Abstract
Chromosomal copy number alterations (aneuploidy) define the genomic landscape of most cancer cells, but identification of the oncogenic drivers behind these imbalances remains an unfinished task. In this study, we conducted a systematic analysis of colorectal carcinomas that integrated genomic copy number changes and gene expression profiles. This analysis revealed 44 highly overexpressed genes mapping to localized amplicons on chromosome 13, gains of which occur often in colorectal cancers. RNAi-mediated silencing identified eight candidates whose loss of function reduced cell viability 20% or more in colorectal cancer cell lines. The functional space of the genes NUPL1, LNX2, POLR1D, POMP, SLC7A1, DIS3, KLF5, and GPR180 was established by global expression profiling after RNAi exposure. One candidate, LNX2, not previously known as an oncogene, was involved in regulating NOTCH signaling. Silencing LNX2 reduced NOTCH levels but also downregulated the transcription factor TCF7L2 and markedly reduced WNT signaling. LNX2 overexpression and chromosome 13 amplification therefore constitutively activates the WNT pathway, offering evidence of an aberrant NOTCH-WNT axis in colorectal cancer.
Keywords: Genomic Amplification, Colorectal Cancer, NOTCH signaling, WNT/β, catenin pathway, RNAi
INTRODUCTION
Colorectal cancer (CRC) has an annual incidence of some 150,000 new cases and a mortality of more than 50,000 in the United States in 2010 (1). Despite the availability of screening programs, colorectal carcinomas are often detected at advanced disease stages, at which surgery alone is no longer a curative therapeutic intervention. The development of colorectal cancer from premalignant precursors to invasive and metastatic disease involves accumulation of mutations in key regulator genes (2), as well as the acquisition of a recurrent pattern of genomic imbalances (3), a hallmark of epithelial cancers (4, 5). Like in many other tumors, these imbalances can be focal (i.e., limited to a chromosomal band or less) or they affect entire chromosome arms or chromosomes, most commonly resulting in low level gains or losses. Once acquired, which can occur as early as in dysplastic polyps, the distribution of chromosome-level imbalances is remarkably stable and consequently results in a pattern of genomic imbalances that is tumor type specific (6, 7). In colorectal cancer such imbalances affect chromosomes 7, 8q, 13q, and 20 as gains, and chromosomes 4, 5q, 8p, 17p, 18 as losses (8). Chromosome-level genomic imbalances directly influence the expression levels of resident genes, therefore resulting in a massive transcriptional deregulation (9–14). It is therefore conceivable that the systematic integration of genomic imbalances with global gene expression profiles has the potential to reveal novel drivers of tumorigenesis (15, 16).
The pattern of genomic imbalances in CRC is peculiar because chromosome 13 is frequently gained or amplified. In many other carcinomas, this chromosome is lost (8), most likely because it contains the tumor suppressor gene RB1. The consistent gain of chromosome 13 in CRC strongly suggests a tissue-derived selection. The identification of driver genes of this selection therefore requires the functional interrogation of potential candidates, which are found among those genes that are consistently overexpressed. In order to identify potential novel oncogenes on chromosome 13, we systematically mapped copy number changes by array CGH and overlaid the gene expression profiles obtained from the same tumors and cell lines. The involvement of candidate genes in growth and survival of CRC was then tested using RNA interference (RNAi), followed by gene expression profiling to define a signature of loss-of-function (LOF). This led to the identification of affected cellular signaling pathways and ultimately to the identification of their role in the cancer cell.
MATERIALS AND METHODS
Patient samples, cell lines and nucleic acid extraction
Fifty-six patients diagnosed with colorectal adenocarcinomas (UICC II/III) recruited as part of the Clinical Research Unit KFO 179 at the Department of General Surgery, University Medicine Göttingen, Germany, were included in this study. This study has been approved by the ethical review committee from the University Medical Center, Göttingen, Germany, and informed consent was obtained from all patients. Figure S1 summarizes the experimental setup and the prioritization of targets.
All of the cell lines were obtained from the ATCC (American Type Culture Collection, Manassas, VA). DNA and RNA were extracted from the cell lines, primary tumors, and normal mucosa samples following standard procedures (17). Nucleic acid quantification was determined using a Nanodrop ND-1000 UV-VIS spectrophotometer (Nanodrop, Rockland, DE), and RNA quality was assessed using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Normal colon RNA from five different donors without a history of colorectal cancer was purchased from Ambion (Applied Biosystems, Foster City, CA).
Array CGH
Oligonucleotide-based array CGH was performed according to the protocol provided by the manufacturer (Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis, protocol version 4.0, June 2006, Agilent Technologies, Santa Clara, CA), with minor modifications.
Gene expression microarrays
Cell line and normal human colon RNA were labeled with Cy3, mixed, and hybridized to an oligonucleotide-based Whole Human Genome Microarray using the manufacture’s recommendations (Agilent). Data were extracted using Agilent Technologies Feature Extraction software (v. 9.1).
Quantification of gene expression
Gene expression levels were validated by quantitative reverse transcription-PCR (qRT-PCR) using Power SYBR Green technology (Applied Biosystems, Inc., Foster City, CA) in the Applied Biosystems Prism 7000 sequence detector. Gene specific PCR primers (Table S1) were obtained from Operon Technologies, Inc. (Huntsville, AL). The genes RAB35, FBXL12 and OTUB1 were used for normalization.
RNAi-based analysis
The target sequences for the synthetic siRNAs (Qiagen Inc., Germantown, MD) are listed in Table S2. Lipid-based reverse transfections were performed using Oligofectamine (Life Technologies, Carlsbad, CA). Briefly, for analysis of the cell viability each siRNA (2 pmol) was added to individual wells in a 96-well plate in 25 µl of serum-free RPMI and complexed with transfection reagent in 25 µl of serum-free RPMI to a final siRNA concentration of 20 nM. Cells were then added in 50 µl RPMI supplemented with 20% FBS. Cellular viability was determined after 72 and 96 hrs post transfection using the CellTiter-Blue® reagent (Promega, Madison, WI).
Whole genome expression profiles were generated for eight genes following RNAi to enable the determination of gene specific LOF RNAi signatures. Reduction in target mRNA levels was confirmed by RT-PCR, and effects on cell viability were verified by functional assays. The gene expression data used to generate the gene specific RNAi signatures have been deposited in the NCBI Gene Expression Omnibus3 (GEO ID: GSE33824).
Immunoblotting
Immunoblotting was performed according to standard procedures. Whole cell lysates were obtained using SDS lysis buffer, sonicated, and denaturated at 95°C for 10 min. Equal amounts of protein from whole cell lysates were loaded in a NuPAGE Bis-Tris electrophoresis gel. The following antibodies were used: anti-KLF5 (Abcam, Cambridge, MA), anti-SOX2 (L73B4; Cell Signaling Technology), anti-LNX2 (Abcam), anti-NOTCH1 (Epitomics, Burlingame, CA), anti-NUMB (C29G11; Cell Signaling Technology), anti-HEY2/HRT2 (Millipore, Temecula, CA), anti-Hes1 (Epitomics), anti-TCF7L2 (EP2033Y; Abcam), anti-CTNNB1 (Abcam), anti-PARP (Abcam), anti-GAPDH (Sigma-Aldrich, St. Louis, MO).
