Abstract
Hepatocellular carcinoma (HCC) is one of the most aggressive cancers and is the third leading cause of all cancer-related death. Limited noninvasive biomarkers are available for HCC detection. Early detection is the key in improving the survival of HCC patients. In this study, we tested the hypothesis that serum miRNAs can be used as a potential biomarker for HCC. Quantitative RT-PCR for miRNA analysis was performed using 70 serum samples. Receiver operating characteristic analysis was performed to measure the prognostic power of the miRNAs. The miRNA expression level was also measured from liver biopsy samples. Our study revealed that two miRNAs, miR-30e and miR-223, were expressed at significantly lower levels (P < 0.003) in the sera of HCC patients compared with healthy volunteers. Furthermore, expression of these miRNAs was compared between sera from chronic liver disease and sera from HCC patients. miR-30e and miR-223 expression was significantly lower in HCC sera compared with sera from chronic liver disease patients. Both miRNA expression levels were lower in HCC liver biopsy specimens compared with normal liver RNA. Taken together, our results suggested that serum miR-30e and miR-223 are useful biomarkers of HCC, irrespective of etiology, and deserve further study for their diagnostic value.
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world, and the third most frequent cause of cancer mortality worldwide, leading to >600,000 deaths each year.1, 2 The American Cancer Society estimates that approximately 35,660 new cases will be diagnosed as primary liver cancer, and approximately 24,550 individuals will die of HCC in the United States in 2015. Several factors are involved in the development of HCC. The most common risk factor worldwide for HCC is chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV). In addition, sex, alcohol abuse, obesity, and type 2 diabetes are associated with an increased risk of HCC. Noninvasive fibroscan is becoming a diagnostic tool; however, it does not predict liver disease progression. Thus, identifying individuals at high risk for developing HCC generates new opportunities for surveillance, therapeutic intervention, and patient management.
miRNAs are a class of small, single-stranded, noncoding RNA of 22 nucleotides with a characteristic hairpin secondary structure.3, 4 They regulate gene silencing by either targeting mRNA directly into degradation or inhibiting translation. Altered expression of miRNAs has been associated with various types of cancers, including HCC.
Serum miRNAs have been implicated for potential biomarkers in several diseases. Approximately 100 circulating miRNAs have been identified as biomarkers for different diseases, and the number is growing.5, 6 Growing evidence indicates that their deregulation plays an important role in cancer onset and progression.7, 8 For example, miR-25 and miR-223 are potential serum biomarkers for lung cancer,9 miR-184 for squamous cell carcinoma,10 miR-92a for leukemia,11 and miR-122 for liver injury.12, 13 miR-141 and miR-375 are the most promising markers correlated with prostate tumor progression.14 Herein, we demonstrated that down-regulation of serum miR-30e and miR-223 is associated with HCC, and they could be used as noninvasive biomarkers for the detection of this disease.
Materials and Methods
Study Design and Patient Samples
Our study was approved by the Saint Louis University (St. Louis, MO) Institutional Review Board, and written informed consent was obtained from all subjects. A total of 70 subjects, including 14 HCV-infected patients with HCC, 14 HBV-infected patients with HCC, 11 non–viral-associated patients with HCC, 17 chronic liver disease (CLD) patients, and 14 healthy volunteers (HVs), were included in this study. All subjects in the HV group had normal aminotransferase activities, no history of liver disease or alcohol abuse, and had test results that were negative for HBV, HCV, and HIV infections. Table 1 shows the characteristics of the serum samples included in this study.
Table 1.
Patient Characteristics
| Characteristics | Serum samples |
||||
|---|---|---|---|---|---|
| HV | CLD | HBV-HCC | HCV-HCC | NV-HCC | |
| No. of samples | 14 | 17 | 14 | 14 | 11 |
| Age, years∗ | 38 ± 9 | 60 ± 13 | 58 ± 12 | 58 ± 10 | 62.5 ± 27 |
| Male/female ratio | 10:4 | 13:4 | 11:3 | 9:5 | 9:2 |
| ALT, IU/L† | 14 (6–37) | 95 (14–149) | 64 (15–301) | 70.8 (5–194) | 41 (24–198) |
| AST, IU/L† | 19 (9–25) | 61.5 (12–169) | 87.5 (35–308) | 114 (4–157) | 43 (16–161) |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; CLD, chronic liver disease; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HV, healthy volunteer; NV, nonviral.
