Abstract
Type 17 helper T-cell cytokines have been implicated in the pathogenesis of inflammatory bowel disease, a chronic condition affecting the gastrointestinal tract, but information regarding their contribution to pathology in different regions of the gut is lacking. By using a murine model of bacteria-induced typhlocolitis, we investigated the role of IL-17A, IL-17F, and IL-22 in cecal versus colonic inflammation. Cecal, but not colonic, pathology in C57BL/6 mice inoculated with Helicobacter hepaticus plus anti–IL-10 receptor (IL-10R) monoclonal antibody was exacerbated by co-administration of anti–IL-17A monoclonal antibody, suggesting a disease-protective role for IL-17A in the cecum. In contrast, anti–IL-17F had no effect on H. hepaticus-induced intestinal pathology. Neutralization of IL-22 prevented the development of colonic, but not cecal, inflammation in H. hepaticus-infected anti–IL-10R–treated mice, demonstrating a pathogenic role for IL-22 in the colon. Analysis of transcript levels revealed differential expression of IL-22R, IL-22 binding protein, and IL-23R between cecum and colon, a finding that may help explain why these tissues respond differently after anti–IL-22 treatment. Analysis of microarray data from healthy human intestine further revealed significant differences in cytokine receptor transcript levels (including IL-22RA1 and IL-23R) in distinct parts of the human gut. Together, our findings demonstrate that individual type 17 helper T-cell cytokines can have proinflammatory or anti-inflammatory effects in different regions of the intestine, an observation that may have implications for interventions against human inflammatory bowel disease.
Crohn disease and ulcerative colitis, the two major forms of inflammatory bowel disease (IBD), each have their own characteristic inflammatory response and anatomical location. Thus, Crohn disease is characterized by a granulomatous, transmural inflammation that can affect any part of the gastrointestinal tract, whereas ulcerative colitis is limited to the mucosa and submucosa of the colon.1 IBD is caused by an inappropriate immune response to intestinal microflora, and genome-wide association studies have now identified >160 IBD loci,2 many of them linked to the immune response.3 A prime example is the IL23R gene, where several single-nucleotide polymorphisms associated with either susceptibility or resistance to IBD have been described.4 Experimental data from murine models of intestinal inflammation have further demonstrated a crucial role for IL-23 in disease pathology.5 IL-23 is produced by activated dendritic cells and macrophages after microbial stimulation,6, 7 and one of its many functions is the enhancement of CD4+ type 1 helper T-cell (Th1) and type 17 helper T-cell (Th17) responses and the induction of Th1/Th17-type cytokines by non–T-cell sources.8 Increased levels of both Th1- [interferon (IFN)-γ] and Th17-type cytokines (IL-17A, IL-17F, and IL-22) are found in inflamed tissues in both human IBD and experimental models of the disease,8 but the importance of these cytokines in disease pathogenesis at different anatomical locations of the gut is not well characterized.
IL-17A, IL-17F, and IL-22 are produced by CD4+ and CD8+ T lymphocytes, γδ T cells, and cells of the innate immune system, including innate lymphoid cells.8, 9 The receptor for IL-17A and IL-17F is composed of IL-17RA and IL-17RC, and is expressed on both hematopoietic and nonhematopoietic cells.10 In contrast, the receptor for IL-22, a heterodimer composed of IL-22 receptor α1 (IL-22Rα1) and IL-10Rβ2,11, 12 is found on nonhematopoietic cells.13, 14 Moreover, a secreted single-chain receptor known as the IL-22 binding protein (IL-22BP), or IL-22Rα2, acts as a naturally occurring IL-22 antagonist.15, 16, 17 IL-17A, IL-17F, and IL-22 may all contribute to inflammatory responses in the gut. Thus, IL-17A and IL-17F are known to induce the expression of neutrophil chemoattractants and proinflammatory cytokines, such as IL-1β, IL-6, tumor necrosis factor-α, granulocyte colony-stimulating factor, and granulocyte macrophage colony-stimulating factor, from fibroblasts, endothelial cells, and epithelial cells.9, 10 IL-22 can induce tumor necrosis factor-α production by intestinal epithelial cells18 and proinflammatory cytokines, chemokines, and matrix metalloproteinases from colonic subepithelial myofibroblasts.19 IL-17A, IL-17F, and IL-22 have also been reported to enhance barrier function and promote antimicrobial defense,9, 20, 21, 22, 23, 24, 25, 26 thus displaying tissue-protective functions at mucosal sites.
We have previously shown that inoculation with Helicobacter hepaticus triggers typhlocolitis in specific pathogen-free IL-10–deficient (Il10−/−) mice and in wild-type (WT) animals treated with anti–IL-10R monoclonal antibody (mAb).27, 28 The development of intestinal pathology after H. hepaticus inoculation is dependent on IL-23 and is associated with up-regulated Th1- and Th17-type cytokine mRNA levels as well as enhanced frequencies and numbers of cecal and colonic lamina propria (LP) CD4+ T cells expressing IFN-γ, IL-17A, IL-17F, and/or IL-22.28, 29 Interestingly, IFN-γ was shown to be necessary for maximal pathology in the colon, but not in the cecum in this model,28 suggesting regional differences in cytokine requirements for disease pathogenesis. The role of Th17-type cytokines in cecal versus colonic inflammation in H. hepaticus–induced T-cell–dependent intestinal pathology has, however, not been examined.
By using mAb blockade of cytokines, we herein demonstrate that IL-17A and IL-22 play distinct roles in different anatomical regions of the intestine during H. hepaticus–induced inflammation. Thus, IL-17A was found to play a disease-protective role in the cecum, whereas IL-22 was shown to be pathogenic in the colon. Analysis of transcript levels revealed significant differences in mRNA accumulation of Il22ra1, Il22ra2, and Il23r between the cecum and colon, a finding that may help explain the difference in IL-22 dependence of the inflammation in these two tissues. Regional differences in cytokine receptor expression may be of relevance to disease pathogenesis also in human IBD, because we observed significant differences in transcript levels for several receptor subunits, including IL-22RA1 and IL-23 receptor (IL-23R), in distinct parts of the healthy human gut. Together, these findings demonstrate that individual Th17-type cytokines may have proinflammatory or anti-inflammatory effects in different parts of the intestine, and highlight the possibility that differential expression of cytokine receptors at those sites may influence disease pathogenesis.
Materials and Methods
Mice
Female C57BL/6 (B6) CD45.1+ and B6 CD45.2+ WT mice were bred and maintained in an accredited specific pathogen-free facility, and experiments were conducted in accordance with the UK Scientific Procedures Act (1986) under a Project License authorized by the UK Home Office and approved by the Animal Welfare and Ethical Review Body at the Department of Biology, University of York (York, UK). Animals used tested negative for antibodies to specific murine viruses, were free of Helicobacter species, as assessed by PCR, and were >6 weeks old when used.
Experimental Infections and mAb Treatment
To induce typhlocolitis, mice were allocated to treatment groups and inoculated intragastrically with 1.5 × 107 H. hepaticus National Cancer Institute–Frederick isolate 1A,30 isolated from the same mouse colony as isolate Hh-131 (strain 51449; ATCC, Manassas, VA) and treated i.p. with 1 mg of anti–IL-10R (clone 1B1.3a) on days 0 and 7 (and in some experiments also day 14) of H. hepaticus infection.28 Age-matched uninfected animals were included as controls. To examine the effect of in vivo neutralization of IL-17A, IL-17F, or IL-22, mice were treated i.p. with anti–IL-17A (clone 50104, 0.5 mg/injection; R&D Systems, Abingdon, UK), anti–IL-17F (clone RK016-17,32 1 mg/injection; Pfizer, Cambridge, MA), anti–IL-22 (clone Ab-01,33 1 mg/injection; Pfizer), or a control rat IgG1 (Pfizer) in 0.5 mL of phosphate-buffered saline (PBS) on days 0 and 7 (and in some experiments also day 14) of H. hepaticus infection. One week after the last mAb injection, mice were sacrificed and intestines were collected for analysis.
Pathology
Cecum and a piece of ascending colon (approximately 1 cm from the cecum) were fixed in buffered 10% formalin, and paraffin-embedded sections were stained with hematoxylin and eosin (Mary Lyon Center at Medical Research Council Harwell, Oxfordshire, UK). Samples (a longitudinal section of the cecum and a cross section through the colon) were scored blindly (D.J. or M.C.K.) using two different scoring systems. Initially, histology sections were evaluated in a blinded manner by the same pathologist (D.J.) for 20 parameters of inflammation. A total score for the whole section was assigned on the basis of a 0 to 5 scoring system for different features of epithelial damage (mucus depletion, crypt length and hyperplasia, crypt loss and irregularity, epithelial flattening, and ulceration), polymorphonuclear cell response (crypt abscesses in the epithelium, and polys in the epithelium, LP, submucosa, and serosa), macrophage-plasma cell-lymphocyte cellularity (in the LP, submucosa, muscularis, and serosa), granulomas (mucosal, submucosal, and lymphatic granulomas), and fibrosis. A total score was calculated by adding the individual scores. Subsequently, a simplified scoring system was adapted on the basis of epithelial hyperplasia and LP cellularity (0 to 3 each), and goblet cell depletion, submucosal inflammation, edema, crypt abscesses, and ulcers (0 to 1 each). A total score was calculated by adding the individual scores, reaching a maximum score of 11. When samples from the experiments were rescored using this latter scoring system, the histology findings were reproduced; thus, the simplified scoring system was used throughout the rest of the study with all histology sections evaluated blinded (M.C.K.).