Reporter assay
TOP-Flash reporter vector contains a Luciferase open reading frame and two sets of three copies of TCF binding sites, upstream of a minimal thymidine kinase promoter. FOP-Flash is a control vector, which is identical to TOP-Flash, but the TCF binding sites are inactive due to mutations. Cells were first transfected with siRNA (day 0) using the standard protocol described above. The same cells were then cotransfected with the reporter DNA 48 hr post siRNA transfection using Lipofectamine 2000 (Life Technologies). Cells were double transfected with 100 ng of the reporter and 100 ng of Renilla construct for normalization. Lysates were harvested using the Dual-Glo Luciferase Assay System (Promega) and the luminescence analyzed with a Tecan i-control microplate reader (Tecan Group Ltd., Männedorf, Switzerland).
Flow Cytometry Cytotoxicity Assay
Cells were trypsinized, harvested, and washed twice in 1X PBS. The cells were then washed with annexin V binding buffer provided with the Annexin V-PE Apoptosis Detection Kit (BD Biosciences, San Diego, CA). Cells were resuspended in 100 ml annexin V binding buffer. Samples were stained with 2.5 ml annexin V-PE and 5 ml 7-AAD, and incubated for 15 minutes at room temperature. Next, 300 ml annexin V binding buffer was added post-incubation. Samples were analyzed by flow cytometry on a FACS Calibur instrument (BD Bioscience) and FlowJo software.
Statistical and bioinformatic analysis
Agilent expression data were quantile normalized using the statistical computing language R (18). Differential expression between siNeg and siRNA treated cells was calculated using empirically modified Bayes T-statistics with the R package Limma. Raw data for GSE14333, GSE17536, and GSE17537 were downloaded from the NCBI Gene Expression Omnibus (GEO). Each dataset was normalized separately using the robust multi-chip averaging (RMA) method from Affymetrix (19). One-way hierarchical clustering (average linkage) was performed using JMP (version 8, SAS, Cary, NC).
Transcription factor target gene lists were derived from peer-reviewed literature (Table S3). All gene lists were cross-referenced to the Ingenuity Knowledge Base as well as NCBI Entrez Gene. Gene signatures were split into up- and down-regulated lists and analyzed separately. Enrichment was calculated using a Benjamini-Hochberg corrected one-tailed Fisher’s exact test using Ingenuity Pathway Analysis (Ingenuity Pathway Analysis, v.8.8, Redwood City, CA).
Gene signatures were loaded into IPA (Ingenuity Pathway Analysis), and each gene was mapped to a unique IPA object. As with the transcription factor analysis, gene signatures were parsed into up- and down-regulated lists and were analyzed separately. A Benjamini-Hochberg corrected one-tailed Fisher’s exact test was used to calculate enrichment.
RESULTS
We report here a systematic functional genomic strategy for the identification of driver genes in CRC. This strategy was based on the hypothesis that genes that reside in regions of recurrent genomic amplification, and are consistently overexpressed, present bona fide oncogenes. Our approach was targeted to chromosome 13 as one of the most frequently and highly amplified chromosome in this disease.
Integration of aCGH and gene expression
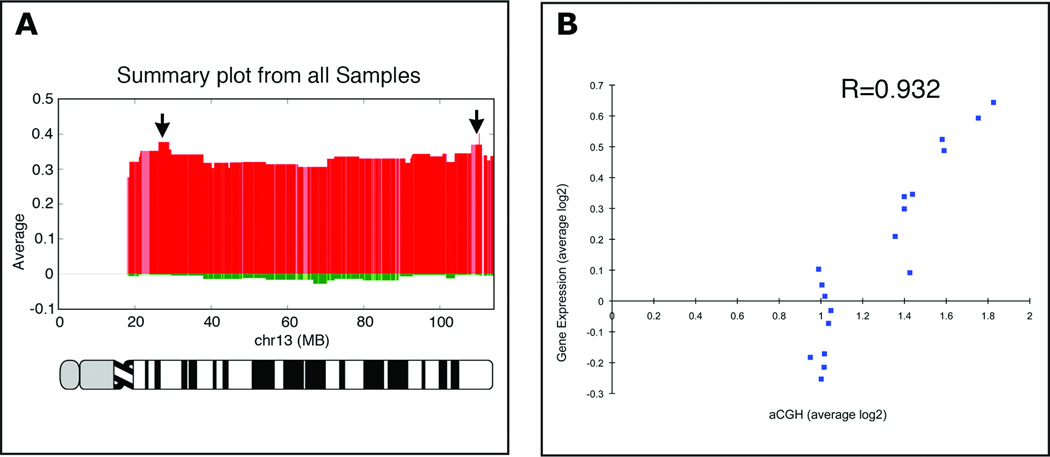
High-resolution aCGH analysis confirmed the consistent gain of chromosome 13 in colorectal carcinomas and derived cell lines. While in most instances the entire chromosome was gained, we observed two common regions of recurrent focal amplifications. The genome coordinates of these amplicons map to chr13:20,856,880–29,466,246 and chr13:103,927,403–114,125,347, for the proximal and distal amplicon, respectively (Fig. 1A). All colon cancer samples and cell lines were also profiled on 44K gene expression arrays using RNA from a pool of five normal colon mucosa samples as reference. We identified 29 (37%) out of 78 genes annotated in the microarray as located in the two amplified regions that were significantly overexpressed in the samples compared to normal colon mucosa (P<0.05), supporting a positive correlation between copy number changes and gene expression (Fig. 1B). While our analysis was focused on genes that mapped to the two amplicons, we included several other genes located on chromosome 13 because they were consistently overexpressed. Overall, the integration of the array-based genomic copy number changes and gene expression datasets from CRC tumors and cell lines generated a list of 67 candidate genes. Functional validation of these candidate CRC genes requires in vitro models. To aid in the identification of suitable model systems for assessment of gene function we further quantified the expression of these 67 candidate genes by qRT-PCR in 25 CRC cell lines (Fig. S2). Overall, out of the 67 genes, overexpression of 44 genes (65%) was validated in 25 cell lines. We therefore conclude, consistent with previous analyses, that the cell lines are adequate in vitro models (20). The list of 44 genes is presented in Table S4. Based on this analysis we chose the cell line SW480 for our functional analysis because it exhibits a gain of chromosome 13. We also used the cell line DLD-1, which does not show a gain of chromosome 13, but the candidate genes were overexpressed in an amplification-independent manner.
Figure 1. Genomic aberration profile, chromosome specific gene expression, and their correlation.