Data are given as means ± SD.
Data are given as medians (ranges).
miRNA-Specific Quantitative Real-Time RT-PCR
Total RNA was isolated from 200 μL of serum by miRVana PARIS kit (Life Technologies, Grand Island, NY), according to the manufacturer's instructions. Synthetic spiked-in Caenorhabditis elegans miR-39 was added to the serum samples before RNA extraction as an internal control. There is no consensus on the use of housekeeping miRNAs, and it was reported that frequently used reference genes, such as U6 snRNA and 5S ribosomal RNA, are easily degraded in plasma/serum samples.15 A large variation of serum U6 levels was also reported in several studies,16 including ours.17 We used TaqMan quantitative RT-PCR assays to examine the expression of miRNAs in serum RNA of all samples. All reagents, primers, and probes were purchased from Life Technologies (TaqMan Gene Expression Master Mix 4369016, TaqMan MicroRNA Reverse Transcription Kit 4366596, miR-30e-5p primer Assay ID–002223 and miR-223-3p primer Assay ID–002295).
Real-time PCR was performed using an ABI 7500 Sequence Detection System (Life Technologies), and fold changes in gene expression were calculated using the 2−ΔΔCt method. The mean miRNA level of the three quantitative real-time PCR experiments was calculated for each case.
Liver biopsy specimens from HCC adult patients for our study were approved by the Saint Louis University Institutional Review Board, and written informed consent was obtained from all subjects. Total RNA was isolated using TRIzol Reagent (Invitrogen, Grand Island, NY). cDNA was synthesized using miR-30e–, miR-223–, or U6-specific primers with the TaqMan MicroRNA Reverse Transcription Kit. Real-time PCR was performed for quantitation using TaqMan universal PCR master mix, and calculated using the 2−ΔΔCt method.
Statistical Analysis
Data were analyzed by nonparametric tests using Wilcoxon test for comparison of paired samples and U test for two nonparametric groups, as we described previously.17 Receiver operating characteristic curves were generated, and the area under the curve (AUC) was calculated to evaluate specificity and sensitivity of predictive value or feasibility of using serum miRNA as a marker for liver disease progression. P < 0.05 was considered statistically significant. All statistical analyses were performed, and graphs were generated using GraphPad Prism version 6.0 (GraphPad Software Inc., La Jolla, CA) and R software version 3.1.3 (The R Project for Statistical Computing, http://www.r-project.org, last accessed September 8, 2015).
Results
Profiling of Serum miRNA Levels in HCV-Infected Patients with HCC
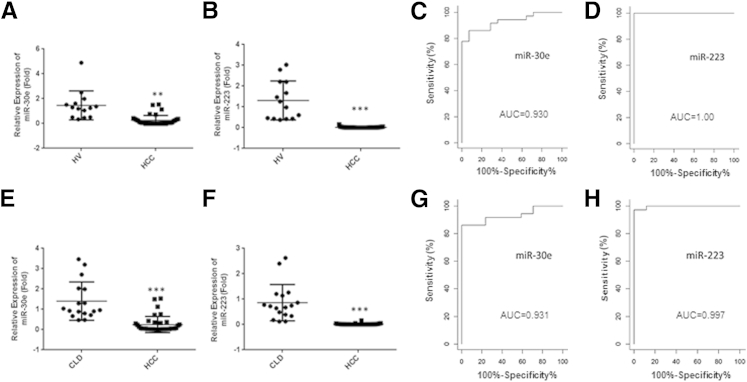
We observed the differential expression of six miRNAs (miR-21, miR-29a, miR-122, and miR-195 are up-regulated, and miR-30e and miR-223 are down-regulated) as a signature for HCC patients, on the basis of our previous array data.17 Among these dysregulated serum miRNAs, miR-21 may be a non-specific biomarker for HCC, because other studies have suggested its potential as a biomarker for other cancers.18 miR-122 has also been shown to be differentially regulated in liver injury irrespective of etiology.12, 19, 20 We chose not to include these two miRNAs as biomarker for HCC. We further validated the remaining four miRNAs individually in a small cohort of sera from HVs and HCC samples. During validation, we did not observe a significant difference in expression of miR-29a and miR-195. On the other hand, we observed that miR-30e and miR-223 expression levels were significantly down-regulated in sera from HCC patients compared with HV sera (Figure 1, A and B). We next determined the predictive value of these miRNAs in identifying the HCC