Isolation of Epithelial Cells and the Nonepithelial Tissue Compartment
Freshly harvested ceca (avoiding the cecal patch) and colons (the proximal half) were cut open and into sections (3 to 5 mm long) and incubated twice in RPMI 1640 medium containing 10 mmol/L HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L glutamine, 1% fetal calf serum (FCS), 1 mmol/L dithiothreitol, and 5 mmol/L EDTA for 20 minutes at 37°C while shaking to release epithelial cells. Cells collected from the first incubation (epithelial cells, generally <10% CD45+ cells by flow cytometry) were spun down, resuspended in RLT buffer (Qiagen, Crawley, UK), and stored at −80°C until processing. The tissue pieces collected after the second incubation (the nonepithelial tissue compartment) were stored in RNAlater (Ambion, Paisley, UK) at 4°C overnight before the RNAlater was removed and samples were stored at −80°C until processing.
Real-Time RT-PCR
Pieces of cecum (avoiding the cecal patch) and ascending colon (an approximately 3-mm piece distal of that used for histology) collected in RNAlater, or the nonepithelial tissue compartment from these tissues, were homogenized in TRIzol (Invitrogen, Paisley, UK), and total RNA was isolated by chloroform extraction, followed by DNase treatment using the DNA-free kit (Applied Biosystems, Paisley, UK). Total RNA was isolated from epithelial cells using the RNeasy mini kit, including DNase treatment as recommended by the manufacturer (Qiagen). RNA was reverse transcribed using SuperScript II (Invitrogen) and random hexamers. cDNA was amplified using either SYBR Green reagents and an ABI Prism 7300 Sequence Detection System (Applied Biosystems) or Fast SYBR Green reagents and a StepOnePlus Real-Time PCR System (Life Technologies, Paisley, UK). Cytokine and cytokine receptor expression levels for each individual sample (run in duplicates) were normalized to hypoxantine-guanine phosphoribosyltransferase (HPRT) using ΔCt calculations. Primer pairs for murine RegIIIg, Il22, Il17ra, Il17rc, Ifngr2, Il10ra, Il10rb, Il22ra1, Il22ra2, and Il23r were from Qiagen. Other specific primer pairs were as follows: Hprt, 5′-GCGTCGTGATTAGCGATGATGAAC-3′ (forward) and 5′-ATCTCCTTCATGACATCTCGAGCAAGTC-3′ (reverse); Il12p40, 5′-CATCAAGAGCAGTAGCAGTTCC-3′ (forward) and 5′-GAATACTTCTCATAGTCCCTTTGG-3′ (reverse); Il12p35, 5′-AAATGAAGCTCTGCATCCTGC-3′ (forward) and 5′-TCACCCTGTTGATGGTCACG-3′ (reverse); Il23p19, 5′-CACATGCACCAGCGGGACAT-3′ (forward) and 5′-CTTTGCAAGCAGAACTGGCTGTTG-3′ (reverse); Ifng, 5′-GGATGCATTCATGAGTATTGC-3′ (forward) and 5′-GCTTCCTGAGGCTGGATTC-3′ (reverse); and Il17a, 5′-GCTCCAGAAGGCCCTCAG-3′ (forward) and 5′-CTTTCCCTCCGCATTGACA-3′ (reverse).
LP Cell Isolation
Ceca and colons were cut open and into sections (3 to 5 mm long) and incubated twice in RPMI 1640 medium containing 10 mmol/L HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L glutamine, 1% FCS, 1 mmol/L dithiothreitol, and 5 mmol/L EDTA for 20 minutes at 37°C while shaking to release epithelial cells. Tissue pieces were then digested with Liberase TL (0.3125 mg/mL; Roche, Burgess Hill, UK) and DNase I (125 U/mL; Sigma-Aldrich, Gillingham, UK) in RPMI 1640 medium containing 10 mmol/L HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L glutamine, and 1% FCS for 1 hour at 37°C while shaking. The resulting tissue suspension was passed through a 100-μm cell strainer, centrifuged, and resuspended in 40% Percoll (Sigma-Aldrich) and underlayed with 80% Percoll. After centrifugation at 600 × g for 20 minutes at 10°C, LP cells were recovered from the 40%/80% interface and resuspended in complete medium (RPMI 1640 medium with 10% heat-inactivated FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L glutamine, 10 mmol/L HEPES, 1 mmol/L sodium pyruvate, 0.1 mmol/L nonessential amino acids, and 50 μmol/L 2-mercaptoethanol).
Intracellular Staining for Cytokines
LP cells (5 × 106/mL) were stimulated with 10 ng/mL of phorbol myristate acetate and 1 μg/mL of ionomycin for 4 hours at 37°C with 10 μg/mL brefeldin A during the last 3 hours. Thereafter, cells were resuspended in PBS containing 5 μg/mL anti-FcγRII/III mAb (2.4G2; BD, Oxford, UK), stained at 4°C with a fixable Aqua Dead Cell stain (Invitrogen), washed with PBS, and fixed in 2% paraformaldehyde for 15 minutes. After a wash in PBS, cells were resuspended in permeabilization buffer (PBS containing Mg2+ and Ca2+, 10 mmol/L HEPES, 0.1% saponin, and 1% FCS) with 10% normal mouse serum and 5 μg/mL anti-FcγRII/III, and incubated for 30 minutes at 4°C. Thereafter, cells were stained for 30 minutes at 4°C with anti–IL-17A–fluorescein isothiocyanate (Mabtech, Nacka Strand, Sweden), anti–IFN-γ-eFluor450 (XMG1.2; eBioscience, Hatfield, UK), anti–IL-22-Alexa Fluor 647 (IL-22-02; labeled in-house), washed with permeabilization buffer, and resuspended in PBS + 0.5% FCS. Cell fluorescence was measured using a CyAn ADP flow cytometer (Beckman Coulter, High Wycombe, UK), and data were analyzed using FlowJo software version 8.8.6 (Tree Star, Ashland, OR).
Analysis of Microarray Data from Human Intestinal Tissue
The National Center for Biotechnology Information Gene Expression Omnibus database was searched for studies measuring gene expression by microarray in human intestinal tissue. Specifically, we were looking for studies in which biopsy specimens had been collected from multiple sampling sites throughout the intestinal tract of healthy individuals. Only one suitable study was found (http://www.ncbi.nlm.nih.gov/geo; accession number GSE55258), with collection of Agilent microarray data from intestinal biopsy specimens of the terminal ileum (TI), ileocecal valve (ICV), appendiceal orifice (AO), and sigmoid colon (SC) from healthy human subjects.34 For our analysis, data were downloaded from Gene Expression Omnibus and processed using the limma library in R (http://www.r-project.org). Samples from five of the six patients in the study were included in the analysis (patients 9, 10, 18, 22, and 23), because all four tissue types were available for these subjects; patient 17 was excluded, because only the TI was available from this subject. In limma, background expression was corrected using the normexp method, with an offset of 16. Quantile normalization was used to normalize between arrays. Data were then fitted to a linear model, and the empirical Bayes method combined with a nested post hoc test was used to assess significance. P values were adjusted for multiple tests with the Benjamini-Hochberg correction.35
Statistical Analysis
Colitis scores were compared using the nonparametric Mann-Whitney test. Murine mRNA expression data were analyzed by one-way analysis of variance when comparing multiple groups, and by Student's two-tailed t-test when comparing cecum and colon data. Differences were considered statistically significant with P < 0.05.
