Abstract
Despite the fact that colorectal cancer (CRC) is one of the most prevalent and deadly cancers in the world, the development of improved and robust biomarkers to enable screening, surveillance, and therapy monitoring of CRC continues to be evasive. In particular, patients with colon polyps are at higher risk of developing colon cancer; however, noninvasive methods to identify these patients suffer from poor performance. In consideration of the challenges involved in identifying metabolite biomarkers in individuals with high risk for colon cancer, we have investigated NMR-based metabolite profiling in combination with numerous demographic parameters to investigate the ability of serum metabolites to differentiate polyp patients from healthy subjects. We also investigated the effect of disease risk on different groups of biologically related metabolites. A powerful statistical approach, seemingly unrelated regression (SUR), was used to model the correlated levels of metabolites in the same biological group. The metabolites were found to be significantly affected by demographic covariates such as gender, BMI, BMI2, and smoking status. After accounting for the effects of the confounding factors, we then investigated potential of metabolites from serum to differentiate patients with polyps and age matched healthy controls. Our results showed that while only valine was slightly associated, individually, with polyp patients, a number of biologically related groups of metabolites were significantly associated with polyps. These results may explain some of the challenges and promise a novel avenue for future metabolite profiling methodologies.
Keywords: seemingly unrelated regression, colorectal polyp, NMR spectroscopy, metabolic profiling, metabolomics
INTRODUCTION
Colorectal cancer (CRC) is one of the most prevalent types of cancer worldwide, and a major cause of human morbidity and mortality.1 As the third most common type of cancer in the U.S. according to the American Cancer Society, over 136 000 new CRC cases and 50 000 deaths are estimated for 2015.2 Several preventive screening and detection methods are suggested for CRC, including the fecal occult blood test (FOBT), fecal immunochemical test (FIT), colonoscopy/sigmoidoscopy, and family-history-based risk assessment.3 More recently, population-wide studies have investigated the repeated use of the faecal immunochemical test (FIT) to measure hemoglobin and the detection of altered DNA associated with colorectal cancer for colorectal cancer screening.4 Patients with colon polyps are at high risk for the development of colon cancer; however, only colonoscopy has suffcient sensitivity to detect polyps. While colonoscopy and sigmoidoscopy remain the gold standards for screening and detection of CRC and polyps, they have major disadvantages, which include invasiveness, potential risks of complications, and high cost.1 Thus, compliance rates are far less than ideal (~48%);5 as a result many patients needlessly develop CRC. Of these CRC patients, only 40% are diagnosed and treated with early stage, localized disease (Stages I–II), which have relatively high (80–90%) 5-year survival rates.6 Therefore, the development of new screening methods that are highly sensitive, specific, and noninvasive is critically needed for the early diagnosis and timely treatment of CRC.
An important characteristic of cancer is its abnormal metabolism (i.e., the Warburg effect7), which causes altered levels of numerous cellular metabolites. Perturbations in important metabolic pathways have therefore been the focus of many cancer studies.8 Metabolomics, the comprehensive study of small molecular weight metabolites and their dynamic changes in biological systems, provides advanced methods to identify changing metabolism and in particular metabolite levels, that has resulted in rapid progress in disease biomarker discovery over the past decade.9 While numerous studies have focused on detecting CRC,10-14 only a few studies have focused on identifying metabolite biomarkers for polyp patients. For instance, a recent study investigated urine specimens combining nuclear magnetic resonance (NMR) spectroscopy and machine learning focused on the prediction of patients with colonic polyps showed a low to moderate sensitivity and specificity of 64% and 65%, respectively.15 A recent study by our group investigated serum from polyp patients along with the samples from CRC and healthy controls using targeted LC–MS analysis and found similar results.16
In this study, we have utilized NMR-based metabolomics combined with a powerful statistical approach, seemingly unrelated regression (SUR),17-19 to investigate differential metabolites between serum samples from polyp patients and healthy controls. Multivariate statistical analysis methods such as logistic regression and partial least-squares discriminant analysis (PLS-DA) offer outstanding capabilities for identifying specific differences in spectral signatures from different sample groups or classes. However, the construction of metabolomic signatures is challenged by the low statistical power due to a large number of metabolites and confounding factors such as gender, age, BMI, diet, smoking status, and so forth. On the other hand, analysis of covariance or linear regression models may be used to model metabolite levels affected by disease risks and confounding demographic factors. An overriding challenge is that the metabolic differences between patients with polyps and healthy controls are typically subtle compared to the more dominant confounding effects. Here, we pool the models of metabolites in biologically related groups, and acknowledge the potential correlation between these metabolites, with the analysis by SUR, which, to our knowledge, has not previously been applied to metabolite profiling. SUR allows the simultaneous investigation and thus aggregation of disease risk effects on metabolites in the same group, and therefore empowers the detection of subtle disease risk effects. Using SUR analysis, we explored several covariates which were statistically significant for all the metabolites and identified several biological groups of metabolites with altered levels that are significantly associated with a polyp diagnosis.
