Summary
The role of gut bacteria in modulating gastrointestinal physiology is increasingly being appreciated. In a recent issue of Cell, Dey, et al. (2015) report how a single dietary ingredient—turmeric—interacts with gut bacteria to alter gastrointestinal motility.
Keywords: curcumin, serotonin, bile acids, deconjugation, microbiota, motility, neurons, Ret
Gastrointestinal (GI) motility enables mixing, storage, anterograde propulsion and absorption of nutrients, and represents one of the most important functions of the GI tract. GI transit time, the time for a food bolus to pass through the GI tract, is often used as a surrogate for GI motility. This seemingly simple process requires coordination among several key cell types in the enteric neuromuscular apparatus including the enteric neurons and glia, interstitial cells of Cajal (ICC), smooth muscle and immune cells. Until recently, the majority of the work on GI motility disorders has focused on the interplay amongst different host cell types, even though it is established that gut bacteria and luminal compounds have a significant impact on GI motility. However, recent studies have started to uncover this additional layer of complexity, exploring how gut microbiota and their products influence host GI physiology.
Studies of gut microbiota and neuromuscular apparatus, including neurotransmitters in the context of GI motility, have shown that gut microbiota and their products have effects on the enteric neurons, enteric muscularis macrophages and enteric glia. Gut microbiota-derived lipopolysaccharide (LPS) improves enteric neuronal survival and influences GI motility (Figure1), acting via the TLR4 and NF-κB pathway (Anitha et al., 2012). Gut microbiota products also influence the crosstalk between enteric neurons and muscularis macrophages, (Figure 1), which plays an important role in maintaining normal GI motility (Muller et al., 2014). Serotonin, an important neurotransmitter in the gut that plays a role in modulating GI motility, can be modulated by gut microbes (Reigstad et al., 2015; Yano et al., 2015). Metabolites resulting from primary and secondary fermentation of dietary nutrients by gut microbes, such as short chain fatty acids and bile acids (Figure1), can increase serotonin biosynthesis and release in the gut, thereby altering GI motility in a diet-dependent manner (Kashyap et al., 2013).
Figure 1. Microbiota-mediated pathways that affect GI transit.
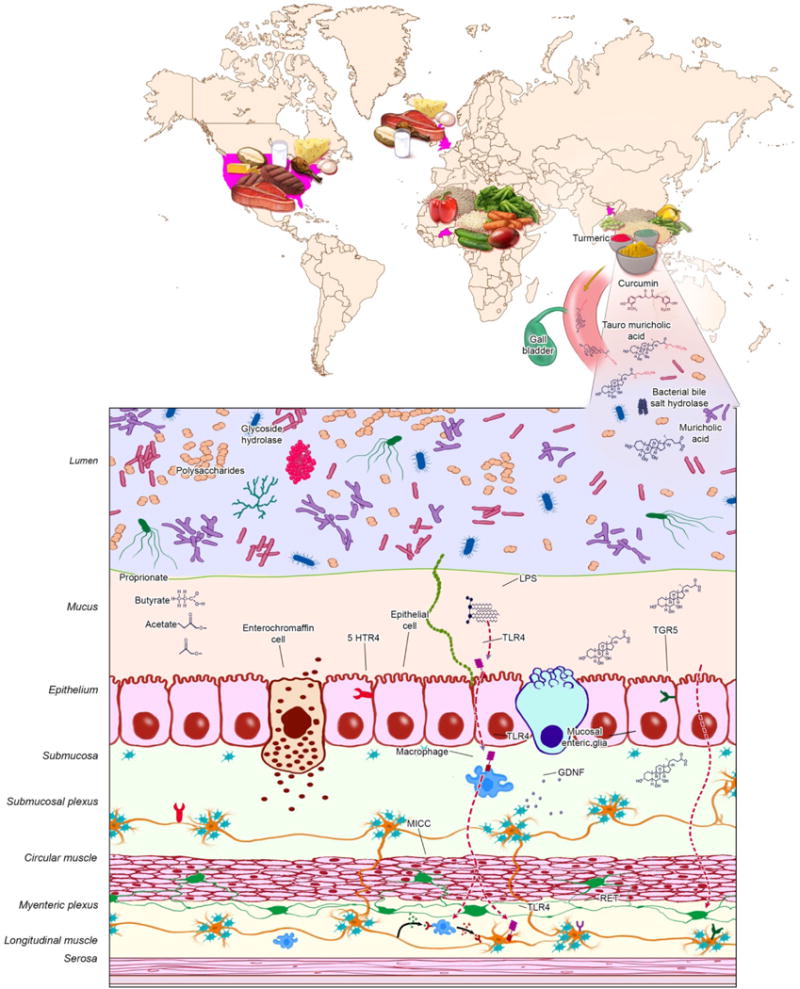
Diet–gut microbiota interaction results in a large array of metabolites. In the presence of dietary turmeric, gut microbial communities with high bile-salt hydrolase activity make higher amounts of unconjugated bile acids, which influence enteric nervous system signaling and lead to faster GI transit. Dietary fermentation by gut microbiota results in formation of short chain fatty acids which affect GI motility by effects on gut neuromuscular apparatus and the host serotonergic pathway. LPS and other bacterial products are implicated in improving neuronal survival as well as affecting enteric neuron–muscularis macrophage crosstalk. Gut microbiota are required for the post-natal development and generation of new enteric glia. LPS, lipopolysaccharide; GDNF, glia derived neurotrophic factor; TLR-4, Toll like receptor 4; 5HTR, 4 5-hydroxytryptamine receptor-4; ICC, interstitial cells of Cajal.