Results
Neutralization of IL-22, But Not IL-17A, Prevents H. hepaticus–Induced Colonic Inflammation
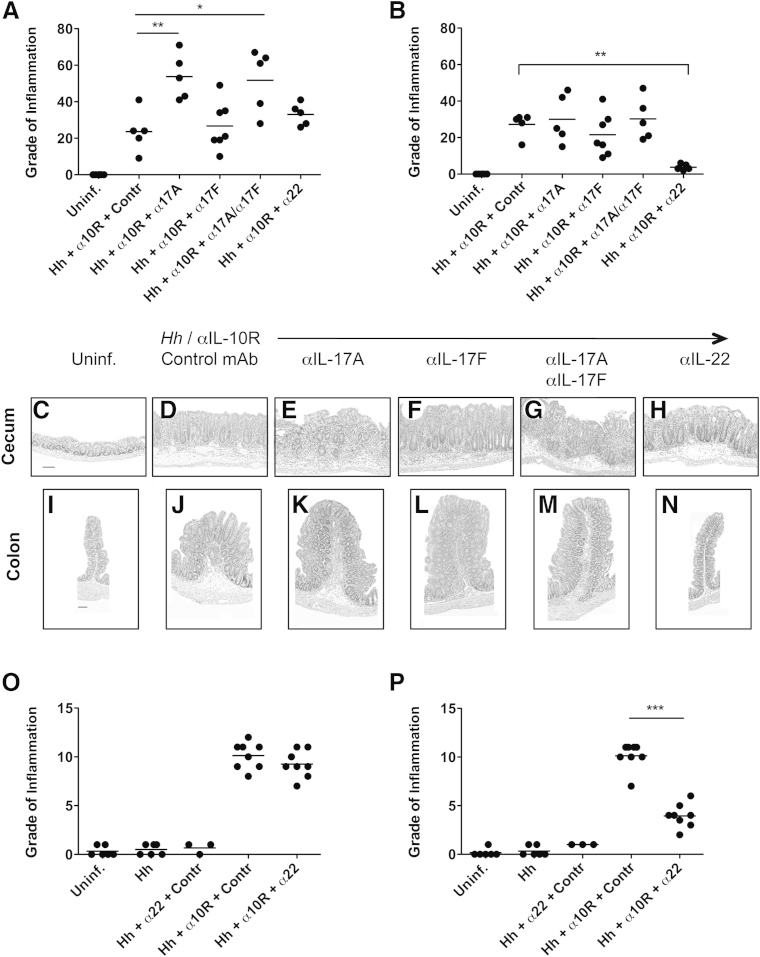
We have previously shown that specific pathogen-free WT mice given H. hepaticus plus anti–IL-10R mAb develop typhlocolitis, a disease dependent on IL-23 and associated with the up-regulation of Th1- and Th17-type cytokines in the large intestine.28 Moreover, IFN-γ was found to be required for maximal pathology in the colon, but not the cecum, in this model.28 To assess the contribution of Th17-type cytokines to disease pathogenesis in different regions of the intestine, parallel groups of H. hepaticus–infected anti–IL-10R–treated WT mice were given weekly injections of neutralizing mAb to IL-17A, IL-17F, IL-17A plus IL-17F, IL-22, or isotype control from the start of the infection, and pathology was examined in the cecum and colon 2 to 3 weeks later. For these initial experiments, histology sections were scored for 20 parameters of inflammation, including measures of epithelial damage, crypt abscesses, granulomas, fibrosis, polymorphonuclear leukocytes, and cellularity (macrophages, plasma cells, and lymphocytes) in the LP, submucosa, muscularis, and serosa. Uninfected mice were included as controls (Figure 1, A–C and I). Our results revealed that cecal pathology, but not colonic inflammation, in WT mice inoculated with H. hepaticus plus anti–IL-10R was exacerbated by co-administration of anti–IL-17A mAb (Figure 1, A, B, D, E, J, and K), suggesting a disease-protective role for IL-17A in the cecum in this model. In contrast, neutralization of IL-17F had no effect on either cecal or colonic inflammation (Figure 1, A, B, F and L). Cecal pathology in mice given anti–IL-17A plus anti–IL-17F was indistinguishable from that seen in H. hepaticus–infected anti–IL-10R–treated animals given anti–IL-17A alone (Figure 1, A, B, G and M). [Of note, in one of the experiments performed, by day 16 postinfection (pi) one mouse was found dead and one mouse had to be sacrificed because of ill health in the two groups given anti–IL-17A mAb. The large intestines of these animals were severely inflamed and enlarged on visible inspection, and histological analysis revealed the tissues were largely autolyzed and necrotic; thus, no histology scores were obtained from these mice.] The increased cecal pathology observed after anti–IL-17A or anti–IL-17A plus anti–IL-17F treatment was not because of a specific feature of inflammation being increased, but most of the 20 parameters examined were scored higher compared with animals receiving H. hepaticus plus anti–IL-10R alone (data not shown). More important, administration of anti–IL-22 mAb revealed a different role for IL-22 in H. hepaticus–induced intestinal pathology. Thus, neutralization of IL-22 significantly reduced pathology in the colon of H. hepaticus–infected anti–IL-10R–treated WT mice, without affecting cecal inflammation (Figure 1, A, B, H and N), a pattern similar to that seen when blocking IFN-γ in the H. hepaticus colitis model.28
Figure 1.
Cecal and colonic pathology after type 17 helper T-cell cytokine blockade in Helicobacter hepaticus (Hh)–induced intestinal inflammation. C57BL/6 wild-type (WT) mice were inoculated with H. hepaticus and treated with anti–IL-10 receptor (IL-10R) monoclonal antibody (mAb) plus neutralizing mAb to IL-17A, IL-17F, IL-17A + IL-17F, IL-22, or isotype control (Contr) on days 0, 7, and (if applicable) 14 postinfection (pi). One week after the last mAb administration, pathology was analyzed in the cecum and ascending colon. A and B: Histology scores of cecum (A) and ascending colon (B) using a scoring system on the basis of 20 parameters of inflammation, as described in Materials and Methods. Each symbol represents an individual mouse. Data are pooled from two separate experiments analyzed at 2 and 3 weeks pi, respectively. C–N: Histology of representative cecal (C–H) and colonic (I–N) sections from the mice in A and B. Uninfected (Uninf.) WT (C and I), H. hepaticus–infected WT plus anti–IL-10R plus control mAb (D and J), H. hepaticus–infected WT plus anti–IL-10R plus anti–IL-17A (E and K), H. hepaticus–infected WT plus anti–IL-10R plus anti–IL-17F (F and L), H. hepaticus–infected WT plus anti–IL-10R plus anti–IL-17A plus anti–IL-17F (G and M), and H. hepaticus–infected WT plus anti–IL-10R plus anti–IL-22 (H and N). Hematoxylin and eosin staining was used. O and P: Histology scores of cecum (O) and ascending colon (P) using a scoring system on the basis of five parameters of inflammation [epithelial hyperplasia, lamina propria (LP) cellularity, goblet cell depletion, crypt abscesses, and ulcers]. Each symbol represents an individual mouse. Data are pooled from two separate experiments analyzed at 2 weeks pi. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bar = 100 μm (C–N).
The finding that IL-22 neutralization reduced colonic, but not cecal, pathology in H. hepaticus–infected anti–IL-10R–treated WT animals was confirmed using a simplified scoring system focusing on seven parameters (epithelial hyperplasia, goblet cell depletion, LP and submucosal cellularity, edema, crypt abscesses, and ulcers) (Figure 1, O and P), validating this more basic scoring system. Moreover, H. hepaticus–infected WT animals given anti–IL-22 showed no increase in pathology compared with mice given H. hepaticus–infected alone or with uninfected controls (Figure 1, O and P). Together, our findings suggest that although IL-17A has a disease-protective role in the cecum, IL-22, similar to IFN-γ,28 is essential for maximal pathology in the colon in H. hepaticus–induced T-cell–dependent colitis.
IL-17A and IL-22 Expression Is Higher in the Cecum than in the Colon
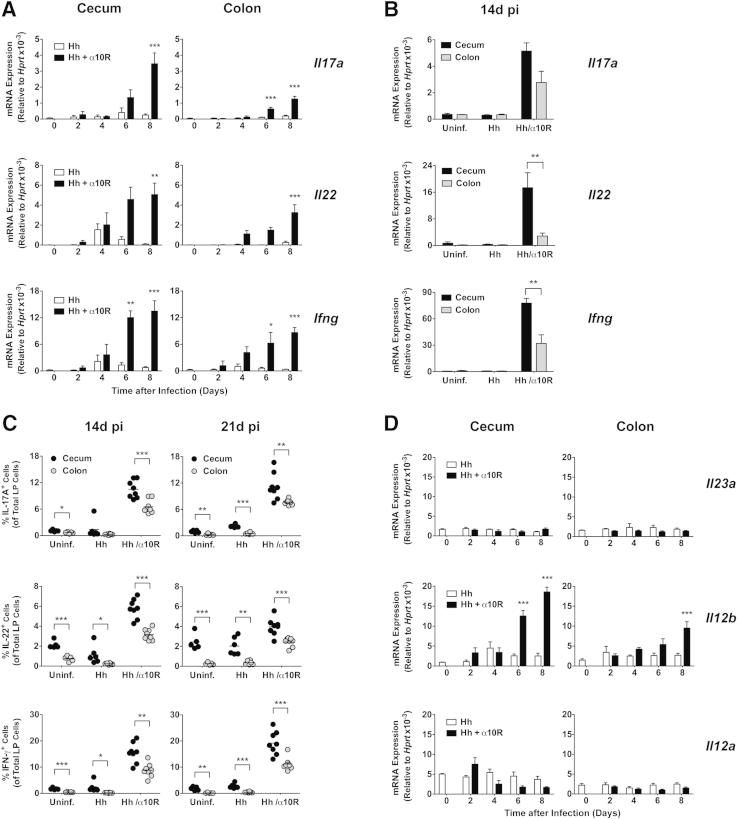
To investigate if there were regional differences in IL-17A and IL-22 expression in the large intestine after H. hepaticus inoculation, we next analyzed mRNA levels of these cytokines in the cecum and colon every other day during the first 8 days after bacterial challenge. This time period was chosen to capture the early kinetics of H. hepaticus–induced typhlocolitis, where visible inflammation is detected by day 7 after H. hepaticus inoculation.29 Compared with uninfected animals, WT mice given H. hepaticus alone showed minimal induction of Il17a and Il22 transcripts in the large intestine (Figure 2A). In contrast, animals given H. hepaticus plus anti–IL-10R displayed enhanced mRNA accumulation of Il17a and Il22 from day 6 and day 4 pi, respectively, in both the cecum and colon, reaching significance by day 6 to 8 pi (Figure 2A). Large intestinal Ifng transcript levels were also enhanced from day 4 pi onwards in H. hepaticus–infected anti–IL-10R–treated WT mice, reaching significance by day 6 pi (Figure 2A). In general, mRNA accumulation of Il17a, Il22, and Ifng was higher in the cecum than in the colon of H. hepaticus–infected anti–IL-10R–treated WT mice (Figure 2A), a pattern seen also at day 14 pi (Figure 2B) when H. hepaticus–induced inflammation and the number of cytokine-secreting CD4+ T cells reach their peak in the large intestine.29 Moreover, when examining cytokine expression at the protein level, the proportion of total LP cells able to secrete IL-17A, IL-22, and IFN-γ in H. hepaticus–infected anti–IL-10R–treated WT mice was higher in the cecum than in the colon at both days 14 and 21 pi (Figure 2C). The frequency of LP cells producing these cytokines was higher in the cecum than in the colon also in uninfected controls and in WT mice given H. hepaticus alone, although the percentages were lower in these groups than in inflamed hosts (Figure 2C). When examining mRNA levels for Th1-/Th17-polarizing cytokines, we noted no major changes in the expression of Il23a transcripts in the cecum or colon over the first 8 days after bacterial inoculation (Figure 2D). However, Il12b mRNA levels were significantly enhanced in H. hepaticus–infected anti–IL-10R–treated WT mice from day 6 and 8 pi in the cecum and colon, respectively, with higher levels in the cecum (Figure 2D). Finally, Il12a transcripts did not change significantly during the course of the experiment (Figure 2D).