MATERIALS AND METHODS
Chemicals
Deuterium oxide (D2O, 99.9%D) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). Trimethylsilylpropionic-2,2,3,3-d4 acid sodium salt (TSP) and sodium azide were purchased from Sigma-Aldrich (Milwaukee, WI).
Serum Samples
Serum samples from patients with polyps (n = 44) and age matched healthy controls (n = 58) were obtained from the Indiana University School of Medicine. Following the IRB protocol approved by both Indiana and Purdue Universities, patients undergoing colonoscopy for CRC screening were evaluated and blood from the consented patients was obtained after overnight fasting and bowel preparation but prior to colonoscopy. Blood samples were allowed to clot at room temperature for 45 min and then centrifuged at 2000 rpm for 10 min. The sera were collected, and aliquoted into separate vials, then transported to Purdue University over dry ice and stored at −80 °C until used for analysis. Polyp patients were compared to age-matched healthy controls. The summary of demographic data for the patients and healthy controls included in this study are shown in Table 1.
Table 1. Summary of Demographic Data for Participants in the Study.
| total n = 102 |
polyps n = 44 |
healthy controls n = 58 |
|
|---|---|---|---|
| Age | Mean | 62.4 | 59.7 |
| SD | 6.3 | 6.6 | |
| Gender | Male | 29 | 24 |
| Female | 15 | 34 | |
| BMI | Mean | 29.1 | 27.9 |
| SD | 5.6 | 6.5 | |
| Ever Smoked | Yes | 22 | 25 |
| No | 22 | 33 | |
| Alcohol Status | Alcohol | 29 | 43 |
| No Alcohol | 15 | 15 |
1H NMR Spectroscopy
Each frozen serum sample was thawed and vortexed; 530 μL aliquots were mixed with 5 μL sodium azide solution (5% in H2O). The resulting solution was centrifuged, and 530 μL was transferred to a 5 mm NMR tube. A coaxial capillary containing 60 μL TSP (20.9 nmol) in D2O was placed into the NMR tube to serve as a chemical shift and quantitative reference. The samples were randomized before performing the NMR experiments. All 1H NMR experiments were carried out at 25 °C on a Bruker DRX 500-MHz NMR spectrometer equipped with an HCN cryogenic probe. 1H NMR data for each sample was acquired using both one-dimensional NOESY and CPMG (Carr–Purcell-Meiboom–Gill) pulse sequences. The water signal was suppressed using a presaturation pulse. For each spectrum, 128 transients were collected and 16K data points were acquired using a spectral width of 6000 Hz. An exponential weighting function corresponding to 0.5 Hz line broadening was applied to the free-induction decay before Fourier transformation. Phasing and baseline correction were applied using Bruker Xwinnmr software version 3.5.
Data Preprocessing
The NMR spectra obtained using the CPMG sequence were devoid of broad peaks from macromolecules and hence were more suitable for investigating altered levels of metabolites. The 1D NOESY spectra were very complex due to contributions from metabolites as well as abundant macromolecules, and hence, 1D NOESY spectra were not used in the analysis. The data sets from CPMG spectra were aligned with reference to the alanine peak at 1.46 ppm using KnowItAll software version 7.8 (Bio-Rad Laboratories Inc., Hercules, CA). Each CPMG NMR spectrum was binned to 4096 frequency buckets of equal size (0.003 ppm). TSP could not be used to align the spectra since it was separately contained in a coaxial capillary tube to avoid TSP’s known interaction with proteins. Thus, TSP’s peak position does not account for small spectral shifts from sample to sample due to variations in bulk magnetic susceptibility.
Subsequent analysis was focused on selected metabolite regions as we have utilized in earlier studies.20 Integrals for 24 regions were obtained and the resulting data were normalized using the total spectral integral (10.0 > δ > 5.2 and 4.7 > δ > 0.40 ppm). Peak integrals were then each standardized to a mean of 0 across the different spectra and their standard deviations normalized to 1 to avoid any signal magnitude bias (autoscaling). In addition, detected signals from 1,2-propanediol were excluded from the analysis since this molecule is one of the ingredients used for bowel preparation prior to colonoscopy. The 24 metabolite signals (see Supporting Information Table S1 for chemical shift ranges) detected in the entire polyp patient and healthy control samples were then subjected to statistical analysis. It may be noted that leucine was not integrated, although it was easily identifiable, since the characteristic peaks of leucine around 1 ppm were masked by broad residual peaks from lipids, which prevented accurate quantitation of leucine.
