MOLECULAR CYTOGENETICS
The screening of tumor genomes using cytogenetic techniques has been an important asset in cancer genetics over the last four decades. Cytogenetic techniques allow detection of chromosomal translocations, both balanced and unbalanced, of numerical aberrations or chromosomal aneuploidies, and to some extent, chromosomal correlates of gene amplification, such as double minute chromosomes (dmin) and heterogeneously staining regions (hsr). The identification of such cytogenetic abnormalities is extremely relevant for cancer genetics, because it reveals sites of recurrent changes, which in turn point to the genomic localization of genes that are involved in tumorigenesis. The comprehensive cytogenetic analysis of complex aberrations of the highly rearranged chromosomes of solid tumors, however, is extremely challenging. It is for these reasons that the relative contribution of conventional karypotype analysis using chromosome banding techniques has been far greater in hematological malignancies, which have, in general, fewer aberrations compared to the carcinomas, including beast cancer [1]. Similar challenges apply to mouse chromosomes: while human chromosomes differ from one another in both size and the position of the centromere, mouse chromosomes are less heterogeneous, and only few laboratories master the challenge of karyotyping mouse tumors. The introduction of molecular cytogenetic techniques that allow genome screening based on fluorescence in situ hybridization was therefore a welcome addition to the armament of tools to comprehensibly describe karyotypic changes in both human carcinomas and their respective murine models. The first such hybridization based genome screening technique, comparative genomic hybridization (CGH), was introduced in 1992 for human chromosomes [2]. The first application to murine tumors was published in 1995 [3]. CGH allows mapping of regions of genomic imbalances onto normal chromosomes. This omits the requirement for metaphase preparations from tumor cells, which is technically demanding. In a first step, the tumor genome and a karytopypically normal reference genome are differentially labeled with discernible fluorochromes (e.g., fluorescein for the tumor DNA and rhodamine for the reference DNA). These two differentially labeled genomes are then combined together with an excess of unlabeled Cot1 DNA and hybridized to normal metaphase chromosomes. The resulting relative fluorescence intensity values reveal genomic imbalances in the tumor genome. An example of a ratio profile used for the identification of genomic imbalances in presented in Fig. 1(c). More recent advancements utilizing the availability of genomic clone collections and the ability to perform fluorescent hybridization to DNA immobilized on glass slides have resulted in the development of array CGH [4, 5]. Array CGH has the advantage that the detection of genomic imbalances can be performed with higher resolution and increased objectivity (it does not require the identification of normal chromosomes).
Fig. 1.
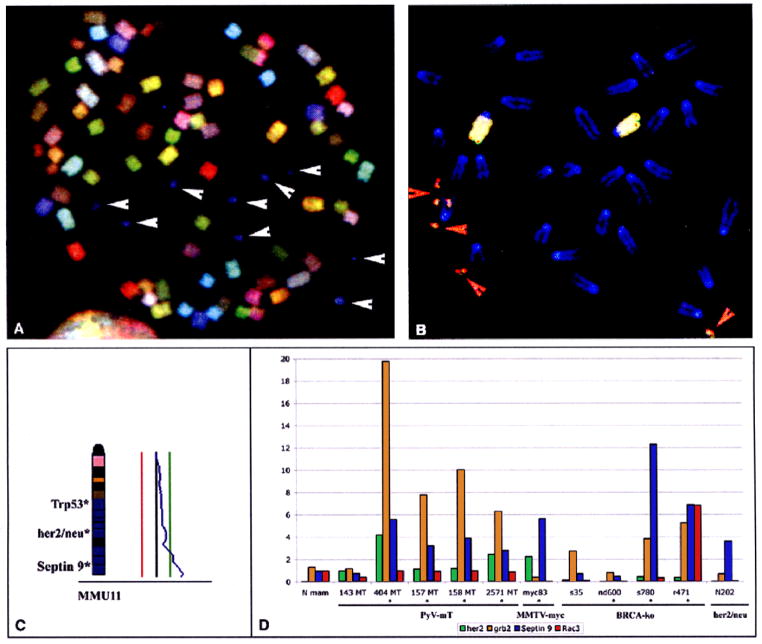