Figure 2.
Kinetics of cecal and colonic cytokine expression after Helicobacter hepaticus (Hh) inoculation. C57BL/6 wild-type (WT) mice were inoculated with H. hepaticus alone or H. hepaticus plus anti–IL-10 receptor (IL-10R) monoclonal antibody (mAb), and ceca and colons collected at different time points postinfection (pi) and processed for real-time RT-PCR analysis of cytokines (A, B, and D) or intracellular cytokine staining of lamina propria (LP) cells (C). Uninfected (Uninf.) mice were included as controls. A and D: Day 2, 4, 6, and 8 pi transcript levels of Il17a (IL-17A), Il22 (IL-22), and Ifng (IFN-γ; A) and Il23a (IL-23p19), Il12b (IL-12p40), and Il12a (IL-12p35; D) in cecum and colon of mice given H. hepaticus alone or H. hepaticus plus anti–IL-10R on day 0. Significance was compared with uninfected controls. B: Day 14 pi transcript levels of Il17a (IL-17A), Il22 (IL-22), and Ifng (IFN-γ) in cecum and colon of uninfected controls and mice given H. hepaticus alone or H. hepaticus plus anti–IL-10R on days 0 and 7 pi. Data are pooled from two independent experiments, except for Il17a where only one experiment was analyzed. C: LP cells were stimulated with phorbol myristate acetate (PMA)/ionomycin, and stained with a fixable viability dye and mAb to IL-17A, IL-22, and IFN-γ. Graphs show the proportion of IL-17A+, IL-22+, and IFN-γ+ cells within the total live LP cells in the cecum and colon at 14 and 21 days pi in mice given H. hepaticus with or without weekly injections of anti–IL-10R. Uninfected controls were included at each time point. Each symbol represents an individual mouse, and data are pooled from two independent experiments with four mice per group and time point. Data represent means ± SEM mRNA levels relative to hypoxantine-guanine phosphoribosyltransferase (HPRT; A, B, and D). n = 4 mice per group and time point (A and D); n = 4 individual mice per group (Il17a) or n = 8 individual mice per group (Il22 and Ifng; B). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
We next examined Il17a and Il22 mRNA levels in the epithelial fraction versus the nonepithelial tissue compartment from the cecum and colon of uninfected controls and of mice given H. hepaticus with or without anti–IL-10R 7 days earlier. As expected, the expression of these cytokines was primarily detected in the nonepithelial tissue compartment in all three groups of mice (Supplemental Figure S1). Moreover, when analyzing the nonepithelial tissue compartment, Il17a and Il22 transcripts were significantly higher in the cecum than in the colon, both at steady state, in mice given H. hepaticus alone, and in mice inoculated with H. hepaticus plus anti–IL-10R mAb (Supplemental Figure S1). Together, this reinforced a difference between these two tissues in steady state and during the early stages of colitis induction. A similar trend was observed for IFN-γ with higher levels of Ifng transcripts in the nonepithelial tissue compartment from the cecum compared with the colon 7 days after H. hepaticus inoculation with or without anti–IL-10R treatment (Supplemental Figure S1).
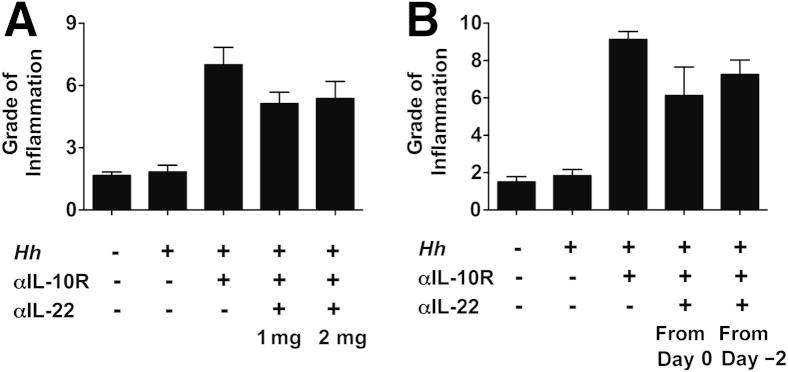
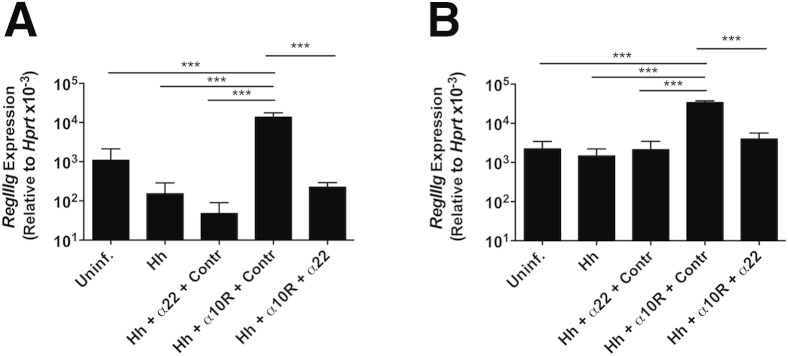
The observation of higher Il22 transcript levels and proportions of LP cells able to secrete IL-22 in the cecum compared with the colon raised the possibility that the anti–IL-22 treatment may have failed to block cecal inflammation because of insufficient neutralization of the cytokine in this tissue. We, however, believe this scenario to be unlikely on the basis of additional experimental data. First, no effect on H. hepaticus/anti–IL-10R–induced cecal inflammation was seen when mice were given a higher dose of anti–IL-22 (2 instead of 1 mg/week) (Figure 3A) or when mAb treatment was initiated 2 days before bacterial inoculation (Figure 3B). Second, transcript levels for Reg3γ, an antimicrobial peptide known to be induced by IL-22,36 were shown to be significantly lower in both the cecum and colon of H. hepaticus–infected anti–IL-10R/anti–IL-22–treated WT mice compared with animals given H. hepaticus plus anti–IL-10R alone (approximately 61-fold and approximately ninefold reduction in the cecum and colon, respectively) (Figure 4, A and B). This demonstrated that the anti–IL-22 mAb was able to efficiently neutralize the antimicrobial peptide-inducing effect of IL-22 in both tissues. Together, these findings suggest mechanisms other than inefficient cytokine neutralization as being responsible for the different effects seen on H. hepaticus–induced pathology in the cecum versus colon after anti–IL-22 treatment.
Figure 3.
Increasing the dose of anti–IL-22 or starting monoclonal antibody (mAb) treatment before Helicobacter hepaticus (Hh)/anti–IL-10 receptor (IL-10R) inoculation does not affect cecal pathology. C57BL/6 wild-type mice were inoculated with H. hepaticus and treated with anti–IL-10R plus anti–IL-22 as indicated. One week after the last mAb administration, pathology was analyzed in the cecum. Uninfected mice were included as controls. A: A total of 1 or 2 mg of anti–IL-22 was given on days 0 and 7 postinfection (pi). B: A total of 1 mg of anti–IL-22 was given on days 0 and 7 pi or on days −2, 0, and 7 pi. Data represent grade of inflammation (using a scoring system on the basis of five parameters of inflammation: epithelial hyperplasia, lamina propria cellularity, goblet cell depletion, crypt abscesses, and ulcers). Data are means ± SEM (A and B). n = 4 mice per group (A and B, H. hepaticus/anti–IL-10R, H. hepaticus/anti–IL-10R/anti–IL-22 1 mg, and H. hepaticus/anti–IL-10R/anti–IL-22 2 mg; and B, uninfected mice); n = 3 mice per group (A and B, H. hepaticus alone; and A, uninfected mice).
Figure 4.
Anti–IL-22 monoclonal antibody (mAb) treatment reduces RegIIIg accumulation in both the cecum and colon of Helicobacter hepaticus (Hh)/anti–IL-10 receptor (IL-10R)–treated wild-type (WT) mice. C57BL/6 WT mice were inoculated with H. hepaticus and treated with anti–IL-10R mAb plus neutralizing mAb to IL-17A, IL-17F, IL-17A + IL-17F, IL-22, or isotype control (Contr) on days 0 and 7 postinfection. One week later, large intestines were collected for real-time RT-PCR analysis of RegIIIg in the cecum (A) and ascending colon (B). Data represent means ± SEM mRNA levels relative to hypoxantine-guanine phosphoribosyltransferase (HPRT; A and B). n = 4 to 5 individual mice from one of the experiments shown in Figure 1 (O and P; A and B). ∗∗∗P < 0.001. Uninf., uninfected.
Differences in IL-22R, IL-22BP, and IL-23R Expression in Steady-State Cecum and Colon
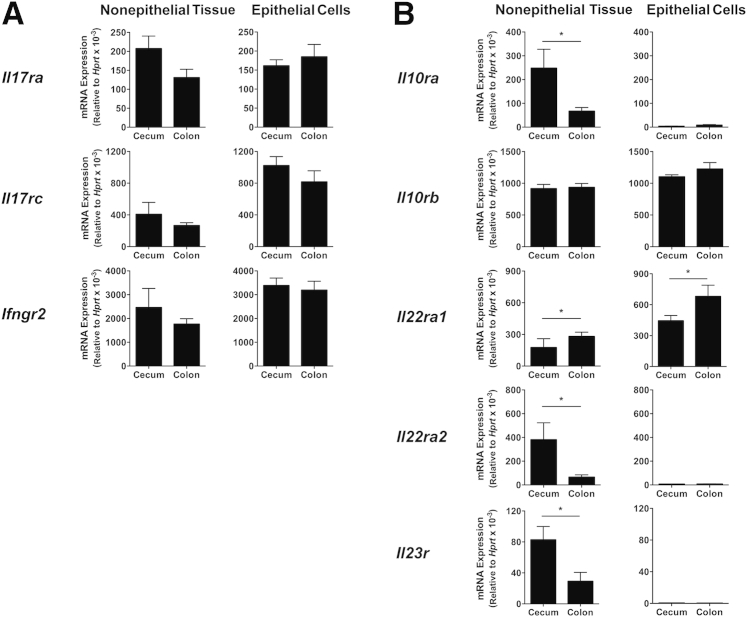
We next characterized the steady-state expression of cytokine receptors in the cecum and colon. Il17ra and Il17rc (which encode the subunits of the receptor for IL-17A, IL-17F, and IL-17A/F10) were expressed at similar levels in the cecum and colon in both the nonepithelial tissue compartment and the epithelial cell fraction (Figure 5A). Similarly, no differences were observed in Ifngr2 transcripts between the cecum and colon (Figure 5A). These results indicate that differential cytokine receptor expression neither accounts for the protective effect of IL-17A in the cecum (Figure 1, A and E) nor the pathogenic role of IFN-γ in the colon.28
Figure 5.