The original data set was filtered to obtain samples that had values for all of the clinical variables under investigation (i.e., no missing values) (see Supporting Information Figure S1 and Table S2). To eliminate any effect of race on the data, only Caucasian subjects were considered, due to the small number of individuals from other populations in the study. The two subject cohorts were age matched by limiting the subject ages to be between 45 and 65. These restrictions reduced the number of samples such that 44 from the polyp group and 58 from the healthy control group were used for further analysis.
Seemingly Unrelated Regression
SUR was proposed by Zellner in 196217 to generalize linear regression models for multiple response variables. SUR consists of multiple regression equations, each equation provides one response variable, and incorporates a correlated error matrix that gives rise to its name. In the current context, SUR assumes that error terms of the same individual (in different response variables) are likely correlated, but error terms of different individuals are independent. Specifically, we assume that p covariates are investigated for their effects on each of m metabolites, with serum samples from n individuals in total. Let Yij denote the level of the j-th metabolite for the i-th individual, and Xik denote the level of the k-th covariate of the i-th individual, i = 1, 2, …, n; j = 1, 2, …, m; k = 1, 2, …, p. The SUR model for each of the m metabolites is,
| (1) |
where εij is the error term which follows N(0, σ2j), and cov(εij, εlk) = σjk. Note that for different individuals i ≠ l, and σjk = 0. The possible correlation between error terms of different response variables makes the SUR model different from a simple pool of linear regression models.
In our study, all equations of the same SUR model include the same set of covariates. With n > p, estimates of the regression coeffcients are equivalent to the Ordinary Least Squares (OLS) estimates.17 However, our primary purpose of using SUR is to test the significance of the identical covariate(s) involved in all equations of the same SUR model. The effcient likelihood ratio test was utilized for this purpose. For example, to investigate the effects of a single covariate (say Xi1) on the levels of m metabolites, we can construct a likelihood ratio test which follows a χ2m distribution under the following null hypothesis,
| (2) |
When the effects of a group of covariates, for example k covariates, on the levels of m metabolites are simultaneously investigated for their significance, the likelihood ratio test statistic follows a χ2km distribution under the null hypothesis that all these effects are zero. Such likelihood ratio tests are presumably more powerful than any test based on a single regression equation as they aggregate the power of multiple regression equations.
Statistical Analysis
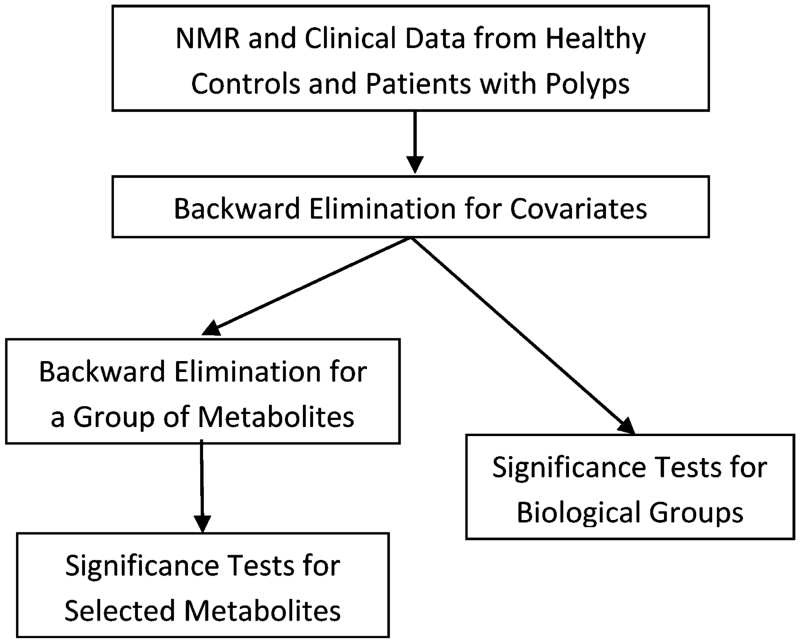
Figure 1 shows the flowchart for the analysis of metabolomics data using SUR. A SUR model17-19 was built to investigate how the levels of 24 metabolites were influenced by the demographic variables including age, age2, gender, BMI, BMI2, smoking status, alcohol status, diagnosis, as well as the interactions between diagnosis and the other covariates, leading to a total of 15 covariates in each of 24 regression equations involved in the SUR model. Backward elimination21 was performed to remove insignificant covariates (i.e., those with p-value ≥ 0.05). For the demographic covariates having significant interactions with diagnosis, we kept their main effects in the model. For explanatory purposes immediately below, and as the results of our analysis will show (see Results section), the selected four demographic covariates (gender, BMI, BMI2, smoking status) are included in all linear regression models on each of the 24 metabolites and in SUR models using groups of biologically related metabolites in the subsequent analysis.
Figure 1.
