sites for beta-catenin and plakoglobin. J Biol Chem. 2002;277:1884–1891. doi: 10.1074/jbc.M110248200. [DOI] [PubMed] [Google Scholar]
- 40.Kolligs F.T., Kolligs B., Hajra K.M., Hu G., Tani M., Cho K.R., Fearon E.R. gamma-Catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of beta-catenin. Genes Dev. 2000;14:1319–1331. [PMC free article] [PubMed] [Google Scholar]
- 41.Aktary Z., Kulak S., Mackey J., Jahroudi N., Pasdar M. Plakoglobin interacts with the transcription factor p53 and regulates the expression of 14-3-3sigma. J Cell Sci. 2013;126:3031–3042. doi: 10.1242/jcs.120642. [DOI] [PubMed] [Google Scholar]
- 42.Seong H.A., Jung H., Ha H. NM23-H1 tumor suppressor physically interacts with serine-threonine kinase receptor-associated protein, a transforming growth factor-beta (TGF-beta) receptor-interacting protein, and negatively regulates TGF-beta signaling. J Biol Chem. 2007;282:12075–12096. doi: 10.1074/jbc.M609832200. [DOI] [PubMed] [Google Scholar]