Difference in cytokine receptor mRNA levels between steady-state cecum and colon of wild-type (WT) mice. Ceca and colons were collected from uninfected C57BL/6 WT mice, and epithelial cells and the nonepithelial tissue compartment were processed for real-time RT-PCR analysis of cytokine receptor subunits. Transcript levels of Il17ra (IL-17RA), Il17rc (IL-17RC), and Ifngr2 (IFN-γR2; A) and Il10ra (IL-10RA), Il10rb (IL-10RB), Il22ra1 (IL-22RA1), Il22ra2 [IL-22RA2/IL-22 binding protein (IL-22BP)], and Il23r (IL-23R; B) in the nonepithelial tissue compartment and epithelial cells of cecum and colon. Data represent means ± SEM mRNA levels relative to hypoxantine-guanine phosphoribosyltransferase (HPRT; A and B). n = 4 individual mice from one experiment (A); n = 8 individual mice from two independent experiments, except for Il10ra, where n = 4 mice were analyzed (B). ∗P < 0.05 when comparing cecum with colon.
When mRNA levels for other cytokine receptors were examined, Il10rb2, the shared subunit between the receptors for IL-10 and IL-22, was expressed ubiquitously and at similar levels in the cecum and colon (Figure 5B). Il10ra1, the IL-10–specific subunit of the IL-10R, was primarily found in the nonepithelial tissue compartment, with significantly higher levels in the cecum compared with the colon (Figure 5B). In contrast, Il22ra1, the IL-22–specific subunit of the IL-22R, was expressed at significantly higher levels in the colon compared with the cecum (Figure 5B). Of interest, Il22ra2, the IL-22BP that acts as a naturally occurring IL-22 antagonist, and Il23r were expressed at significantly higher levels in the cecal compared with the colonic nonepithelial tissue compartment (Figure 5B). Together, these data indicate that the colon may be more likely to respond to IL-22 because of higher expression of membrane-bound IL-22R and lower abundance of the IL-22BP.
Differences in IL-22R, IL-22BP, and IL-23R Expression in Inflamed Cecum and Colon
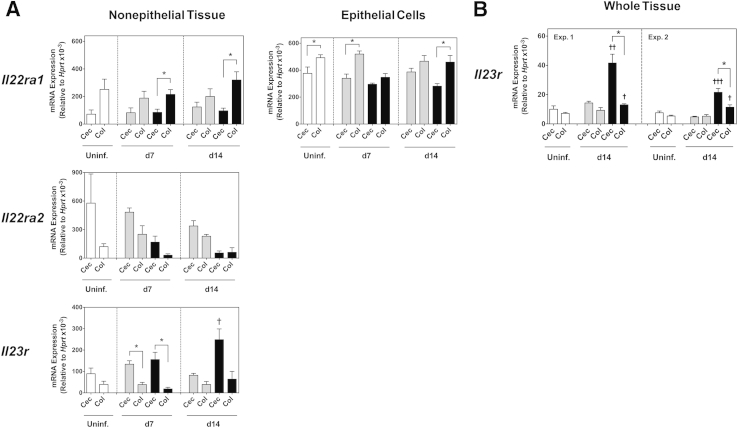
To examine whether the expression of IL-22R, IL-22BP, and IL-23R changed after bacterial challenge and/or during inflammation, we next analyzed Il22ra1, Il22ra2, and Il23r mRNA levels in the cecal and colonic nonepithelial tissue compartment, and Il22ra1 mRNA levels in cecal and colonic epithelial cells from 7- and 14-day H. hepaticus–infected mice with and without anti–IL-10R treatment. We compared the pattern of expression between cecum and colon at each time point, and also whether there was a change in expression levels compared with tissues from uninfected mice. Inoculation with H. hepaticus alone did not significantly alter the mRNA accumulation of the three cytokine receptor subunits. Thus, as in uninfected mice, Il22ra1 levels were higher in the colon, whereas Il22ra2 and Il23r levels were higher in the cecum, of mice given H. hepaticus alone, and there was no change in expression levels compared with uninfected controls (Figure 6A). When examining mice with H. hepaticus/anti–IL-10R–induced intestinal inflammation, Il22ra1 levels were generally higher in the colon, and Il22ra2 and Il23r levels were generally higher in the cecum also in these hosts; however, the expression patterns for two of the receptor subunits were altered in colitic mice compared with uninfected controls. First, although not reaching statistical significance, Il22ra2 accumulation in the nonepithelial tissue compartment was lower in both the cecum and colon of 7- and 14-day H. hepaticus–infected anti–IL-10R–treated mice compared with uninfected controls (Figure 6A). Second, Il23r transcript levels in the cecal nonepithelial tissue compartment were significantly higher in 14-day H. hepaticus–infected anti–IL-10R–treated mice compared with uninfected animals (Figure 6A). Moreover, when analyzing whole tissue samples from 14-day H. hepaticus–infected anti–IL-10R–treated mice, Il23r transcript levels were significantly higher in inflamed cecum compared with inflamed colon (Figure 6B), and the accumulation of Il23r in both these tissues was significantly higher than that found in uninfected controls (Figure 6B), suggesting that IL-23R is up-regulated in both tissues during inflammation. Whether this up-regulation represents an increased expression per cell and/or an increase in the actual number of IL-23R+ cells still remains to be determined.
Figure 6.
Cytokine receptor expression in the cecum (Cec) and colon (Col) after Helicobacter hepaticus (Hh)–induced intestinal inflammation. C57BL/6 wild-type (WT) mice were inoculated with H. hepaticus alone (gray bars) or H. hepaticus plus anti–IL-10 receptor (IL-10R; black bars) monoclonal antibody (mAb) on days (d) 0 and 7 postinfection (pi). One week after the last mAb administration, ceca and colons were collected, and epithelial cells and the nonepithelial tissue compartment (A) or whole tissue (B) were processed for real-time RT-PCR analysis of cytokine receptor subunits. Uninfected mice were included as controls. A: Day 7 and 14 pi transcript levels of Il22ra1 (IL-22RA1), Il22ra2 [IL-22RA2/IL-22 binding protein (IL-22BP)], and Il23r (IL-23R) in the nonepithelial tissue compartment and epithelial cells of Cec and Col. B: Day 14 pi transcript levels of Il23r in whole Cec and Col from two independent experiments (Exp.). Data represent means ± SEM mRNA levels relative to hypoxantine-guanine phosphoribosyltransferase (HPRT; A and B). n = 4 individual mice per group from one experiment (A); n = 3 individual mice per group [Uninfected (uninf.; white bars) and H. hepaticus alone] or n = 4 individual mice per group (H. hepaticus/anti–IL-10R) per experiment (B). ∗P < 0.05 when comparing Cec with Col; †P < 0.05, ††P < 0.01, and †††P < 0.001 when compared with Cec or Col from uninfected hosts.
Differential Expression of Cytokine Receptor Transcripts in Different Regions of the Healthy Human Intestine
To examine whether regional variations in cytokine receptor expression were seen also in the healthy human gut, we mined microarray data from a study by Wolff et al34 in which biopsy specimens had been collected from four different parts of the intestine of healthy individuals, followed by transcriptional profiling analysis. We chose to focus on this study, because it had been designed to compare gene expression in different regions of the gut in the same individual (rather than collecting a single biopsy specimen per patient). The regions that underwent biopsy in the study by Wolff et al34 were the TI, ICV, AO, and SC, with the TI representing the small intestine, the ICV and AO corresponding to the most proximal part of the large intestine (ie, the cecum), and the SC defined as 20 cm from the anal verge.34 Samples from five of the six individuals in the study were included in the analysis, because microarray data were available from all four regions of the gut for these subjects.