Flowchart for the analysis of metabolomics data using seemingly unrelated regression.
We first employed the following multiple linear regression to study the differential profile of each metabolite, i.e., for the i-th individual,
| (3) |
where Yi refers to the metabolite level of the i-th individual; Gi, Bi, Si, and Di represent, respectively, gender, BMI, smoking status, and diagnosis (0 for controls and 1 for polyps) of the i-th individual. We then tested the hypothesis
| (4) |
to investigate whether levels of the metabolite were different among polyps and control groups. Likelihood ratio tests were employed to calculate the p-values.
We further applied SUR analysis to 15 different groups of biologically related metabolites. For each group, the SUR model consists of multiple regression equations, one for each metabolite following eq 3. Specifically, for the j-th metabolite of the i-th individual,
| (5) |
We studied the differential profile of the grouped metabolites by testing the hypothesis
| (6) |
to investigate whether levels of the metabolites were different among polyps and control groups. Likelihood ratio tests were again utilized to calculate the p-values. We used the Benjamini-Hochberg procedure to control the false discovery rate (FDR)22 and calculated adjusted p-values accordingly. The FDR was controlled at 0.05.
RESULTS
In the SUR analysis of all 24 metabolites, we started with the following demographic variables: age, age2, gender, BMI, BMI2, smoking status, alcohol status, diagnosis, as well as the interactions between diagnosis and the 7 covariates. Cubic terms of age and BMI were not significant, and thus were ignored. Following backward elimination to remove insignificant covariates (see detailed information presented in Supporting Information), we selected four demographic covariates, i.e., gender, BMI, BMI2, and smoking status (Table 2). The covariates were then grouped and used in combination with the 24 metabolites to differentiate polyp patients from healthy controls. As shown in Table 3, metabolite profiles are significantly different between males and females (with a p-value of 2.2 × 10−7 for testing both gender and its interaction with diagnosis); indeed, the p-value for testing the main effect of gender is 9.9 × 10−8 (Table 2). BMI also significantly affects the metabolite profiles (with a p-value of 2.8 × 10−5 for testing BMI, BMI2, diagnosis × BMI, and diagnosis × BMI2 simultaneously), while metabolite profiles are also significantly different between smokers and nonsmokers (with a p-value of 0.041 for testing both smoking and diagnosis × smoking, and a p-value of 0.0049 for diagnosis × smoking only).
Table 2. Results of Backward Elimination for Predictors Including Age, Age2, Gender, BMI, BMI2, Smoking Status, Alcohol Status, Diagnosis, as Well as the Interactions between Diagnosis and Other Covariates.
| selected effects | p-value |
|---|---|
| Gender | 9.9 × 10−8 |
| BMI | 0.0023 |
| BMI2 | 0.15 |
| Smoking | 0.56 |
| Diagnosis | 0.39 |
| Diagnosis × Gender | 0.045 |
| Diagnosis × BMI | 0.012 |
| Diagnosis × BMI2 | 0.17 |
| Diagnosis × Smoking | 0.0049 |
Table 3. Significance of Grouped Covariates in Evaluating Differential Profiles of All 24 Metabolites.
| selected effects | p-value | adjusted p- value |
|---|---|---|
| Gender, Diagnosis × Gender | 2.2 × 10−7 | 1.5 × 10−6 |
| BMI, BMI2 | 1.7 × 10−4 | 4.0 × 10−4 |
| BMI2, Diagnosis × BMI2 | 0.066 | 0.066 |
| Diagnosis × BMI, Diagnosis × BMI2 | 0.010 | 0.014 |
| BMI, BMI2, Diagnosis × BMI, Diagnosis × BMI2 |
2.8 × 10−5 | 9.8 × 10−5 |
| Smoking, Diagnosis × Smoking | 0.041 | 0.048 |
| Diagnosis, Diagnosis × Gender, Diagnosis × BMI, Diagnosis × BMI2, Diagnosis × Smoking |
0.0012 | 0.0021 |
The SUR analysis of all 24 metabolites also suggests that metabolite profiles are significantly different between patients with polyps and healthy controls with a p-value of 0.0012 for testing diagnosis and its interaction with all other demographic covariates (Table 3). We therefore proceeded to analyze each of the 24 metabolites separately with a multiple linear regression as shown in eq 3, using the same covariates as in the aforementioned SUR model. The p-values for testing diagnosis and its interaction with other demographic covariates are shown in Table 4. The levels of valine are slightly different (with an uncorrected p-value of 0.010), and none of the other individual metabolites show significantly different levels between the patients with polyps and healthy controls as a result of the SUR modeling. The insignificance of diagnosis in the multiple linear regression analysis of each metabolite is due in part to our somewhat limited sample numbers.