patients in a cohort of 14 HVs and 39 HCC patients' sera. The expression levels of miR-30e and miR-223 in these serum samples were measured, and receiver operating characteristic analysis was performed on individual miRNAs. miR-30e had an AUC of 0.930 ± 0.067 (95% CI, 0.864–0.997), with a sensitivity of 91.67% and a specificity of 71.43%, and miR-223 had an AUC of 1.00, with a sensitivity of 100% and a specificity of 100% in separating the healthy controls from HCC patients (Figure 1, C and D). Next, we compared the expression profile of miR-30e and miR-223 between sera from CLD and HCC patients. Our results demonstrated that both miRNAs are significantly reduced in HCC sera compared with sera from CLD (Figure 1, E and F). miR-30e had an AUC of 0.931 ± 0.068 (95% CI, 0.864–0.999), with a sensitivity of 91.67% and a specificity of 70.59%, and miR-223 had an AUC of 0.9945 ± 0.0055 (95% CI, 0.989–1.000), with a sensitivity of 97.22% and a specificity of 94.12% in separating the CLD from all HCC patients (Figure 1, G and H).
Figure 1.
Down-regulation of serum levels of miR-30e and miR-223. Scatter plot of serum miRNA levels of miR-30e (A) and miR-223 (B) in healthy volunteers (HVs) and hepatocellular carcinoma (HCC) patients. The line indicates the median value per group. Fold-regulation values are expressed as relative quantification on the basis of 2ΔΔCt method. A U test was used to determine statistical significance. Receiver operating characteristic (ROC) curve was analyzed using serum miR-30e and miR-223 for discriminating HCC patients. ROC curves with corresponding area under the ROC curve (AUC) for miR-30e (C) and miR-223 (D) in discriminating HCC patients from HVs. Scatter plot of serum levels of miR-30e (E) and miR-223 (F) in chronic liver disease (CLD) and HCC patients. ROC curves with corresponding AUC for miR-30e (G) and miR-223 (H) in discriminating HCC patients from CLD. Data are presented as means ± SD. n = 14 (A and B, healthy volunteers); n = 39 (A, B, E, and F, HCC patients); n = 17 (E and F, CLD patients). ∗∗P < 0.01, ∗∗∗P < 0.001.
Expression Level of miR-30e and miR-223 Is Significantly Reduced in HCC Patients Irrespective of Etiology
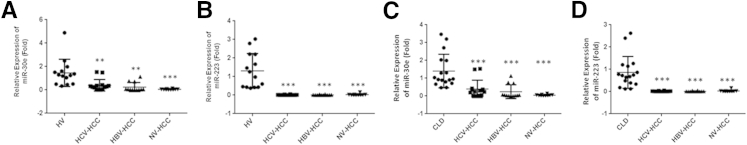
We next stratified the data on the basis of the etiology of the sera samples. Our data indicated that expression of miR-30e and miR-223 was down-regulated in HCC sera samples from HCV/HBV-associated HCC and nonviral HCC compared with sera from HVs (Figure 2, A and B). Similar results were obtained when we compared the expression level of miR-30e and miR-223 from CLD patients and HCC sera from different etiology (Figure 2, C and D).
Figure 2.
Down-regulation of serum levels of miR-30e and miR-223 in different etiologies of hepatocellular carcinoma (HCC) patients. A: Scatter plot of serum levels of miR-30e in healthy volunteers (HVs), hepatitis B virus (HBV)–induced HCC, hepatitis C virus (HCV)–induced HCC patients, and non–viral (NV)-induced HCC. B: Scatter plot of serum levels of miR-223 in HVs, HBV-induced HCC, HCV-induced HCC patients, and NV-induced HCC. C and D: Scatter plot of serum levels of miR-30e (C) and miR-223 (D) in chronic liver disease (CLD), HBV-induced HCC, HCV-induced HCC patients, and NV-induced HCC. The line indicates the median value per group. Fold-regulation values are expressed as relative quantification on the basis of 2ΔΔCt method. One-way analysis of variance was used to determine statistical significance. Data are presented as means ± SD (A–D). n = 14 (A and B, healthy volunteers and HBV- and HCV-induced HCC patients; C and D, HBV- and HCV-induced HCC patients); n = 11 (A, C, and D, NV-induced HCC patients); n = 17 (C and D, CLD patients). ∗∗P < 0.01, ∗∗∗P < 0.001.