(A) Genomic aberration profile of chromosome 13 in primary colorectal carcinomas established using aCGH. This chromosome is consistently gained. Note two distinct peaks of genomic amplification (arrows). Numbers on the x-axis denote genomic coordinates, and on the y-axis shows the values of average copy number changes in log2 ratios. (B) Correlation between aCGH and resident gene expression levels. Plotted here are 18 samples for which we had concordant data on genomic copy number and gene expression levels. Gene expression levels were correlated with genomic copy number for samples with no gain of chromosome 13 (those that map in the range of 1 on the x-axis), and in a linear fashion for those with increasing genomic amplification (R=0.932).
Functional analysis: Loss-of-function compromises cell viability
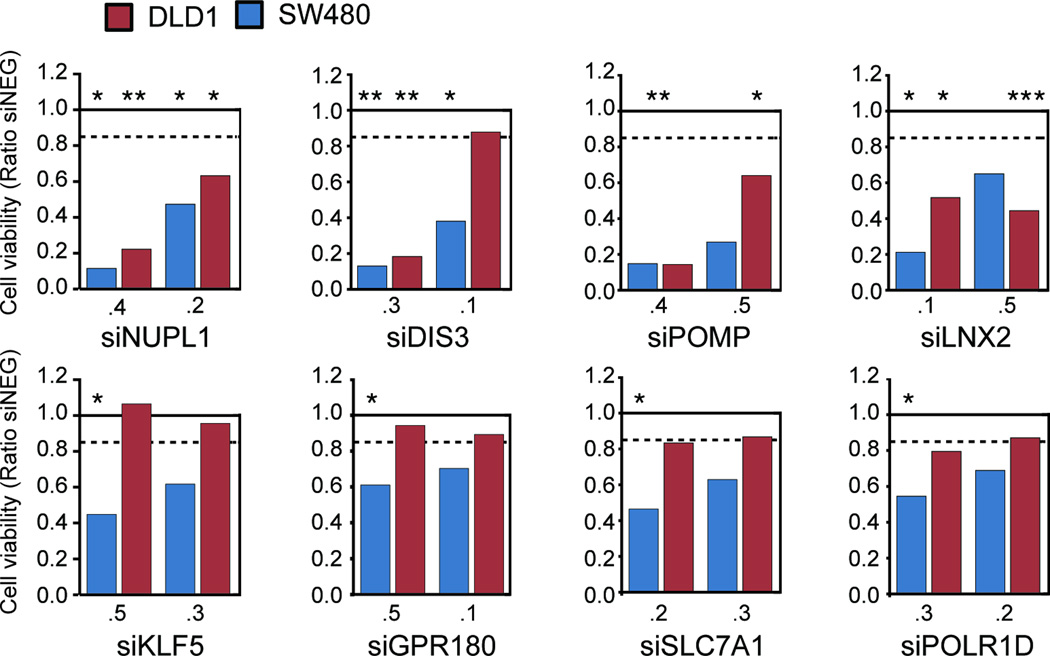
To test the hypothesis that one or more genes located on chromosome 13, that are frequently amplified and overexpressed in CRC, are required for tumor survival and/or growth we used RNAi to identify genes whose LOF reduces the viability of either SW480 and/or DLD-1 CRC cells. We initially assessed viability by using two siRNAs for each of the 44 overexpressed genes (Fig. S3A). Based on the results of the viability assays, 17 genes were selected for analysis with two additional siRNAs per gene. For nine of those 17 genes, the additional siRNAs failed to recapitulate a loss of viability following gene silencing (Fig. S3B). We therefore selected eight genes that significantly induced a 15% or greater decrease in viability (P<0.05) in one or more cell lines for further experimentation (Fig. 2). The LOF of NUPL1, DIS3, POMP and LNX2 resulted in a substantial reduction of viability of both SW480 and DLD-1 cells with at least two siRNAs. SW480 cells were more sensitive to the LOF of KLF5, GPR180, SLC7A1 and POLR1D than the DLD-1 cells; this may be attriutable to slight differences of the expression levels of these genes (Table S5). However, in all instances the observed decrease of viability was paralleled by an efficient reduction of the target mRNA compared to control transfections with siNeg, as determined by quantitative RT-PCR (Fig. S4). Since only KLF5 has been functionally associated with CRC (21), we thus decided to use an unbiased approach to query the potential functional role of these eight genes in the context of CRC.
Figure 2. Viability of the cell lines SW480 and DLD-1 after silencing of candidate genes.
The graph shows the results of silencing of the eight candidate genes (as a ratio of siNeg transfected cells) following RNAi (here shown the results for the two siRNAs used for subsequent experimentation) in SW480 and DLD-1. The solid line indicates the normalization against siNeg transfected cells, the dotted line the threshold of 15% reduction in viability as measured with the CellTiter-Blue assay. The statistical comparison (t-test) is based on the comparison to siNeg transfected cells (* P≤0.05, ** P≤0.01, *** P≤0.001).
Transcriptional signatures associated with loss-of-function by RNAi
Whole genome expression profiles of cells in which RNAi has been applied to silence a single gene can be used to identify, in an unbiased manner, genes and pathways perturbed as a consequence of this silencing and thus the potential function of a specific gene. Gene expression profiles were generated from SW480 cells transfected with siRNAs corresponding to NUPL1, LNX2, POLR1D, POMP, SLC7A1, DIS3, KLF5, and GPR180 (two different siRNAs corresponding to each gene, triplicate independent transfections for each siRNA), and with a negative control siRNA (siNeg). To define gene specific expression profiles following RNAi, i.e., an RNAi signature, we first considered a significant change following silencing of each target gene as a probe that showed a ≥ ±0.6 Log2 ratio fold change in expression (corresponding to ~1.5 fold linear change in expression) with a q-value ≤0.05 when the siNeg expression profile was compared to the expression profiles generated for each gene specific siRNA. This resulted in a remarkable correlation by the two siRNAs corresponding to each gene (Fig. S5). All expression profiles showed a significant decrease in the expression levels of the target gene and the number of genes within each signature varied considerably from fewer than 50 genes to nearly 4,000 genes (Fig. S6 and Table S6).