Table 4. Overall p-Value for Diagnosis for Each Individual Metabolite.
| metabolite | p-value | adjusted p-value |
|---|---|---|
| 3-Hydroxybutyric acid | 0.59 | 0.95 |
| Acetic acid | 0.98 | 0.98 |
| Acetoacetate | 0.66 | 0.95 |
| Acetone | 0.22 | 0.95 |
| Alanine | 0.35 | 0.95 |
| Choline/Phosphocholine | 0.78 | 0.95 |
| Citric acid | 0.75 | 0.95 |
| Creatinine | 0.46 | 0.95 |
| Dimethylglycine | 0.91 | 0.95 |
| Formate | 0.91 | 0.95 |
| Glucose | 0.070 | 0.78 |
| Glutamic acid | 0.84 | 0.95 |
| Glutamine | 0.86 | 0.95 |
| Glycine | 0.60 | 0.95 |
| Histidine | 0.67 | 0.95 |
| Isoleucine | 0.54 | 0.95 |
| Lactate | 0.81 | 0.95 |
| Lysine | 0.51 | 0.95 |
| Phenylalanine | 0.26 | 0.95 |
| Threonine | 0.78 | 0.95 |
| Tyrosine | 0.55 | 0.95 |
| Unsaturated-Lipids | 0.39 | 0.95 |
| Urea | 0.098 | 0.78 |
| Valine | 0.010 | 0.24 |
On the other hand, the analysis of grouped metabolites can enhance the power of hypothesis tests by combining related metabolites in terms of the effects of diagnosis and its interaction with other demographic covariates. We therefore employed the SUR model in eq 5 to investigate 15 different groups of biologically related metabolites, and test whether levels of all metabolites in the same group were different among patients with polyps and healthy controls. Each metabolite group was chosen based on known or inferred biological relationships among the measured metabolites. In particular, we used the KEGG metabolic pathways23 to map the links among different metabolites detected by NMR. Metabolites that shared a common metabolic pathway or pathways that are connected to each other were considered biologically related and grouped together for analysis. Mainly, the metabolites were grouped in terms of glucose metabolism, amino acid metabolism and lipid metabolism. For example, NMR detected metabolites of group 1 represent glucose metabolism, both glycolysis and gluconeogenesis; group 2 metabolites are part of branched chain amino acid metabolism; and group 3 metabolites are part of alanine and glutamate metabolism (Table 5). Separately, NMR detected metabolites were also grouped by combining two or more groups of metabolites. See groups 10, 13 and 14, for example, in Table 5.
Table 5. Results of Testing the Effects of Diagnosis on 15 Groups of Biologically Related Metabolites.
| biological groups | associated metabolic pathway(s) | p-value | adjusted p- value |
|---|---|---|---|
| Group 1: acetate, glucose, lactate | Glycolysis/gluconeogenesis | 0.014 | 0.023 |
| Group 2: isoleucine, valine | Valine/Leucine/Isoleucine biosynthesis | 0.0046 | 0.012 |
| Group 3: alanine, glutamic acid, glutamine | Alanine/Aspartate/Glutamate metabolism | 0.060 | 0.069 |
| Group 4: creatinine, glutamine, urea | Arginine/Proline metabolism | 0.0010 | 0.0050 |
| Group 5: glutamic acid, histidine | Histidine metabolism | 0.058 | 0.072 |
| Group 6: acetoacetate, acetone, lactate | Propionate metabolism | 0.33 | 0.33 |
| Group 7: acetoacetate, citric acid, tyrosine | Tyrosine metabolism | 0.0011 | 0.0041 |
| Group 8: citric acid, formate, glutamic acid, glutamine | Glyoxalate and Dicarboxylate metabolism | 0.23 | 0.25 |
| Group 9: phenylalanine, tyrosine | Phenylalanine/Tyrosine/Tryptophan metabolism | 0.0021 | 0.0063 |
| Group 10: alanine, glutamic acid, glutamine, glycine, histidine, isoleucine, lysine, phenylalanine, threonine, tyrosine, valine |
Combination of Alanine/Aspartate/Glutamate, Glycine/Serine/Threonine, Valine/Leucine/Isoleucine, Phenylalanine/Tyrosine/Tryptophan, Histidine and Lysine metabolism |
1.5 × 10−7 | 2.3 × 10−6 |
| Group 11: alanine, citric acid, glucose, lactate | Glycolysis/TCA cycle | 0.021 | 0.032 |
| Group 12: glycine, threonine | Glycine/Serine/Threonine metabolism | 0.0051 | 0.011 |
| Group 13: alanine, glutamic acid, glycine, threonine | Combination of Glycolysis, TCA cycle and Glycine/Serine/Threonine metabolism |
0.031 | 0.042 |
| Group 14: alanine, glutamic acid, glycine, isoleucine, threonine, valine |
Combination of Glycolysis, TCA cycle, Glycine/Serine/Threonine and Valine/Leucine/Isoleucine metabolism |
1.2 × 10−5 | 9.1 × 10−5 |
| Group 15: choline/phosphocholine, glycine, threonine | Glycine/Serine/Threonine metabolism | 0.0057 | 0.011 |
The results of hypothesis tests on diagnosis and its interaction with other demographic covariates are summarized in Table 5, in which the adjusted p-values were calculated for controlling the overall FDR at 0.05. Eleven groups of metabolites (all but group 3, 5, 6, and 8) show significant profiles between patients with polyps and healthy controls. Indeed, metabolites in Group 10 and Group 14 show extremely strong evidence (with adjusted p-values of 2.3 × 10−6 and 9.1 × 10−5, respectively) that levels of the grouped metabolites are very different between patients with polyps and healthy controls.