miR-30e and miR-223 Expression Levels Are Highly Reduced in Liver Biopsy Samples of HCC Patients
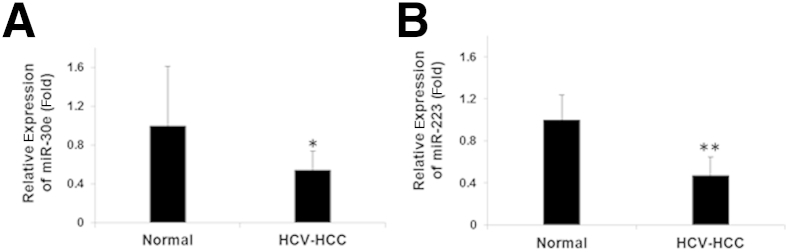
We further verified the expression level of miR-30e and miR-223 in HCC liver biopsy specimens by quantitative RT-PCR. We observed the down-regulation of miR-30e in liver samples from HCC patients (Figure 3A). In agreement with an earlier report,21 we also observed reduced expression of miR-223 in these samples (Figure 3B). In fact, Wong et al21 used HCC tissues from T2-T4 stage from different etiology, and observed inhibition of miR-223 expression. A recent report also suggested the reduced miR-223 expression in HCV-mediated HCC sera.22 However, miR-30c was slightly up-regulated (1.069-fold) as opposed to our study (although we examined miR-30e), and this difference could be because of different HCV genotype–infected HCC sera or demographic location.
Figure 3.
Reduced expression of miR-30e and miR-223 of liver biopsy samples compared with normal liver RNA. Relative expression of miR-30e (A) and miR-223 (B) from liver biopsy samples collected from hepatocellular carcinoma (HCC) patients. Normal liver RNA was purchased from Life Technologies. Fold-regulation values are expressed as relative quantification on the basis of the 2ΔΔCt method. Data are presented as means ± SD (A and B). ∗P < 0.05, ∗∗P < 0.01. HCV, hepatitis C virus.
Discussion
HCC is one of the most common malignant tumors, with poor prognosis and a major cause of death worldwide.1, 2 Serum miRNAs have been implicated as potential biomarkers in several diseases and represent a promising area of research for clinical diagnostics. Growing evidence indicates that their deregulation plays an important role in cancer onset and progression. In this study, we found two serum miRNAs, miR-30e and miR-223, were present at lower levels in HCC patient sera compared with healthy controls. Several miRNAs have been reported to be aberrantly overexpressed or underexpressed in liver disease.8 However, there is no report describing a common serum miRNA as a biomarker in HCC from different etiology. Herein, we demonstrated that miR-30e and miR-223 are down-regulated in HCC sera from different etiology, although our sample size was not powered for this study.
miR-30e has been implicated as a subtype-specific prognostic marker in breast cancer.23, 24 miR-30e expression is lower in chronic myeloid leukemia.25 miR-30 has been implicated for the development of the hepatobiliary system in embryonic and postnatal development26 and lipid biosynthesis.27 A recent study indicated that miR-30 helps to protect against chemokine ligand 4–induced liver fibrosis in a transforming growth factor–mediated pathway.28 However, the role of miR-30e in HCC remains unknown.
miR-223 is responsible for regulating cytochrome B5. Cytochrome B5 is a hemoprotein that transfers electrons to several enzymes to fulfill functions in fatty acid desaturation.29 Down-regulation of miR-223 can activate cell proliferation via insulin-like growth factor 1 receptor, and helps to maintain cholesterol homeostasis.30 The overexpression of miR-223 in HCC increases the sensitivity of anticancer drugs in HCC cell lines.31 miR-223 expression is reduced in HCC from adjacent nontumoral liver, irrespective of viral and nonviral associations,21 in agreement with our results. On the contrary, miR-223 expression has been shown to be elevated in patients with HCC or chronic hepatitis.32 We found that miR-223 is significantly reduced in HCC patient sera irrespective of the etiology compared with HVs.