The RNAi signatures for KLF5, NUPL1, POMP and LNX2 show altered expression of specific CRC associated genes
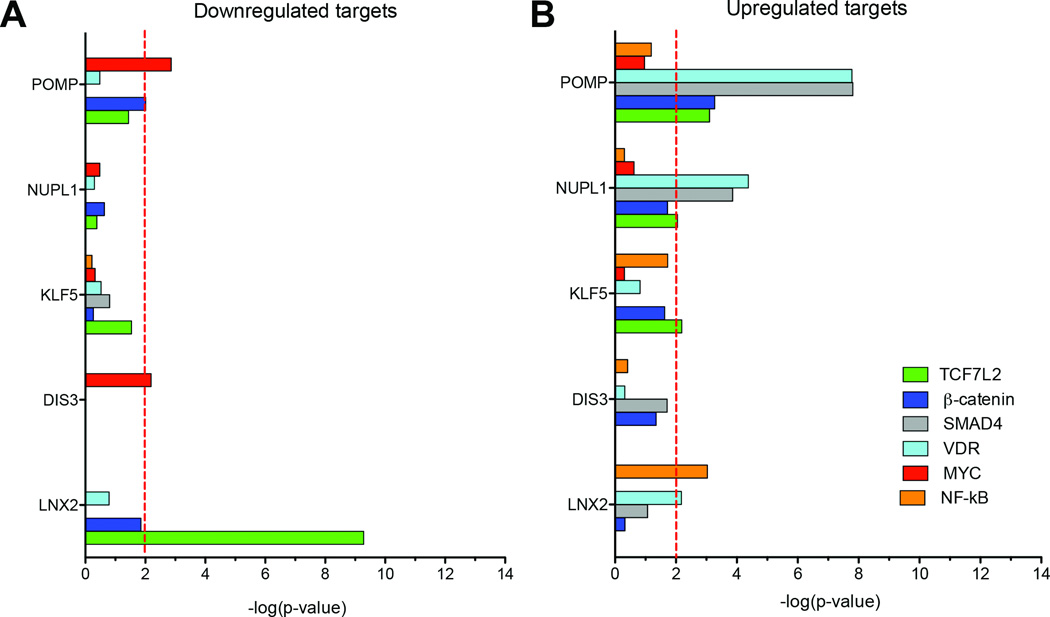
To understand the biological significance of these more extensive changes to the transcriptome we begun by investigating whether the LOF signatures were enriched for targets of transcription factors associated with colorectal cancer, including the Vitamin D receptor (VDR), NF-κB, SMAD4, TCF7L2, β-catenin, and MYC (Fig. 3). The enrichment for the targets of CRC associated transcription factors was very different for each of the five genes; silencing of POMP showed enrichment for the upregulation of targets of SMAD4 and VDR, as did NUPL1, while the LNX2 RNAi signature showed highly significant enrichment for the downregulation of targets of TCF7L2. The silencing of DIS3 showed slight enrichment for gene targets of MYC within the downregulated portion of its RNAi signature, while silencing of KLF5 was associated with enrichment for the up-regulation of TCF7L2 and NF-κB targets.
Figure 3. Comparison of RNA signatures derived from candidate gene silencing with gene expression modules of transcription factors involved in CRC.
Determination of the overlap of signature genes after RNAi-mediated silencing of POMP, NUPL1, KLF5, DIS3, and LNX2 with genes known to be either downregulated (A) or upregulated (B) by colorectal cancer-associated transcription factors TCF7L2, β-catenin, SMAD4, VDR, MYC, and κ (Color code at the bottom of the Figure). The red-dashed line indicates a significance threshold of P<0.01.
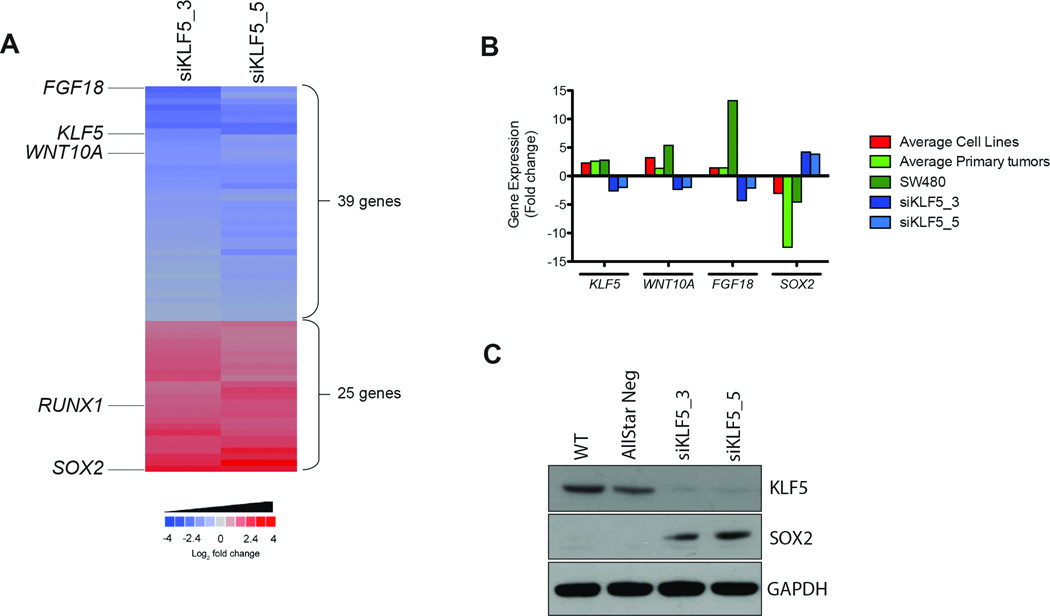
Consistent with the enrichment for NF-κB targets within those genes showing altered expression following LOF of the KLF5 transcription factor, pathway analysis of the KLF5 RNAi signature generated a network centered upon NF-κB (Fig. S7A). This network included the most downregulated gene in the KLF5 RNAi signature, FGF18, a gene involved in CRC (Fig. 4A and Table S6) (22–24). We have consistently observed increased expression of FGF18 in primary colon tumors and derived cell lines. Upon silencing of KLF5, the expression of FGF18 showed a 4-fold linear reduction in expression (Fig. 4B), suggesting that potentially the amplification and overexpression of KLF5 is contributing to the growth stimulation mediated by FGF18. The most up-regulated gene within the KLF5 RNAi signature was the transcription factor SOX2 (Fig. 4A and Table S6). Both in cell lines and in primary tumors, SOX2 expression is depleted when compared to normal colon mucosa, however, RNAi against KLF5 induced expression of SOX2 both at the mRNA and protein levels (Fig. 4B and 4C). SOX2 and KLF5 have both been linked to embryonic stem cell self-renewal with SOX2 the better characterized of these transcription factors (25, 26). These data though suggest a direct transcriptional link between suppression of KLF5 expression and upregulation of SOX2 that may have implications for both stem cell and cancer biology.
Figure 4. Analysis of the RNAi signature of the candidate gene KLF5.
(A) Heatmap of the commonly deregulated genes upon silencing of KLF5 in SW480. Sixty-four genes were significantly deregulated in the KLF5-specific RNAi signature. Note that two different siRNAs against this gene result in a very reproducible deregulation of the same signature genes. Among the most downregulated (blue) and upregulated (red) genes are the transcription factors FGF18, RUNX1, and SOX2, as well as the WNT-pathway member WNT10A. (B) Average expression levels of KLF5, FGF18, WNT10A, and SOX2 in cell lines, primary tumors and in SW480 before and after silencing of KLF5. Color code on the right.
(C) Western blot analysis of expression levels of KLF5 and SOX2 after silencing of KLF5 with two different siRNAs. Controls: untransfected cells (WT) and cells transfected with AllStarNeg. The profound downregulation of KLF5 and upregulation of SOX2 are consistent with the gene expression analysis.