DISCUSSION
Significantly altered metabolic reprogramming in CRC is reflected in major changes to metabolite levels; as a result, a high classification accuracy relative to healthy controls can be achieved.10-14,16 However, it is more challenging to distinguish high risk polyps patients and healthy controls because of the subtle metabolic differences between the two cohorts. This challenge becomes more severe when the effects of confounding variables such as age, BMI, etc., are comparable or larger than the differences between the two cohorts of samples. Focusing on this challenge, we have employed NMR spectroscopy, a highly reproducible and quantitative method for analyzing metabolites in complex biological mixtures, with SUR analysis that represents a powerful approach to take such confounding variables into account. Originally developed for economic analysis, SUR has been previously applied to genomic and nutrition analysis,24,25 but not previously to metabolomics. Given the inherent correlations observed among many metabolites as well as clinical variables, SUR may provide a good model to capture the relationships among measured variables as well as residual, unmeasured components or confounding factors that may also contribute to the model or even error rates in the model.
With blood samples collected from healthy controls and patients with colorectal polyps, we investigated how the levels of individual metabolites and groups of metabolites are affected by clinical variables including age, age2, gender, BMI, BMI2, smoking status, alcohol status, and diagnosis. Exploratory data analysis showed that several clinical covariates, including gender, BMI, BMI2, and smoking status are significantly associated with the levels of many metabolites. Each of the 24 metabolites was fitted using multiple linear regression to identify metabolites that are significantly associated with diagnosis. The results indicate that while only one of the individual metabolites (valine) is slightly associated with diagnosis (Table 4), the effects of diagnosis on the metabolic activities of several groups of biologically related metabolites are indeed quite significant. A better understanding of these differences is expected to provide new insight into the early development of CRC and potentially provide new targets for therapy.
The groups of metabolites found to significantly distinguish (p < 0.05) polyps patients from healthy subjects represent numerous metabolic pathways including glycolysis, the Krebs cycle, as well as amino acid and lipid metabolism (see Table 5). Numerous metabolomics investigations have shown that metabolites associated with these pathways are significantly altered in CRC,11,16,26 It is well-known that altered glycolysis is the hallmark of virtually all types of cancer and altered glycolysis in CRC has been shown in a number of earlier studies including our own recent investigation.16 The Warburg effect in cancer,7 the phenomenon of a high rate of conversion of glucose to lactate even in the presence of oxygen (aerobic glycolysis), is associated with metabolic reprogramming, which involves utilizing alternative metabolite sources as substrates for the Krebs cycle. Amino acids including glutamine, glutamic acid, alanine, histidine, isoleucine, lysine, phenylalanine, tyrosine, valine, and threonine all fuel the Krebs cycle either directly by their conversion to Krebs cycle metabolites such as α-keto glutarate, succinyl CoA, fumarate, and oxaloacetate or indirectly through pyruvate and acetyl CoA. Correlation of these amino acids with the detection of polyps patients in this study indicates their potential association with CRC. Utilization of amino acids as energy sources in cancer is well-known,27 and a recent study found that a majority of these amino acids are altered significantly in CRC.26,28 The catabolism of amino acids is associated with conversion of their amino groups to ammonia, which is used for the synthesis of urea. Thus, in addition to the many altered amino acid levels, the association of urea with polyp patients in this study appears to agree with this phenomenon. The association of choline/phosphocholine with polyps also agrees with the altered lipid metabolism observed in CRC. It is well-known that altered lipid metabolism is observed in many cancers and recent metabolomics investigations of CRC have indeed shown altered lipid metabolism.11,13
In conclusion, a powerful SUR modeling approach that combines the result of NMR based metabolomics and the analysis of confounding factors has resulted in the identification of a number of metabolic pathways that are significantly associated with the presence of colon polyps. While the data were collected from a limited number of subjects, and thus further investigations are needed to validate the findings, the results show that the modeling of metabolite levels using confounding variables has potential for developing improved diagnostic tests and for better understanding disease development. Additional analyses including the colon cancer group will be carried out when more samples are available.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge the support of the Cancer Care Engineering (CCE) project, a joint effort between the Oncological Sciences Center (Purdue Center for Cancer Research, NCI P30CA23168) in the Purdue University Discovery Park and the Indiana University Melvin and Bren Simon Cancer Center (NCI P30CA082709). Support for the CCE project is gratefully acknowledged from the Walther Cancer Foundation, NIH (UL1RR025761), DOD (USAMRMC (CDMRP) W81XWH-008-1-0065, 9107003), and the Regenstrief Foundation. Grant support from the National Basic Research Program of China (2014CB744600), Beijing Municipal Commission of Education (KM201410025013) and Beijing Institute for Brain Disorders (BIBDPXM2014_014226_000016) are also gratefully acknowledged. The authors would like to thank Dr. Li Yuan Bermel for assistance with the CCE project sample collection bank and Dr. Ann Christine Catlin for helping with data storage as well as transfer. D.R. thanks the University of Washington for startup funding. The China Scholarship Council is also gratefully acknowledged by L.D. for funding her Visiting Student fellowship.