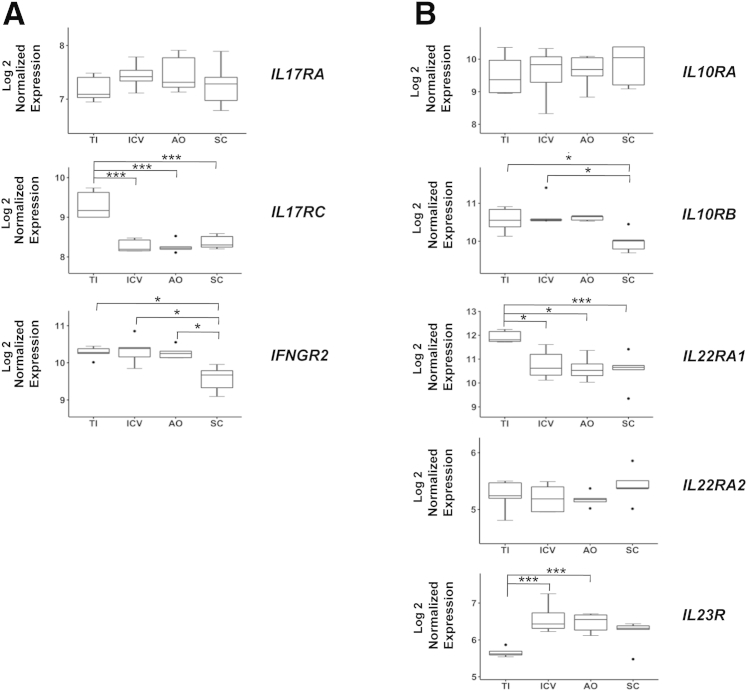
We first analyzed normalized expression values for the cytokine receptor transcripts we had previously examined in the mouse (Figure 5). Significant differences were observed between the small (TI) and large (ICV, AO, and SC) intestine for IL17RC, IL22RA1, and IL23R. Thus, the TI displayed higher levels of IL17RC and IL22RA1, but lower levels of IL23R than those found in the ICV, AO, and SC (Figure 7, A and B). Moreover, there were differences in transcript accumulation also within the large intestine itself, with SC expressing significantly lower levels of IFNGR2 and IL10RB than those detected in ICV and AO (Figure 7A) and ICV (Figure 7B), respectively. Moreover, analysis of mRNA accumulation for other cytokine receptors revealed additional differences in expression levels between different regions of the gut, including those of IL1R2, IL17RD, IL13RA1, IL20RA, IL28RA, and IL3RA (Supplemental Table S1). Together, these findings indicate that, similar to the mouse intestine, regional variations in cytokine receptor expression exist also in the healthy human gut.
Figure 7.
Differential cytokine receptor expression in different regions of the healthy human intestine. Microarray data from biopsy specimens of the terminal ileum (TI), ileocecal valve (ICV), appendiceal orifice (AO), and sigmoid colon (SC) from five healthy human subjects (http://www.ncbi.nlm.nih.gov/geo; accession number GSE55258; Wolff et al34) were analyzed as described in Materials and Methods. Normalized expression of IL17RA, IL17RC, and interferon γ receptor 2 (IFNGR2; A) and IL10RA, IL10RB, Il22RA1, Il22RA2 [IL-22 binding protein (IL-22BP)], and IL23R (B). Box-and-whisker plots represent median log 2 expression levels with boxes showing data between the 25th and 75th percentiles, whiskers extending to 1.5× the interquartile range, and outliers being >1.5× the interquartile range. ∗P < 0.05, ∗∗∗P < 0.001.
Discussion
IL-23 plays an essential role in intestinal inflammation, with IL-23–induced Th17-type cytokines being implicated in disease pathogenesis, but so far no information is available regarding the contribution of specific Th17-type cytokines to pathology in different regions of the gut. By using the H. hepaticus typhlocolitis model, we herein show that IL-17A and IL-22 play distinct roles in different anatomical locations of the intestine. Thus, IL-17A was found to play a disease-protective role in the cecum, whereas IL-22 was shown to be pathogenic in the colon, together demonstrating that different Th17-type cytokines may have proinflammatory or anti-inflammatory effects in different regions of the gut during H. hepaticus–induced pathology. We further provide evidence that cytokine receptor expression may help explain the difference in IL-22 dependence of the inflammation in these two tissues.
Many autoimmune and chronic inflammatory conditions in both humans and rodents are associated with elevated expression of Th17-type cytokines.37 This is particularly true for inflammatory disorders at mucosal sites and in the skin.38 Despite increased levels of Th17-associated cytokines in various diseases, a consensus view of whether these molecules are pathogenic or protective has not yet been reached, most likely because of the pleiotropic effects of cytokines and the fact that inflammation occurs in different tissues with distinct cellular compositions at steady state. With respect to the gastrointestinal tract, elevated levels of IL-17A, IL-17F, IL-21, and IL-22 have been found in both the inflamed intestine of humans with IBD and in experimental models of the disease.8 Although IL-17A can promote proinflammatory responses through the induction of neutrophil-attracting chemokines and matrix metalloproteinases,9 we herein demonstrate that H. hepaticus–induced typhlitis is exacerbated when IL-17A is neutralized, suggesting an anti-inflammatory role for this cytokine in T-cell–driven bacteria-induced intestinal inflammation. Consistent with our findings, previous studies have reported exacerbation of dextran sulphate sodium colitis in the absence of IL-17A, through either anti–IL-17A mAb treatment or the use of IL-17A−/− mice.39, 40 IL-17A has been reported to induce the expression of claudins in intestinal epithelial cells and promote mucin production,20, 21 effects known to increase barrier function. Thus, IL-17A may exert its tissue-protective role through fortification of the epithelial barrier. The reason for the different responses to anti–IL-17A treatment in the cecum versus colon in our study is less clear. Both tissues expressed similar levels of Il17ra and Il17rc transcripts; however, mRNA levels for IL-17A (and IL-22 and IFN-γ) were higher in the cecum than the colon, possibly affecting responses down-stream of the receptor. Moreover, differences in H. hepaticus loads41 and/or microflora composition between the cecum and colon are two additional factors that may influence the role of IL-17A in these tissues in the H. hepaticus colitis model. Although IL-17A neutralization exacerbated typhlitis in H. hepaticus–infected anti–IL-10R–treated WT mice, preliminary data from our laboratory indicate that WT and IL-17A–deficient animals given H. hepaticus plus anti–IL-10R display similar pathology (P.J.M. and M.C.K., unpublished data). These findings emphasize that experiments using genetic ablation of a cytokine may not necessarily yield the same result as mAb neutralization in a cytokine-sufficient setting. As such, the use of mAb neutralization may reveal proinflammatory or anti-inflammatory effects of a cytokine that are not observed when using cytokine-deficient mice where compensatory mechanisms have been allowed to emerge.
Although anti–IL-17A treatment revealed a disease-protective role of this cytokine in the cecum, blockade of IL-17F had no effect on either cecal or colonic inflammation in the H. hepaticus colitis model. Although IL-17A and IL-17F signal through the same IL-17RA/IL-17RC complex and are thought to have proinflammatory functions, such as expression of chemokines by epithelial cells and fibroblasts with subsequent neutrophil recruitment, in common,10 distinct biological effects have also been reported. Thus, IL-17A appears more efficient than IL-17F in inducing antimicrobial peptides by primary human keratinocytes and proinflammatory mediators from mouse embryonic fibroblasts in vitro.33, 40 Moreover, the use of gene-knockout animals has revealed that these two cytokines can have distinct functions in inflammatory responses in the gut. Thus, although IL-17A−/− animals develop exacerbated dextran sulphate sodium colitis compared with WT controls, IL-17F−/− mice display reduced disease with greatly diminished cell infiltrates in the LP.40 Besides IL-17A and IL-17F homodimers, there are also reports of IL-17F/A heterodimers,32, 42, 43 adding to the complexity by which these cytokines exert their effects on target cells. Assuming complete in vivo neutralization by the anti–IL-17A and anti–IL-17F mAbs used in the present study, one may deduce from our findings that the tissue-protective effect revealed by the anti–IL-17A mAb is because of homodimeric IL-17A, and not IL-17F/A heterodimers.
Unlike IL-17A, IL-22 was shown to be proinflammatory in H. hepaticus–induced intestinal inflammation. The literature has revealed both protective and pathogenic roles for IL-22 in the intestine. Thus, IL-22 has been reported to have protective effects against Citrobacter rodentium–induced inflammation,36 in the spontaneous colitis arising in T-cell receptor-α−/− mice,44 and in colitis induced by dextran sulphate sodium or by transfer to Rag−/− mice of CD45RBhigh CD4+ T cells.45 Conversely, IL-22 has been demonstrated to contribute to the colitic response in Rag−/− mice given T-regulatory cell–depleted CD45RBlow CD4+ T cells,46 in anti–CD40-induced colitis,47 and in the inflammatory response in the small intestine after Toxoplasma gondii infection.48, 49 Thus, IL-22 may have either beneficial or pathological roles in the intestine, depending on the nature of the immune insult.25 Interestingly and more important, we observed that IL-22 neutralization prevented the development of inflammation in the colon, but not the cecum, of H. hepaticus–infected anti–IL-10R–treated WT mice. This finding is similar to our previous observation that IFN-γ is needed for maximal pathology in the colon, but not the cecum, in the H. hepaticus colitis model.28 Thus, IL-22 and IFN-γ display similar region-specific effects in H. hepaticus–triggered disease pathogenesis.
In an attempt to understand the difference in IL-22 dependence in cecal versus colonic inflammation, we examined IL-22 and IL-22R expression in the two tissues. Although Il22 transcript levels were higher in the cecum compared with the colon, both at steady state and after H. hepaticus inoculation, we believe we have ruled out the possibility that incomplete neutralization of cecal IL-22 by the anti–IL-22 mAb is the reason for the lack of an effect in this tissue. This conclusion is on the basis of no change in cecal histology scores when increasing the dose of anti–IL-22 or when starting mAb treatment 2 days before H. hepaticus inoculation, and an even larger reduction in the accumulation of RegIIIg in the cecum compared with the colon after anti–IL-22 treatment, suggesting efficient neutralization of IL-22 activity at the former site. Interestingly, analysis of various IL-22R subunits in the large intestine indicated that the cecum and colon may exhibit different sensitivities to IL-22 signaling. Thus, the steady-state cecum had significantly higher levels of Il22ra2 transcripts, which encode the soluble IL-22BP that antagonizes IL-22 signaling through its 20- to 1000-fold higher affinity for the cytokine compared with that of the membrane-bound IL-22Rα1.15, 16, 17, 50, 51 Moreover, the colon displayed higher levels of Il22ra1 transcripts compared with the cecum, both at steady state and in inflammation. Thus, one explanation for the differential effect of anti–IL-22 treatment on cecal versus colonic inflammation could be that IL-22 is less involved in cecal inflammation because of it being sequestered by IL-22BP in this tissue, whereas the colon is more sensitive to IL-22 signaling owing to the higher levels of membrane-bound IL-22R. Further studies are required to examine this possibility.