Silencing of NUPL1 decreased POU2AF1 expression (4-fold linear change), as well as another CRC associated gene KIAA1199, a downstream target of VDR. Others and we have noted upregulation of KIAA1199 in our primary tumor dataset and in early colonic lesions (27). The targets of the VDR and SMAD4 transcription factor were also significantly enriched in the POMP RNAi upregulated gene signature (P<1E-7), which overall was the largest RNAi signature including nearly 4,000 genes. Although the expression of TCF7L2 was decreased in the POMP transcriptomic signature, only slightly significant enrichment for this specific transcription factor was observed in the downregulated gene set (P=0.036) (Fig. 3A). Interestingly, significant enrichment for MYC was identified among the downregulated genes following LOF of POMP (P=0.001).
Loss of LNX2 function results in changes in NOTCH and WNT/β-catenin pathway
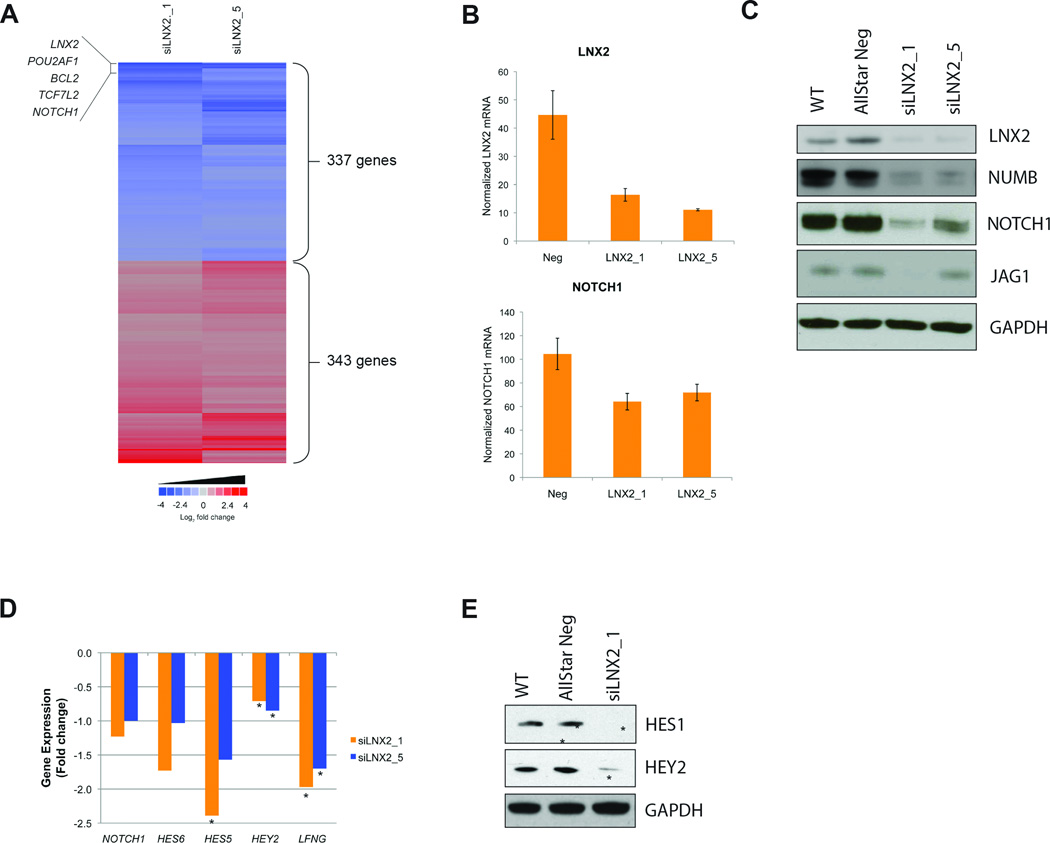
Silencing of LNX2, ligand of numb-protein X 2, had a profound effect on viability and resulted in the deregulation of some 680 genes (Fig. 5A). LNX2 is thought to serves as a scaffold for the membrane protein NUMB. NUMB was originally identified as an important cell fate determinant that is asymmetrically inherited during mitosis and controls the fate of sibling cells by inhibiting the NOTCH signaling pathway in neural tissue. The LNX2 RNAi signature showed downregulation of NOTCH1. To confirm that silencing of LNX2 reduces the expression of NOTCH1, we conducted an independent set of transfections and used a different mRNA assay (Fig. 5B) and also examined NOTCH1 protein levels (Fig. 5C). We consistently observed a downregulation of NOTCH1 expression following LNX2 silencing. While we observed no effect on NUMB mRNA levels, NUMB protein levels were decreased following LNX2 LOF, as too was the expression of JAGGED1, the ligand for the receptor NOTCH1 (Fig. 5C). In addition, pathway analysis of the LNX2 RNAi signature identified NOTCH1 as part of a large molecular network (Fig. S7B). To assess whether the effects on the levels of NOTCH1 were associated with a decrease in the NOTCH signaling pathway, we inquired known downstream targets of NOTCH1 within the LNX2 RNAi signature. Interestingly, several NOTCH1 signaling targets, including HES5, HES6, HEY2, and LFNG were downregulated (P<0.05) (Fig. 5D). Further, Western blot analysis confirmed the reduction of HEY2 and also identified a decrease of HES1 (Fig. 5E), demonstrating that the expression of downstream targets of the NOTCH signaling pathways is reduced. These data suggest a direct link between LNX2 expression and NOTCH1 signaling in SW480 cells.
Figure 5. Analysis of the RNAi signature of the candidate gene LNX2.
(A) Heatmap of the commonly deregulated genes upon silencing of LNX2 in SW480. Six hundred-eighty genes were significantly deregulated in the LNX2-specific RNAi signature. Note that two different siRNAs against this gene result in a very reproducible deregulation of the same signature genes. Among the most downregulated genes (blue) are the transcription factors NOTCH1 and TCF7L2, and the anti-apoptotic gene BCL2. (B) qRT-PCR for LNX2 and NOTCH1 expression levels after silencing of LNX2. The successful silencing of LNX2 correlates with reduced expression of NOTCH1, confirming the expression levels measured on the arrays. (C) Western Blot analysis for the confirmation of silencing on the protein level for LNX2, NOTCH1, NUMB, and JAG1 after silencing LNX2 with two independent siRNAs. Controls: untransfected cells (WT) and cells transfected with AllStarNeg. The results confirm the gene expression data. (D) Gene expression changes of NOTCH1 and the NOTCH downstream targets HES6, HES5, HEY2, and LFGN based on microarray data after silencing LNX2 with two independent siRNAs (* P<0.05). (E) Western Blot analysis demonstrating significant downregulation of NOTCH regulated proteins HES1 and HEY2, indicating negative regulation of NOTCH signaling after silencing LNX2, in accordance with the gene expression data presented in (D).