Footnotes
ASSOCIATED CONTENT
NMR data from polyp patients and healthy controls filtered from the original data set for SUR analysis, in the current study; chemical shift regions used for obtaining integrals for the 24 metabolites used in this study; NMR data from polyp patients and healthy controls filtered from the original data set; the results of backward elimination for predictors. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b00059.
The authors declare the following competing financial interest(s): Daniel Raftery reports holding equity and an executive position at Matrix-Bio, Inc.
REFERENCES
- (1).Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365(9454):153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- (2).Siegel R, Miller KD, Jemal A. Cancer statistics, 2015. CA— Cancer J. Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- (3).Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. American college of gastroenterology guidelines for colorectal cancer screening 2008. Am. J. Gastroenterol. 2009;104(3):739–750. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- (4).(a) Stegeman I, de Wijkerslooth TR, Mallant-Hent RC, de Groot K, Stroobants AK, Fockens P, Mundt M, Bossuyt PM, Dekker E. Implementation of population screening for colorectal cancer by repeated Fecal Immunochemical Test (FIT): third round. BMC Gastroenterol. 2012:12–73. doi: 10.1186/1471-230X-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370(4):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- (5).Taylor DP, Cannon-Albright LA, Sweeney C, Williams MS, Haug PJ, Mitchell JA, Burt RW. Comparison of compliancefor colorectal cancer screening and surveillance by colonoscopy based on risk. Genet. Med. 2011;13(8):737–743. doi: 10.1097/GIM.0b013e3182180c71. [DOI] [PubMed] [Google Scholar]
- (6).Cancer Facts & Figures 2013. American Cancer Society; Atlanta, GA: 2013. [Google Scholar]
- (7).Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- (8).(a) Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]; (c) Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Munoz-Pinedo C, El Mjiyad N, Ricci JE. Cancer metabolism: current perspectives and future directions. Cell Death Dis. 2012;3:e248. doi: 10.1038/cddis.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).(a) Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gu H, Nagana Gowda GA, Raftery D. Metabolic profiling: are we en route to better diagnostic tests for cancer? Future Oncol. 2012;8(10):1207–1210. doi: 10.2217/fon.12.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Nagana Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 2008;8(5):617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal B, Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5(4):435–458. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491(7424):384–392. doi: 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]; (f) Fan T-M, Lane A. NMR-based stable isotope resolved metabolomics in systems biochemistry. J. Biomol. NMR. 2011;49(3-4):267–280. doi: 10.1007/s10858-011-9484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Reaves ML, Rabinowitz JD. Metabolomics in systems microbiology. Curr. Opin. Biotechnol. 2011;22(1):17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes. 2009;58(11):2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Anal. Chem. 2011;83(6):2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).(a) Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W. Serum metabolite profiling of human colorectal cancer using GC–TOFMS and UPLC–QTOFMS. J. Proteome Res. 2009;8(10):4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]; (b) Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, Okuno T, Hayashi N, Kawano S, Takenawa T, Azuma T, Yoshida M. Novel Serum, A. Metabolomics-based diagnostic approach for colorectal cancer. PLoS One. 2012;7(7):e40459. doi: 10.1371/journal.pone.0040459. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tan B, Qiu Y, Zou X, Chen T, Xie G, Cheng Y, Dong T, Zhao L, Feng B, Hu X, Xu LX, Zhao A, Zhang M, Cai G, Cai S, Zhou Z, Zheng M, Zhang Y, Jia W. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 2013;12(6):3000–3009. doi: 10.1021/pr400337b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, Fiehn O. Metabolite profiling of human colon carcinoma—deregulation of TCA cycle and amino acid turnover. Mol. Cancer. 2008;7(1):72. doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chan ECY, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS) J. Proteome Res. 2008;8(1):352–361. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- (12).Ma Y-L, Qin H-L, Liu W-J, Peng J-Y, Huang L, Zhao X-P, Cheng Y-Y. Ultra-high performance liquid chromatographymass spectrometry for the metabolomic analysis of urine in colorectal cancer. Dig. Dis. Sci. 2009;54(12):2655–2662. doi: 10.1007/s10620-008-0665-4. [DOI] [PubMed] [Google Scholar]