The role of miR-30e and miR-223 in the development of HCC is poorly understood. To define potential targets of miR-30e and miR-223, we performed an in silico search for candidate genes that were commonly predicted by three publicly available algorithms, miRanda (http://www.microrna.org), miRDB (http://mirdb.org/miRDB), and TargetScan (http://www.targetscan.org; all last accessed March 4, 2015). Autophagy plays a role in pathophysiology of liver, and the autophagy pathway can be a novel therapeutic target for liver disease.33 miR-30e targets autophagy-related genes (ATG5 and ATG12), and inhibition of miR-30e in HCC may enhance autophagy. miR-30e also targets homeobox A1, a transcription factor that enhances STAT3/5 expression,34 which, in part, promotes cell growth.
miR-223 targets insulin-like growth factor 1 receptor. High insulin-like growth factor 1 receptor expression correlated with the liver tumor grade and cirrhosis.35 miR-223 also targets Stathmin1, a key microtubule-regulatory protein that controls the microtubule dynamics, cellular proliferation, and S-phase of the cell cycle.21 Because miRNA-mediated gene regulation is involved in gene regulatory pathways, it is possible that miR-30e and miR-223 are involved in HCC. However, future work is needed to elucidate the mechanism.
In conclusion, we demonstrated that down-regulation of miR-30e and miR-223 is associated with HCC, with high sensitivity and specificity. We also observed that expression levels of miR-30e and miR-223 were reduced in HCC sera and liver biopsy specimens, irrespective of their etiology, suggesting that miR-30e and miR-223 have potential as a noninvasive biomarker for HCC. Further studies are needed for their diagnostic value using a larger cohort.
Acknowledgments
We thank Anupam Mukherjee for initiation of this work and Patricia Osmack for helping us with the serum samples.
Footnotes
Supported by NIH research grant DK081817 and Saint Louis University Liver Center Grant 292287.
Disclosures: None declared.
References
- 1.El-Serag H.B., Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: where are we? where do we go? Hepatology. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Matzke M., Ruvkun G., Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 5.Weiland M., Gao X.H., Zhou L., Mi Q.S. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9:850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 6.Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75–93. doi: 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Giordano S., Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 8.Liu H.S., Xiao H.S. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol. 2014;20:12007–12017. doi: 10.3748/wjg.v20.i34.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Zhang Y., Wang J., Jiang X., Xiang Y., Xu C., Zheng P., Zhang J., Li R., Zhang H., Shang X., Gong T., Ning G., Wang J., Zen K., Zhang J., Zhang C.Y. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 10.Wong T.S., Liu X.B., Wong B.Y., Ng R.W., Yuen A.P., Wei W.I. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka M., Oikawa K., Takanashi M., Kudo M., Ohyashiki J., Ohyashiki K., Kuroda M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Zhang S., Marzolf B., Troisch P., Brightman A., Hu Z., Hood L.E., Galas D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding X., Ding J., Ning J., Yi F., Chen J., Zhao D., Zheng J., Liang Z., Hu Z., Du Q. Circulating microRNA-122 as a potential biomarker for liver injury. Mol Med Rep. 2012;5:1428–1432. doi: 10.3892/mmr.2012.838. [DOI] [PubMed] [Google Scholar]
- 14.Brase J.C., Johannes M., Schlomm T., Fälth M., Haese A., Steuber T., Beissbarth T., Kuner R., Sültmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 15.Peltier H.J., Latham G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi R., Weiland M., Gao X.H., Zhou L., Mi Q.S. Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology. 2012;55:1640–1642. doi: 10.1002/hep.25558. [DOI] [PubMed] [Google Scholar]