The other notable feature of the LNX2 RNAi signature was the highly significant enrichment for targets of the transcription factor TCF7L2 (Fig. 3A). TCF7L2 is the main effector of canonical WNT signaling. Of the 272 genes downregulated following silencing of LNX2, 78 are putative TCF7L2 target genes (P=1.96E-8, Benjamini-Hochberg corrected Fisher’s exact test). These 78 genes included the proliferative and anti-apoptotic genes MYB, CCND1, BCL2, POU2AF1, and ERBB4. Similarly, target genes for the TCF7L2 binding partner and transcriptional co-activator β-catenin (CTNNB1) were also enriched within this downregulated set (28 of 272, P=0.049). Eleven downregulated targets of β-catenin were also targets of TCF7L2. After combining the downregulated TCF7L2 and β-catenin gene targets into a single list (95 total), we found it to be enriched for functional ontologies such as gastrointestinal disease (P=0.001), cancer (P=0.002), and cell cycle (P=0.008).
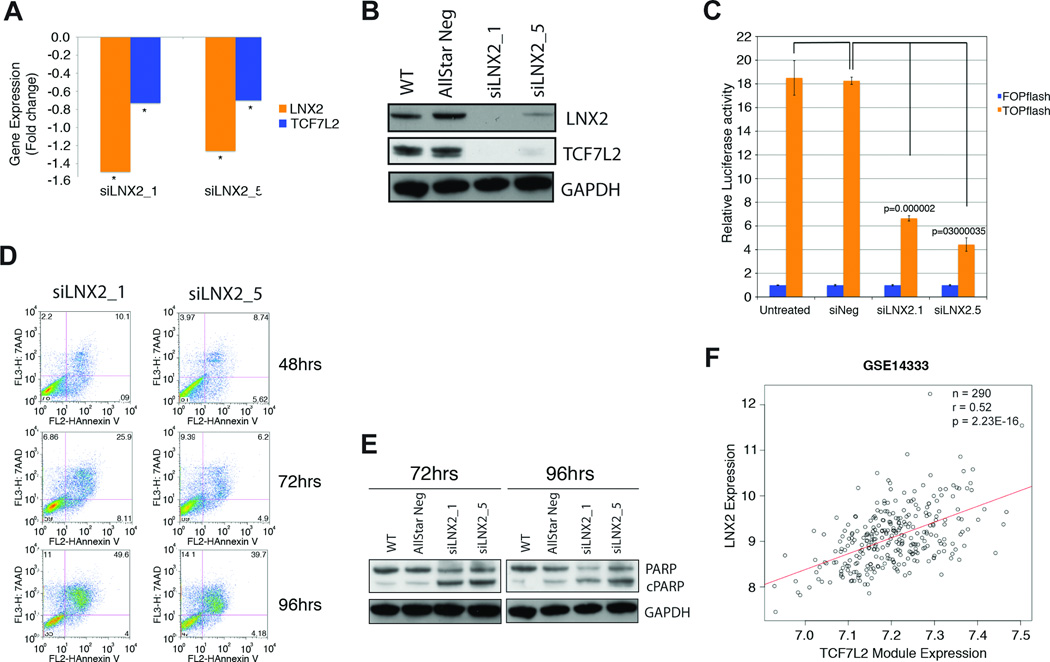
The transcription factor TCF7L2 is the dominant effector of the WNT pathway and responsible for transcriptional regulation of genes involved in WNT signaling. The enrichment for TCF7L2 gene targets in the LNX2 RNAi signature suggests a potential link between the WNT pathway and LNX2 function, and as an extension of this, between chromosome 13 amplification and activation of WNT signaling. In order to test this hypothesis, we first examined the expression levels of TCF7L2 in the expression profiles obtained following LNX2 silencing and did observe a decrease in TCF7L2 expression (Fig. 6A). This was confirmed at the protein level 96 hrs post LNX2 siRNA transfection (Fig. 6B). Further, TCF7L2 is present in one of the top networks generated by pathway analysis of the LNX2 RNAi signature (Fig. S7C). Importantly, we next investigated the role of LNX2 in regulating WNT pathway activity using a reporter assay. Using the TOP-FOP-Flash reporter system we demonstrated a significantly decreased WNT/β-catenin activity in SW480 cells after gene silencing of LNX2 when compared to negative control (P<0.0001) (Fig. 6C). Additionally, by Western-blot analysis we observed a decrease of the protein levels of CTNNB1 upon treatment with siRNAs against LNX2 (Fig. S8).
Figure 6. Functional and bioinformatic analysis of the role of LNX2 in CRC.
Confirmation of downregulation of LNX2 and TCF7L2 after silencing of LNX2 with two independent siRNAs by qRT-PCR. We observed significantly reduced expression of the transcription factor TCF7L2, both (A) on the RNA (* P<0.05) and (B) protein level. (C) Functional validation of reduced WNT/β-catenin signaling using the TOP-FOP-Flash reporter assay. Reduced expression of TCF7L2 results in markedly lower canonical WNT/β-catenin activity as measured using a TOP-FOP-Flash reporter assay after silencing LNX2 with two independent siRNAs. (D) Silencing of LNX2 with both siRNAs triggers increased apoptosis in SW480 cells as measured by Annexin V vs. 7-AAD using flow cytometry. The Annexin V positive cells at 96 hrs after transfection reached almost 50%. (E) Western blot analysis measuring cleaved PARP at two different time points, 72 and 96 hrs, confirmed an increased rate of apoptosis as a consequence of silencing LNX2 for both siRNAs. (F) Correlation of the expression of a module of genes regulated by the transcription factor TCF7L2 and downregulated signature genes after silencing LNX2 in a set of 290 primary colorectal cancer samples. Y-axis: expression levels of LNX2 in primary colorectal carcinomas; x-axis: expression levels of the TCF7L2 module. The correlation indicates a direct influence of LNX2 on the expression of TCF7L2-regulated genes and consequently the genomic amplification of chromosome 13 and activation of WNT-signaling.
Finally, towards a complete characterization of the mechanism underlying the reduction in viability following LNX2 silencing, we assessed two cellular markers of apoptosis; annexin V levels by flow cytometry analysis and cleavage of PARP using Western blot analysis. A time-course at 48, 72, and 96 hrs post-transfection showed a sequential activation of apoptosis, which on average for the two siRNAs reached some 45% of Annexin V positive cells at 96 hrs (Fig. 6D). This activation perfectly correlates with increased levels of cleaved PARP and the absence of LNX2 protein (Fig. 6E). Our analysis therefore suggests that silencing of LNX2 results in altered NOTCH1 and WNT signaling and activation of apoptosis.