- (13).Li F, Qin X, Chen H, Qiu L, Guo Y, Liu H, Chen G, Song G, Wang X, Li F, Guo S, Wang B, Li Z. Lipid profiling for early diagnosis and progression of colorectal cancer using direct infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2013;27(1):24–34. doi: 10.1002/rcm.6420. [DOI] [PubMed] [Google Scholar]
- (14).(a) Ritchie S, Ahiahonu P, Jayasinghe D, Heath D, Liu J, Lu Y, Jin W, Kavianpour A, Yamazaki Y, Khan A, Hossain M, Su-Myat K, Wood P, Krenitsky K, Takemasa I, Miyake M, Sekimoto M, Monden M, Matsubara H, Nomura F, Goodenowe D. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. BMC Med. 2010;(8):13. doi: 10.1186/1741-7015-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ritchie SA, Tonita J, Alvi R, Lehotay D, Elshoni H, Myat S, McHattie J, Goodenowe DB. Low-serum GTA-446 anti-inflammatory fatty acid levels as a new risk factor for colon cancer. Int. J. Cancer. 2013;132(2):355–362. doi: 10.1002/ijc.27673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Eisner R, Greiner R, Tso V, Wang H, Fedorak RN. A machine-learned predictor of colonic polyps based on urinary metabolomics. Biomed. Res. Int. 2013;2013:303982. doi: 10.1155/2013/303982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome. Res. 2014;13(9):4120–30. doi: 10.1021/pr500494u. [DOI] [PubMed] [Google Scholar]
- (17).Zellner A. An efficient method of estimating seemingly unrelated regressions and tests for aggregation bias. J. Am. Stat. Assoc. 1962;57(298):348–368. [Google Scholar]
- (18).Aiken LS, West SG. Multiple Regression: Testing and Interpreting Interactions. SAGE Publications, Inc.; Newbury Park, CA: 1991. [Google Scholar]
- (19).Krishnaiah AH, Paruchuri R, editors. Multivariate Analysis. Academic Press, Inc.; New York: 1971. p. 1967. [Google Scholar]
- (20).Zhang J, Liu L, Wei S, Nagana Gowda GA, Hammoud Z, Kesler KA, Raftery D. Metabolomics study of esophageal adenocarcinoma. J. Thorac. Cardiovasc. Surg. 2011;141:469–475. doi: 10.1016/j.jtcvs.2010.08.025. [DOI] [PubMed] [Google Scholar]
- (21).Kohavi R, John G. Wrappers for feature selection. Artif. Intell. 1997;97(1-2):273–324. [Google Scholar]
- (22).Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B. 1995;57(1):289–300. [Google Scholar]
- (23).Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Saint-Pierre A, Kaufman JM, Ostertag A, Cohen-Solal M, Boland A, Toye K, Zelenika D, Lathrop M, de Vernejoul MC, Martinez M. Bivariate association analysis in selected samples: application to a GWAS of two bone mineral density phenotypes in males with high or low BMD. Eur. J. Hum Genet. 2011;19(6):710–716. doi: 10.1038/ejhg.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Carroll RJ, Midthune D, Freedman LS, Kipnis V. Seemingly unrelated measurement error models, with application to nutritional epidemiology. Biometrics. 2006;62(1):75–84. doi: 10.1111/j.1541-0420.2005.00400.x. [DOI] [PubMed] [Google Scholar]
- (26).Leichtle AB, Nuoffer JM, Ceglarek U, Kase J, Conrad T, Witzigmann H, Thiery J, Fiedler GM. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics. 2012;8(4):643–653. doi: 10.1007/s11306-011-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Argilés J, Azcón-Bieto J. The metabolic environment of cancer. Mol. Cell. Biochem. 1988;81(1):3–17. doi: 10.1007/BF00225648. [DOI] [PubMed] [Google Scholar]
- (28).Miyagi Y, Higashiyama M, Gochi A, Akaike M, Ishikawa T, Miura T, Saruki N, Bando E, Kimura H, Imamura F, Moriyama M, Ikeda I, Chiba A, Oshita F, Imaizumi A, Yamamoto H, Miyano H, Horimoto K, Tochikubo O, Mitsushima T, Yamakado M, Okamoto N. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS One. 2011;6(9):e24143. doi: 10.1371/journal.pone.0024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.