An alternative explanation for the differential role of IL-22 in cecal versus colonic inflammation relates to the context in which IL-22 is expressed and how that regulates the functional consequences of its production. For example, IL-22 can act in a synergistic manner with cytokines, such as IL-1α and tumor necrosis factor-α, to promote inflammation in the skin.52 Furthermore, in bleomycin-induced airway inflammation, IL-22 has been reported to be pathogenic in the presence of IL-17A, but tissue protective in the absence of this cytokine.53 With our finding of IL-17A being more highly expressed in the cecum than in the colon, it is possible that IL-17A expression may influence the outcome of IL-22 signaling in H. hepaticus–induced disease. However, our finding of more severe cecal inflammation in H. hepaticus–infected anti–IL-10R–treated WT mice co-administered anti–IL-17A seems contradictory to IL-22 being tissue protective in the absence of IL-17A in the H. hepaticus colitis model.
IL-23, which is crucial for driving both cecal and colonic inflammation in H. hepaticus–infected hosts,28 has multiple functions in the immune system, including promoting Th17-type cytokine secretion by T- and non–T-cell sources, inhibiting the differentiation of induced T-regulatory cells, and indirectly restraining IL-10 production by CD4+ T cells.8 As such, IL-23 may be an additional driver of pathology in the cecum that overrides the effect of IL-22 blockade. Although we did not find any difference in Il23a transcript levels between the cecum and colon, we observed enhanced mRNA levels of both Il12b and Il23r in the cecum compared with the colon, suggesting that the former tissue may be more responsive to IL-23. Thus, it is possible that more overt IL-23 signaling in the cecum may prevent anti–IL-22 treatment from reducing H. hepaticus–induced pathology.
Taken together, our study provides novel insights into the roles of IL-17A and IL-22 in bacterial-induced T-cell–dependent intestinal inflammation. We demonstrate that individual Th17-type cytokines act in a region-specific manner in the gastrointestinal tract, providing either tissue-protective or proinflammatory effects. Although the definite mechanism is still unknown, these findings may potentially be explained partly by underlying differences in the expression of cytokine receptors in different anatomical locations of the intestine. The relevance of these findings to humans is highlighted by the fact that regional variations in cytokine receptor transcripts were observed also in the healthy human gut. Our data further indicate that IL-17A is not an attractive target for therapeutic intervention in intestinal inflammation. Indeed, anti–IL-17A treatment has been found to have insufficient therapeutic effect and may even increase the risk of serious adverse events in patients with severe Crohn disease.54 Whether targeting of IL-22 could have beneficial effects in patients with IBD is still unknown.26 Preliminary data from our laboratory, however, indicate that although IL-22 neutralization was able to prevent colonic inflammation when treatment was delayed to 7 days after H. hepaticus/anti–IL-10R inoculation, anti–IL-22 had no effect on pathology when treatment was initiated 14 days after H. hepaticus challenge (data not shown). Thus, in the case of already established disease, resolution of intestinal inflammation may depend on blocking additional proinflammatory pathways in addition to that down-stream of IL-22. In this regard, we previously demonstrated that anti–IL-12p40 mAb was able to reverse H. hepaticus–induced large intestinal inflammation in Il10−/− mice,55 a treatment likely to affect multiple pathways down-stream of IL-12 and/or IL-23. It is also plausible that IL-22 may display proinflammatory and/or tissue-protective effects at different phases of the inflammatory response. With increased IL-22 levels in active lesions of IBD patients,18, 19 and increased serum levels of IL-22 correlating with disease activity in Crohn disease,56 additional research is required to establish what factors control the outcome of IL-22 signaling in the intestine. Such information will be essential to help establish whether this cytokine can be exploited for clinical benefit.
Acknowledgments
We thank Allison Green and Paul Kaye for critical reading of the manuscript. Mouse Contract Services were provided by the Mary Lyon Center at Medical Research Council Harwell (Oxfordshire, UK).
Footnotes
Supported by Medical Research Council grant G0700806 (M.C.K.), Royal Society grant scheme 2007/R1 (M.C.K.), the Crohn's in Childhood Research Association (M.C.K.; doctoral studentship for P.J.M.), and Wellcome Trust grant 097829/Z/11/A [salary support for S.J.M. through the Centre for Chronic Diseases and Disorders (C2D2)].
Disclosures: J.F.W. is currently employed at Pfizer Inc. L.A.F. was employed at Pfizer Inc. at the time of this work but has since left the company.
Current address of P.J.M., The Francis Crick Institute Mill Hill Laboratory, The Ridgeway, Mill Hill, London, UK; of L.A.F., L A Fouser Consulting, LLC, Acton, MA.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2015.08.015.
Supplemental Data
Enhanced cytokine expression in the cecal compared with colonic nonepithelial tissue compartment in steady-state and during early stages of colitis. C57BL/6 wild-type (WT) mice were inoculated with Helicobacter hepaticus (Hh) alone or H. hepaticus plus anti–IL-10 receptor (IL-10R) monoclonal antibody. Ceca and colons were collected 7 days later, and epithelial cells and the nonepithelial tissue compartment were processed for real-time RT-PCR analysis of cytokines. Uninfected (Uninf.) mice were included as controls. Cecal and colonic mRNA levels of Il17a (IL-17A), Il22 (IL-22), and Ifng (IFN-γ) 7 days postinfection (pi) in the nonepithelial tissue compartment and epithelial cells (note y axis in two segments). Data are representative of two independent experiments using tissues collected between days 4 and 7 pi. Data represent means ± SEM mRNA levels relative to hypoxantine-guanine phosphoribosyltransferase (HPRT). n = 4 mice per group and time point. ∗P < 0.05 when comparing colon with cecum.
References
- 1.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maloy K.J., Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 4.Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., Dassopoulos T., Bitton A., Yang H., Targan S., Datta L.W., Kistner E.O., Schumm L.P., Lee A.T., Gregersen P.K., Barmada M.M., Rotter J.I., Nicolae D.L., Cho J.H. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maloy K.J., Kullberg M.C. IL-23 and Th17 cytokines in intestinal homeostasis. Mucosal Immunol. 2008;1:339–349. doi: 10.1038/mi.2008.28. [DOI] [PubMed] [Google Scholar]
- 6.Langrish C.L., McKenzie B.S., Wilson N.J., de Waal Malefyt R., Kastelein R.A., Cua D.J. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 7.Kamada N., Hisamatsu T., Okamoto S., Chinen H., Kobayashi T., Sato T., Sakuraba A., Kitazume M.T., Sugita A., Koganei K., Akagawa K.S., Hibi T. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest. 2008;118:2269–2280. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison P.J., Ballantyne S.J., Kullberg M.C. Interleukin-23 and T helper 17-type responses in intestinal inflammation: from cytokines to T-cell plasticity. Immunology. 2011;133:397–408. doi: 10.1111/j.1365-2567.2011.03454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwakura Y., Ishigame H., Saijo S., Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Gaffen S.L. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie M.H., Aggarwal S., Ho W.H., Foster J., Zhang Z., Stinson J., Wood W.I., Goddard A.D., Gurney A.L. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J Biol Chem. 2000;275:31335–31339. doi: 10.1074/jbc.M005304200. [DOI] [PubMed] [Google Scholar]
- 12.Kotenko S.V., Izotova L.S., Mirochnitchenko O.V., Esterova E., Dickensheets H., Donnelly R.P., Pestka S. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J Biol Chem. 2001;276:2725–2732. doi: 10.1074/jbc.M007837200. [DOI] [PubMed] [Google Scholar]
- 13.Tachiiri A., Imamura R., Wang Y., Fukui M., Umemura M., Suda T. Genomic structure and inducible expression of the IL-22 receptor alpha chain in mice. Genes Immun. 2003;4:153–159. doi: 10.1038/sj.gene.6363934. [DOI] [PubMed] [Google Scholar]
- 14.Wolk K., Kunz S., Witte E., Friedrich M., Asadullah K., Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Dumoutier L., Lejeune D., Colau D., Renauld J.C. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J Immunol. 2001;166:7090–7095. doi: 10.4049/jimmunol.166.12.7090. [DOI] [PubMed] [Google Scholar]
- 16.Kotenko S.V., Izotova L.S., Mirochnitchenko O.V., Esterova E., Dickensheets H., Donnelly R.P., Pestka S. Identification, cloning, and characterization of a novel soluble receptor that binds IL-22 and neutralizes its activity. J Immunol. 2001;166:7096–7103. doi: 10.4049/jimmunol.166.12.7096. [DOI] [PubMed] [Google Scholar]