The new study by Dey, et al. (Dey et al., 2015) provides an in depth investigation of the interaction between diet and gut microbiota to elucidate the mechanisms by which these interactions influence GI motility. Travelers’ diarrhea is common during travel to the developing world and is often attributed to acquisition of pathogenic bacteria or viruses. However, an additional aspect of travel is short-term exposure to new diets. Diet is a dominant factor in shaping gut microbial communities (Wu et al., 2011) and has been shown to influence GI motility in both a microbiota-dependent and independent manner (Kashyap et al., 2013). In a series of elegant experiments, the authors subject gnotobiotic mice colonized with microbiota from healthy human donors (humanized) from across the world to multiple cycles of short-term dietary interventions, representing native and non-native diets. The authors were able to identify several bacterial taxa present in different groups of humanized mice that have diet-dependent effects on GI transit. Some of these taxa (eg. E. desmolans) had opposite correlations with transit time depending on supplementation with native or non-native diet. As expected, diet also showed effects on gut microbiota that did not correlate with GI transit time.
In order to investigate bacterial mediators of the dietary effects on GI transit, Dey et al. (2015) quantified bile acid metabolites in the stool. They found that faster transit time was associated with an increase in unconjugated bile acids, resulting from the activity of bacterial bile salt hydrolases. In order to determine how a single dietary ingredient can interact with gut microbiota and change bile acid composition in the gut, the authors used turmeric, a commonly used spice in south Asia. While turmeric contains the natural anti-inflammatory phytochemical, curcumin, it is also known for its cholekinetic effect, or the ability to stimulate bile acid secretion into the gut lumen. Gnotobiotic mice, colonized with a clonally arrayed bacterial culture collection from a child from Bangladesh were found to have significantly higher conjugated bile acids (taurohyodeoxycholic acid and tauromuricholic acid sulphate) and longer GI transit time in the presence of turmeric, compared to unsupplemented animals, highlighting the effect of a single dietary ingredient on gut microbial products and GI transit (Figure1). The authors screened members of this clonally arrayed bacterial culture collection for their bile salt hydrolase activity and assembled two bacterial consortia, one with low bile salt hydrolase (Enterococcus, Eggerthella and a member of Enterobacteriaceae) and the other with high bile salt hydrolase activity (Enterococcus and three members of Bifidobacterium) to examine the effect of turmeric on microbial communities with differing capacities to deconjugate bile acids and on GI transit time. Interestingly, in the presence of turmeric, gnotobiotic mice colonized with a cultured microbial community with low bile salt hydrolase had significantly longer GI transit time, lower bile salt hydrolase expression and lower levels of unconjugated bile acids as compared to gnotobiotic mice colonized with a microbial community with high bile salt hydrolase activity.
The role of bile acids in GI physiology has been well studied. Bile acids can stimulate intestinal secretion of fluid and electrolytes, increase mucosal permeability by acting as emulsifying agents, and induce high amplitude propagated contractions thereby accelerating colonic transit (Camilleri, 2014). The recently described membrane bound bile acid receptor TGR5, which is expressed in enteric neurons and colonic epithelial cells, can regulate smooth muscle contraction and intestinal secretion. Bile acid malabsorption is associated with diarrhea, and can be seen in ileal Crohn’s disease, post-cholecystectomy, small intestinal bacterial overgrowth, irritable bowel syndrome (IBS) and other similar disorders. Interestingly, while gut microbiota play a key role in deconjugating bile acids and are important regulators of the total bile acid pool as well as their composition, only recently has the role of gut microbiota in bile acid metabolism been described in GI diseases. In fact, disturbances in bile acid metabolism with an increase in primary bile acids, in association with alteration in gut microbial communities (eg. increase in E.coli and decrease in Leptum and Bifidobacterium in patients with diarrhea predominant IBS) was recently reported in IBD and IBS (Duboc et al., 2012; Duboc et al., 2013).