- 17.Shrivastava S., Petrone J., Steele R., Lauer G.M., Di Bisceglie A.M., Ray R.B. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58:863–871. doi: 10.1002/hep.26296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brase J.C., Wuttig D., Kuner R., Sültmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trebicka J., Anadol E., Elfimova N., Strack I., Roggendorf M., Viazov S., Wedemeyer I., Drebber U., Rockstroh J., Sauerbruch T., Dienes H.P., Odenthal M. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2013;58:234–239. doi: 10.1016/j.jhep.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Starkey Lewis P.J., Dear J., Platt V., Simpson K.J., Craig D.G., Antoine D.J., French N.S., Dhaun N., Webb D.J., Costello E.M., Neoptolemos J.P., Moggs J., Goldring C.E., Park B.K. Circulating microRNAs as potential markers of human drug induced liver injury. Hepatology. 2011;54:1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 21.Wong Q.W., Lung R.W., Law P.T., Lai P.B., Chan K.Y., To K.F., Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Oksuz Z., Serin M.S., Kaplan E., Dogen A., Tezcan S., Aslan G., Emekdas G., Sezgin O., Altintas E., Tiftik E.N. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015;42:713–720. doi: 10.1007/s11033-014-3819-9. [DOI] [PubMed] [Google Scholar]
- 23.D'Aiuto F., Callari M., Dugo M., Merlino G., Musella V., Miodini P., Paolini B., Cappelletti V., Daidone M.G. miR-30e* is an independent subtype-specific prognostic marker in breast cancer. Br J Cancer. 2015;113:290–298. doi: 10.1038/bjc.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasparini P., Cascione L., Fassan M., Lovat F., Guler G., Balci S., Irkkan C., Morrison C., Croce C.M., Shapiro C.L., Huebner K. MicroRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5:1174–1184. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hershkovitz-Rokah O., Modai S., Pasmanik-Chor M., Toren A., Shomron N., Raanani P., Shpilberg O., Granot G. miR-30e induces apoptosis and sensitizes K562 cells to imatinib treatment via regulation of the BCR-ABL protein. Cancer Lett. 2015;356:597–605. doi: 10.1016/j.canlet.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Hand N.J., Master Z.R., Eauclaire S.F., Weinblatt D.E., Matthews R.P., Friedman J.R. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soh J., Iqbal J., Queiroz J., Fernandez-Hernando C., Hussain M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu X., Zheng X., Li H., Cao Z., Chang H., Luan S., Zhu J., Chen J., Zang Y., Zhang J. MicroRNA-30 protects against carbon tetrachloride-induced liver fibrosis by attenuating transforming growth factor beta signaling in hepatic stellate cells. Toxicol Sci. 2015;146:157–169. doi: 10.1093/toxsci/kfv081. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi K., Oda Y., Toyoda Y., Fukami T., Yokoi T., Nakajima M. Regulation of cytochrome b5 expression by miR-223 in human liver: effects on cytochrome P450 activities. Pharm Res. 2014;31:780–794. doi: 10.1007/s11095-013-1200-7. [DOI] [PubMed] [Google Scholar]
- 30.Jia C.Y., Li H.H., Zhu X.C., Dong Y.W., Fu D., Zhao Q.L., Wu W., Wu X.Z. miR-223 suppresses cell proliferation by targeting IGF-1R. PLoS One. 2011;6:e27008. doi: 10.1371/journal.pone.0027008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang T., Zheng Z.M., Li X.N., Li Z.F., Wang Y., Geng Y.F., Bai L., Zhang X.B. miR-223 modulates multidrug resistance via downregulation of ABCB1 in hepatocellular carcinoma cells. Exp Biol Med (Maywood) 2013;238:1024–1032. doi: 10.1177/1535370213497321. [DOI] [PubMed] [Google Scholar]
- 32.Xu J., Wu C., Che X., Wang L., Yu D., Zhang T., Huang L., Li H., Tan W., Wang C., Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 33.Wang K. Autophagy and apoptosis in liver injury. Cell Cycle. 2015;14:1631–1642. doi: 10.1080/15384101.2015.1038685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee A., Shrivastava S., Chowdhury J.B., Ray R., Ray R.B. Transcriptional suppression of miR-181c by hepatitis C virus enhances homeobox A1 expression. J Virol. 2014;88:7929–7940. doi: 10.1128/JVI.00787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang G.H., Lee B.S., Lee E.S., Kim S.H., Lee H.Y., Kang D.Y. Prognostic significance of p53, mTOR, c-Met, IGF-1R, and HSP70 overexpression after the resection of hepatocellular carcinoma. Gut Liver. 2014;8:79–87. doi: 10.5009/gnl.2014.8.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]