- 17.Xu W., Presnell S.R., Parrish-Novak J., Kindsvogel W., Jaspers S., Chen Z., Dillon S.R., Gao Z., Gilbert T., Madden K., Schlutsmeyer S., Yao L., Whitmore T.E., Chandrasekher Y., Grant F.J., Maurer M., Jelinek L., Storey H., Brender T., Hammond A., Topouzis S., Clegg C.H., Foster D.C. A soluble class II cytokine receptor, IL-22RA2, is a naturally occurring IL-22 antagonist. Proc Natl Acad Sci U S A. 2001;98:9511–9516. doi: 10.1073/pnas.171303198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S.T., Otte J.M., Diepolder H., Marquardt A., Jagla W., Popp A., Leclair S., Herrmann K., Seiderer J., Ochsenkuhn T., Goke B., Auernhammer C.J., Dambacher J. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 19.Andoh A., Zhang Z., Inatomi O., Fujino S., Deguchi Y., Araki Y., Tsujikawa T., Kitoh K., Kim-Mitsuyama S., Takayanagi A., Shimizu N., Fujiyama Y. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 20.Kinugasa T., Sakaguchi T., Gu X., Reinecker H.C. Claudins regulate the intestinal barrier in response to immune mediators. Gastroenterology. 2000;118:1001–1011. doi: 10.1016/s0016-5085(00)70351-9. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y., Thai P., Zhao Y.H., Ho Y.S., DeSouza M.M., Wu R. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. J Biol Chem. 2003;278:17036–17043. doi: 10.1074/jbc.M210429200. [DOI] [PubMed] [Google Scholar]
- 22.Wolk K., Witte E., Wallace E., Docke W.D., Kunz S., Asadullah K., Volk H.D., Sterry W., Sabat R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36:1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 23.Aujla S.J., Chan Y.R., Zheng M., Fei M., Askew D.J., Pociask D.A., Reinhart T.A., McAllister F., Edeal J., Gaus K., Husain S., Kreindler J.L., Dubin P.J., Pilewski J.M., Myerburg M.M., Mason C.A., Iwakura Y., Kolls J.K. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H.A., Hirth S., Weigmann B., Wirtz S., Ouyang W., Neurath M.F., Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnenberg G.F., Fouser L.A., Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 26.Sabat R., Ouyang W., Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 27.Kullberg M.C., Ward J.M., Gorelick P.L., Caspar P., Hieny S., Cheever A., Jankovic D., Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kullberg M.C., Jankovic D., Feng C.G., Hue S., Gorelick P.L., McKenzie B.S., Cua D.J., Powrie F., Cheever A.W., Maloy K.J., Sher A. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison P.J., Bending D., Fouser L.A., Wright J.F., Stockinger B., Cooke A., Kullberg M.C. Th17-cell plasticity in Helicobacter hepaticus-induced intestinal inflammation. Mucosal Immunol. 2013;6:1143–1156. doi: 10.1038/mi.2013.11. [DOI] [PubMed] [Google Scholar]
- 30.Ward J.M., Anver M.R., Haines D.C., Benveniste R.E. Chronic active hepatitis in mice caused by Helicobacter hepaticus. Am J Pathol. 1994;145:959–968. [PMC free article] [PubMed] [Google Scholar]
- 31.Fox J.G., Dewhirst F.E., Tully J.G., Paster B.J., Yan L., Taylor N.S., Collins M.J., Jr., Gorelick P.L., Ward J.M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang S.C., Long A.J., Bennett F., Whitters M.J., Karim R., Collins M., Goldman S.J., Dunussi-Joannopoulos K., Williams C.M., Wright J.F., Fouser L.A. An IL-17F/A heterodimer protein is produced by mouse Th17 cells and induces airway neutrophil recruitment. J Immunol. 2007;179:7791–7799. doi: 10.4049/jimmunol.179.11.7791. [DOI] [PubMed] [Google Scholar]
- 33.Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., Fouser L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolff M.J., Leung J.M., Davenport M., Poles M.A., Cho I., Loke P. TH17, TH22 and TReg cells are enriched in the healthy human cecum. PLoS One. 2012;7:e41373. doi: 10.1371/journal.pone.0041373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 36.Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 37.Korn T., Oukka M., Kuchroo V., Bettelli E. Th17 cells: effector T cells with inflammatory properties. Semin Immunol. 2007;19:362–371. doi: 10.1016/j.smim.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver C.T., Elson C.O., Fouser L.A., Kolls J.K. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogawa A., Andoh A., Araki Y., Bamba T., Fujiyama Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin Immunol. 2004;110:55–62. doi: 10.1016/j.clim.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Yang X.O., Chang S.H., Park H., Nurieva R., Shah B., Acero L., Wang Y.H., Schluns K.S., Broaddus R.R., Zhu Z., Dong C. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maloy K.J., Salaun L., Cahill R., Dougan G., Saunders N.J., Powrie F. CD4+CD25+ TR cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med. 2003;197:111–119. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright J.F., Guo Y., Quazi A., Luxenberg D.P., Bennett F., Ross J.F., Qiu Y., Whitters M.J., Tomkinson K.N., Dunussi-Joannopoulos K., Carreno B.M., Collins M., Wolfman N.M. Identification of an interleukin 17F/17A heterodimer in activated human CD4+ T cells. J Biol Chem. 2007;282:13447–13455. doi: 10.1074/jbc.M700499200. [DOI] [PubMed] [Google Scholar]
- 43.Chang S.H., Dong C. A novel heterodimeric cytokine consisting of IL-17 and IL-17F regulates inflammatory responses. Cell Res. 2007;17:435–440. doi: 10.1038/cr.2007.35. [DOI] [PubMed] [Google Scholar]
- 44.Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A.K., Blumberg R.S., Xavier R.J., Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zenewicz L.A., Yancopoulos G.D., Valenzuela D.M., Murphy A.J., Stevens S., Flavell R.A. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamanaka M., Huber S., Zenewicz L.A., Gagliani N., Rathinam C., O'Connor W., Jr., Wan Y.Y., Nakae S., Iwakura Y., Hao L., Flavell R.A. Memory/effector (CD45RBlo) CD4 T cells are controlled directly by IL-10 and cause IL-22-dependent intestinal pathology. J Exp Med. 2011;208:1027–1040. doi: 10.1084/jem.20102149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eken A., Singh A.K., Treuting P.M., Oukka M. IL-23R+ innate lymphoid cells induce colitis via interleukin-22-dependent mechanism. Mucosal Immunol. 2014;7:143–154. doi: 10.1038/mi.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munoz M., Heimesaat M.M., Danker K., Struck D., Lohmann U., Plickert R., Bereswill S., Fischer A., Dunay I.R., Wolk K., Loddenkemper C., Krell H.W., Libert C., Lund L.R., Frey O., Holscher C., Iwakura Y., Ghilardi N., Ouyang W., Kamradt T., Sabat R., Liesenfeld O. Interleukin (IL)-23 mediates Toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J Exp Med. 2009;206:3047–3059. doi: 10.1084/jem.20090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson M.S., Feng C.G., Barber D.L., Yarovinsky F., Cheever A.W., Sher A., Grigg M., Collins M., Fouser L., Wynn T.A. Redundant and pathogenic roles for IL-22 in mycobacterial, protozoan, and helminth infections. J Immunol. 2010;184:4378–4390. doi: 10.4049/jimmunol.0903416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Logsdon N.J., Jones B.C., Josephson K., Cook J., Walter M.R. Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J Interferon Cytokine Res. 2002;22:1099–1112. doi: 10.1089/10799900260442520. [DOI] [PubMed] [Google Scholar]
- 51.Wolk K., Witte E., Hoffmann U., Doecke W.D., Endesfelder S., Asadullah K., Sterry W., Volk H.D., Wittig B.M., Sabat R. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: a potential systemic role of IL-22 in Crohn's disease. J Immunol. 2007;178:5973–5981. doi: 10.4049/jimmunol.178.9.5973. [DOI] [PubMed] [Google Scholar]
- 52.Guilloteau K., Paris I., Pedretti N., Boniface K., Juchaux F., Huguier V., Guillet G., Bernard F.X., Lecron J.C., Morel F. Skin inflammation induced by the synergistic action of IL-17A, IL-22, oncostatin M, IL-1alpha, and TNF-alpha recapitulates some features of psoriasis. J Immunol. 2010;184:5263–5270. doi: 10.4049/jimmunol.0902464. [DOI] [PubMed] [Google Scholar]
- 53.Sonnenberg G.F., Nair M.G., Kirn T.J., Zaph C., Fouser L.A., Artis D. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J Exp Med. 2010;207:1293–1305. doi: 10.1084/jem.20092054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hueber W., Sands B.E., Lewitzky S., Vandemeulebroecke M., Reinisch W., Higgins P.D., Wehkamp J., Feagan B.G., Yao M.D., Karczewski M., Karczewski J., Pezous N., Bek S., Bruin G., Mellgard B., Berger C., Londei M., Bertolino A.P., Tougas G., Travis S.P. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kullberg M.C., Rothfuchs A.G., Jankovic D., Caspar P., Wynn T.A., Gorelick P.L., Cheever A.W., Sher A. Helicobacter hepaticus-induced colitis in interleukin-10-deficient mice: cytokine requirements for the induction and maintenance of intestinal inflammation. Infect Immun. 2001;69:4232–4241. doi: 10.1128/IAI.69.7.4232-4241.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmechel S., Konrad A., Diegelmann J., Glas J., Wetzke M., Paschos E., Lohse P., Goke B., Brand S. Linking genetic susceptibility to Crohn's disease with Th17 cell function: IL-22 serum levels are increased in Crohn's disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis. 2008;14:204–212. doi: 10.1002/ibd.20315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Enhanced cytokine expression in the cecal compared with colonic nonepithelial tissue compartment in steady-state and during early stages of colitis. C57BL/6 wild-type (WT) mice were inoculated with Helicobacter hepaticus (Hh) alone or H. hepaticus plus anti–IL-10 receptor (IL-10R) monoclonal antibody. Ceca and colons were collected 7 days later, and epithelial cells and the nonepithelial tissue compartment were processed for real-time RT-PCR analysis of cytokines. Uninfected (Uninf.) mice were included as controls. Cecal and colonic mRNA levels of Il17a (IL-17A), Il22 (IL-22), and Ifng (IFN-γ) 7 days postinfection (pi) in the nonepithelial tissue compartment and epithelial cells (note y axis in two segments). Data are representative of two independent experiments using tissues collected between days 4 and 7 pi. Data represent means ± SEM mRNA levels relative to hypoxantine-guanine phosphoribosyltransferase (HPRT). n = 4 mice per group and time point. ∗P < 0.05 when comparing colon with cecum.