The LNX2-TCF7L2 module is recapitulated in clinical samples and correlates with expression of LNX2
In order to detect biologically relevant modularity using pre-defined ontologies and gene expression data (28), we constructed a LNX2-TCF7L2 module from the 78 TCF7L2 target genes, listed in Table S7, that were significantly downregulated after silencing LNX2. In order to further investigate LNX2 association with this gene module, we analyzed the correlation between LNX2 expression and the average expression of all module genes using three independent colorectal cancer gene expression datasets containing more than 500 patient samples. This meta-analysis resulted in highly significant correlations in all three datasets (GSE14333: N=290, R=0.52, P=2.2E-16; GSE17536: N=177, R=0.44, P=5.6E-10; GSE17537: N=49, R=0.39, P=0.006) (shown for dataset GSE14333 in Fig. 6F). Next, using an additional unpublished dataset we proved that this correlation was also present in normal samples (R=0.53, P=7.01E-06) and indeed found that the gene module could form distinct clusters separating normal and cancer specimens (Fig. S9). Taken together, these data strongly support the assertion that LNX2 serves as a regulator of TCF7L2 transcriptional activity and of subsequent downstream WNT signaling. This is entirely consistent with a central involvement of this gene in CRC and with the frequent gain and amplification of chromosome 13.
DISCUSSION
One major mechanism to constitutively overexpress cancer-promoting genes is through genomic amplification of their respective locus. Genes that reside in regions of recurrent genomic amplification and are overexpressed are prime candidates for bona fide oncogenes (29). The gain of chromosome 13 is one of the most common aberrations in CRC and is found in some 60–80% of cases, often in the form of a high level gain of the entire chromosome (3, 8, 30). It is now abundantly clear that gene expression levels are positively correlated with genomic copy number (13). Gains or losses of entire chromosomes or chromosome arms therefore result in aberrant transcript levels of many, if not most, of the resident genes (11, 12, 14, 31). This, of course, hampers the identification of candidates that drive the acquisition of such recurrent genomic imbalances. It has also been shown that the low level upregulation of genes on commonly gained chromosomes often involves genes that favor metabolic activity, and therefore may act in concert with specific oncogenes to promote growth and tumorigenesis (10). While we explicitly do not exclude the possibility that additional driver genes on chromosome 13 play a role in CRC (for instance, CDK8 (32) and CDX2 (33)), we considered it a reasonable strategy to initially concentrate on genes that reside in two focal amplification regions, and are strongly overexpressed in both primary tumors and derived cell lines. To study which of the amplified and overexpressed genes are required for viability of colorectal cancer cells we applied an RNAi-based LOF strategy. This strategy led us to focus on the following eight genes: NUPL1, LNX2, POLR1D, POMP, SLC7A1, DIS3, KLF5, and GPR180. We then, in an unbiased manner, explored the function of these candidates in colorectal cancer cells using genome-wide expression profiling after gene silencing.
Only two of the candidate genes studied, KLF5 and POLR1D, had been previously implicated in CRC tumorigenesis (21, 34, 35); however, limited functional association with molecular pathways involved in colorectal carcinogenesis has been reported. High-resolution DNA copy number profiles of stage II colon cancers identified KLF5, which encodes a Zn-finger transcription factor of the Kruppel-like family, as residing in a distinct focal amplification (34). The presence of this amplification is associated with poor overall survival (36). In our study, silencing of KLF5 strongly induced the expression of SOX2, a transcription factor involved in the maintenance of the undifferentiated state in embryonic stem cells. Although SOX2 has been proposed as an amplified lineage-survival oncogene in squamous cell lung cancer (37) and esophageal squamous cell carcinomas (38), its role in CRC remains to be elucidated.
The LOF of LNX2 resulted in a striking reduction of viability of colorectal cancer cell lines, and LNX2 showed the highest enrichment of TCF7L2 target genes (P<10E-12) and evidence of disruption of NOTCH signaling. LNX2 was a particularly interesting candidate because we and others have shown that it localizes in a minimal region of amplification at chromosome band 13q12.2 (chr13:26,694,966–27,905,729) (Fig. S10) (30). The important role of LNX2 is also supported by the overexpression observed in tumors compared to normal mucosa (Fig. S11), and by data from the Protein Atlas, where it is shown to be strongly and exclusively overexpressed in CRC (39). One role of LNX proteins is to serve as a molecular scaffold that guides NUMB to specific sites in the cell (40). The LNX2 family-related member, LNX1, is required for NUMB ubiquitylation causing proteasome-dependent degradation and enhanced NOTCH signaling (41). The tight regulation of WNT and NOTCH signaling is essential for the development and homeostasis of intestinal epithelial cells (42–44). Aberrant NOTCH signaling, on the other hand, is intricately involved in colorectal tumorigenesis (45). Our data show that silencing of LNX2 results in downregulation of NOTCH1 and several downstream targets, including HES1, HES5, HES6, HEY2, and LFNG. In fact, the effect of NOTCH1 on cell proliferation depends on active WNT (its effect on differentiation does not) (43, 46). The crosstalk between the WNT/β-catenin pathway and NOTCH has been shown to be involved in the control of cell proliferation and colorectal tumorigenesis in mice (46). One of the candidates to mediate such a crosstalk is JAGGED1 (47, 48). Interestingly, expression of JAGGED1 was decreased after silencing of LNX2, suggesting that LNX2 may operate upstream of JAGGED1 in its contribution to the control of NOTCH and WNT/β-catenin pathway cooperation. However, we cannot exclude that deregulation of JAGGED1 was a direct effect of deregulation of the WNT/β-catenin pathway (47). Our observation that silencing of LNX2 not only resulted in reduced NOTCH activity, but also in a significant reduction of the expression of TCF7L2, is entirely consistent with the interplay of NOTCH and WNT signaling. Reduced expression of TCF7L2 was accompanied by the intuitive depression of WNT/β-catenin activity as shown using the TOP-FOP-Flash reporter assay subsequent to LNX2-silencing. β-catenin and TCF7L2 are the downstream effectors of the WNT signaling cascade (49), and according to our data the expression of these two molecules is modulated by LNX2. This is the first data that suggest that the WNT and NOTCH signaling pathways are coregulated in CRC. As a result of LNX2 upregulation, mediated by the amplification of chromosome 13, there is an increase in WNT/β-catenin signaling, thus promoting proliferation through TCF7L2 transcriptional targets (Fig. S12). We believe that the significant reduction of viability as a consequence of LOF of LNX2 is a reflection of the fact that it stimulates two critical cancer pathways simultaneously. The correlation of the expression of LNX2-TCF7L2 signature genes with LNX2 levels was validated in a large set of clinical samples, indicating that genes that depend on expression of the transcription factor TCF7L2 correlate with those genes that are affected when suppressing the expression of an upstream regulator of the WNT signaling pathway.
The finding that there is such an abundance of chromosome 13 genes involved in crucial CRC signaling pathways is entirely consistent with the frequent genomic amplification of chromosome 13 in CRC at the transition from dysplastic adenomas to invasive disease (3). This supports the hypothesis of tumor-specific pathway addiction. It appears indeed to be the case that not only one, but several, if not many, genes on that chromosome activate pathways required for colorectal carcinogenesis (32, 33), a notion that reinforces the idea that this chromosome needs to be present in extra copies in the majority of CRC in a tissue specific manner. In essence, chromosome 13 is a colorectal cancer chromosome.