The enteric nervous system plays an important role in maintaining normal GI motility and hence the authors next went on to test if the diet-microbiota interaction affects GI transit by modulating enteric nervous system signaling (Figure1). Interestingly they found the difference in GI transit between the high and low bile salt hydrolase communities seen on diet supplemented with turmeric was lost when these communities were introduced into germ free RET+/− mice but was retained in wild type animals. RET encodes a transmembrane protein which forms part of the receptor for glia-derived neurotrophic factor (GDNF) family signaling peptides. Loss of function mutations in RET account for the largest proportion of cases of Hirschsprung’s disease, a hereditary developmental disorder where peristalsis is absent in the colon due to lack of neuronal development. Interestingly, a recent paper investigating the effect of gut microbiota on enteric glia (Figure 1) found that the gut microbiota plays an important role both in postnatal development as well sustaining generation of new mucosal enteric glial cells (Kabouridis et al., 2015). Given that the GDNF family of peptides from enteric glia are ligands for RET on enteric neurons, one may speculate the effect of gut microbiota products on RET proposed in the current study is potentially downstream of their effect on enteric glia. These findings which implicate the gut microbiota in potential interactions between key cell types affecting GI motility open several new avenues to be explored in future studies.
In summary, the study by Dey et al. (2015) provides novel insights into the mechanisms by which interaction of dietary ingredients and gut microbiota influence the enteric nervous system to alter GI transit time. This finding has important implications not only for variations in transit time seen with travel and rapid dietary cycling, but also for developing microbiota-targeted therapies in chronic GI disorders associated with microbial dysbioses including IBS and IBD, wherein some symptoms are commonly attributed to dietary intolerance. This and the other recent studies highlight the layers of complexity with cross-kingdom signaling in the regulation of GI motility as we start to investigate the role of gut microbiota in this important process. As the field advances we hope to have a better understanding of the effects of bacteria and their products on the key cell types in the enteric neuromuscular apparatus and the interactions among them. The findings of Dey et al. (2015) raise several additional questions which will need to be addressed in future studies including the mechanisms by which bacteria-mediated processes influence the i) RET gene function and downstream signaling pathways, ii) other key cells types involved in GI motility and iii) development and interaction among different cell types in the gut such as enteric glia, enteric neurons, ICC and macrophages.
Acknowledgments
We would like to thank Michael A. King, senior medical illustrator at Mayo Clinic, Rochester, MN for the illustration. This work was made possible by funding from NIH K08 DK100638, Minnesota Partnership for Biotechnology and Genomics and Center for Individualized Medicine (CIM; Mayo Clinic).
References
- Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology. 2012;143:1006–1016 e1004. doi: 10.1053/j.gastro.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri M. Advances in understanding of bile acid diarrhea. Expert Rev Gastroenterol Hepatol. 2014;8:49–61. doi: 10.1586/17474124.2014.851599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey N, Wagner VE, Blanton LV, Cheng J, Fontana L, Haque R, Ahmed T, Gordon JI. Regulators of Gut Motility Revealed by a Gnotobiotic Model of Diet-Microbiome Interactions Related to Travel. Cell. 2015;163:95–107. doi: 10.1016/j.cell.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc H, Rainteau D, Rajca S, Humbert L, Farabos D, Maubert M, Grondin V, Jouet P, Bouhassira D, Seksik P, et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:513–520. e246–517. doi: 10.1111/j.1365-2982.2012.01893.x. [DOI] [PubMed] [Google Scholar]
- Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, Pettersson S, Pachnis V. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron. 2015;85:289–295. doi: 10.1016/j.neuron.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Koscso B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, Mortha A, Leboeuf M, Li XM, Mucida D, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reigstad CS, Salmonson CE, Rainey JF, 3rd, Szurszewski JH, Linden DR, Sonnenburg JL, Farrugia G, Kashyap PC. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. Faseb J. 2015;29:1395–1403. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]