In summary, we have identified a set of genes that are highly and consistently amplified and overexpressed in colorectal cancer, and we have inferred the mechanism of action by which these genes might have a role in the cellular viability of colorectal cancer cells. By analyzing the gene expression signatures after RNAi mediated gene silencing, followed by targeted functional analysis, we identified LNX2, a gene that was not associated with cancer before, as mediating the crosstalk between WNT and NOTCH signaling cascades and regulating WNT/β-catenin activity.
Supplementary Material
Acknowledgments
The authors are grateful to Buddy Chen for editorial assistance, and Drs. Reinhard Ebner, Robert Callahan, and Jeff Rubin for critical comments on the manuscript and helpful discussions. The TOP-FOP-Flash vectors were kindly provided by Dr. Randall Moon (University of Washington, Seattle, WA) and the renilla luciferase construct was a generous gift from Dr. Silvio Gutkind (NIDCR/NIH).
GRANT SUPPORT
This project was supported by the Intramural Research Program of the NIH, National Cancer Institute, and through the KFO 179 from the Deutsche Forschungsgemeinschaft.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 5.Ried T. Homage to Theodor Boveri (1862–1915): Boveri's theory of cancer as a disease of the chromosomes, and the landscape of genomic imbalances in human carcinomas. Environ Mol Mutagen. 2009;50:593–601. doi: 10.1002/em.20526. [DOI] [PubMed] [Google Scholar]
- 6.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 8.Heim S, Mitelman F. Cancer Cytogenetics. 3rd. Hoboken, NJ: John Wiley & Sons; 2009. [Google Scholar]
- 9.Camps J, Grade M, Nguyen QT, Hormann P, Becker S, Hummon AB, et al. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–1295. doi: 10.1158/0008-5472.CAN-07-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Grade M, Hormann P, Becker S, Hummon AB, Wangsa D, Varma S, et al. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 2007;67:41–56. doi: 10.1158/0008-5472.CAN-06-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ried T, Hu Y, Difilippantonio MJ, Ghadimi BM, Grade M, Camps J. The consequences of chromosomal aneuploidy on the transcriptome of cancer cells. Biochim Biophys Acta. 2012;1819:784–793. doi: 10.1016/j.bbagrm.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 15.Grade M, Hummon AB, Camps J, Emons G, Spitzner M, Gaedcke J, et al. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer. 2011;128:1069–1079. doi: 10.1002/ijc.25453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laboratory of Thomas Ried. [cited; Available from: http://www.riedlab.nci.nih.gov/index.php/protocols.
- 18.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 20.Camps J, Nguyen QT, Padilla-Nash HM, Knutsen T, McNeil NE, Wangsa D, et al. Integrative genomics reveals mechanisms of copy number alterations responsible for transcriptional deregulation in colorectal cancer. Genes Chromosomes Cancer. 2009;48:1002–1017. doi: 10.1002/gcc.20699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, et al. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimokawa T, Furukawa Y, Sakai M, Li M, Miwa N, Lin YM, et al. Involvement of the FGF18 gene in colorectal carcinogenesis, as a novel downstream target of the beta-catenin/T-cell factor complex. Cancer Res. 2003;63:6116–6120. [PubMed] [Google Scholar]
- 23.Sonvilla G, Allerstorfer S, Heinzle C, Stattner S, Karner J, Klimpfinger M, et al. Fibroblast growth factor receptor 3-IIIc mediates colorectal cancer growth and migration. Br J Cancer. 2010;102:1145–1156. doi: 10.1038/sj.bjc.6605596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonvilla G, Allerstorfer S, Stattner S, Karner J, Klimpfinger M, Fischer H, et al. FGF18 in colorectal tumour cells: autocrine and paracrine effects. Carcinogenesis. 2008;29:15–24. doi: 10.1093/carcin/bgm202. [DOI] [PubMed] [Google Scholar]
- 25.Nandan MO, Yang VW. The role of Kruppel-like factors in the reprogramming of somatic cells to induced pluripotent stem cells. Histol Histopathol. 2009;24:1343–1355. doi: 10.14670/hh-24.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, et al. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121:2629–2634. doi: 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- 27.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 28.Segal E, Yelensky R, Koller D. Genome-wide discovery of transcriptional modules from DNA sequence and gene expression. Bioinformatics. 2003;19(Suppl 1):273–282. doi: 10.1093/bioinformatics/btg1038. [DOI] [PubMed] [Google Scholar]
- 29.Myllykangas S, Bohling T, Knuutila S. Specificity, selection and significance of gene amplifications in cancer. Semin Cancer Biol. 2007;17:42–55. doi: 10.1016/j.semcancer.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, et al. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salari K, Spulak ME, Cuff J, Forster AD, Giacomini CP, Huang S, et al. CDX2 is an amplified lineage-survival oncogene in colorectal cancer. Proc Natl Acad Sci U S A. 2012;109:E3196–E3205. doi: 10.1073/pnas.1206004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulak AM, Schumacher S, van Lieshout J, Imamura Y, Fox C, Shim B, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brosens RP, Haan JC, Carvalho B, Rustenburg F, Grabsch H, Quirke P, et al. Candidate driver genes in focal chromosomal aberrations of stage II colon cancer. J Pathol. 2010;221:411–424. doi: 10.1002/path.2724. [DOI] [PubMed] [Google Scholar]
- 37.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gen Y, Yasui K, Zen Y, Zen K, Dohi O, Endo M, et al. SOX2 identified as a target gene for the amplification at 3q26 that is frequently detected in esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2010;202:82–93. doi: 10.1016/j.cancergencyto.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 39.The Human Protein Atlas. [cited; Available from: http://www.proteinatlas.org/ENSG00000139517/cancer.
- 40.Rice DS, Northcutt GM, Kurschner C. The Lnx family proteins function as molecular scaffolds for Numb family proteins. Mol Cell Neurosci. 2001;18:525–540. doi: 10.1006/mcne.2001.1024. [DOI] [PubMed] [Google Scholar]
- 41.Nie J, McGill MA, Dermer M, Dho SE, Wolting CD, McGlade CJ. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002;21:93–102. doi: 10.1093/emboj/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T, Torimura T, Sakamoto M, Hashimoto O, Taniguchi E, Inoue K, et al. Significance and therapeutic potential of endothelial progenitor cell transplantation in a cirrhotic liver rat model. Gastroenterology. 2007;133:91–107. e1. doi: 10.1053/j.gastro.2007.03.110. [DOI] [PubMed] [Google Scholar]
- 44.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vooijs M, Liu Z, Kopan R. Notch: architect, landscaper, and guardian of the intestine. Gastroenterology. 2011;141:448–459. doi: 10.1053/j.gastro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci U S A. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guilmeau S, Flandez M, Mariadason JM, Augenlicht LH. Heterogeneity of Jagged1 expression in human and mouse intestinal tumors: implications for targeting Notch signaling. Oncogene. 2010;29:992–1002. doi: 10.1038/onc.2009.393. [DOI] [PubMed] [Google Scholar]
- 48.Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.