Examples of molecular cytogenetic analysis of mouse mammary gland adenocarcinomas. A: SKY of metaphase chromosomes from PyV-mT induced tumors. Note that aneuploidy (aberrant number of chromosomes) is common, yet structural chromosomal aberrations are rare. SKY also suggests that the double minute chromosomes were derived from chromosome 11 (blue labeling, white arrowheads). B: Identification of septin 9 as the target of genomic amplification. Dual color FISH experiment with a painting probe for chromosome 11 (yellow) and a BAC clone containing sequences for septin 9 (red). Note the localization of the normal copies of septin 9 on distal chromosome 11, and the presence of amplified copies in the double minute chromosomes 9 (red arrowheads). C: CGH analysis of a PyV-mT induced tumor. The average ratio profile indicates copy number increases that map to distal chromosome 11. The location of several breast cancer associated genes on mouse chromosome 11 is provided on the left side of the chromosome ideogram. D: Quantitative real time PCR for the assessment of septin 9 expression levels in different mouse models of breast cancer. Note that septin 9 amplification is not restricted to the PyV-mT induced tumors, but is also found in BRCA1 and her2/neu-associated carcinomas. The expression levels of her2/neu, grb2 (known to be highly overexpressed in PyV-mT induced tumors), and rac3 were used as controls.
Another major advance for the comprehensive characterization of cytogenetic aberrations in cancer genomes was published in 1996, when it became possible to paint all mouse chromosomes in different colors [6]. This technique, termed spectral karyotyping or SKY, is based on the simultaneous hybridization of differentially labeled whole chromosome painting probes and a combination of spectroscopy and digital imaging to discern all chromosomes according to their specific spectral signature [7]. SKY is particularly useful for the identification of structural chromosomal aberrations, such as translocations and insertions, and chromosomal aneuploidies and assists in the identification of the chromosomal origin of amplifications, e.g., those in the form of double minute chromosomes. Figure 1a shows and example of SKY for the analysis of chromosomal aberrations in metaphase chromosomes from a PyV-mT induced mammary gland adenocarcinoma. Together, CGH and SKY now afford the characterization of cytogenetic abnormalities in cancer genomes with unprecedented precision. Both techniques have found widespread application for the mapping of genome aberrations in cancer cells [8, 9] and http://www.helsinki.fi/cmg/cgh_data.html and http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi.
MOUSE MODELS
Cytogenetic and molecular cytogenetic analysis of cancer genomes has generated ample evidence for non-random, tumor specific patterns and distributions of chromosomal aberrations. In hematological malignancies, such aberrations often involve chromosomal translocations, many of which are balanced, that point to the localization of genes whose dysregulation causes malignant transformation. In solid tumors of epithelial origin, i.e., carcinomas, including breast cancers, such balanced translocations are relatively less frequent: the main consequences of cytogenetic abnormalities are genomic imbalances [10]. It is obviously an intriguing question whether these different pathways of tumorigenesis apply to murine models as well. The first step towards an answer requires the establishment of comparative maps of karyotypic abnormalities in human tumors and their mouse models. We have therefore applied CGH and SKY to a large number of primary mouse tumors and derived cell lines (including models for breast, colon, and skin cancer) and to diverse models of hematological malignancies. Table 1 provides a summary of the mammary gland adenocarcinomas analyzed. Our first comprehensive study of chromosomal imbalances in a transgenic mouse model of breast cancer was reported in 1999 [11]. Weaver and colleagues used CGH and SKY to analyze the karyotypes of eight cases of mammary gland adenocarcinomas induced by tissue specific overexpression of c-myc under control of the MMTV-promotor [12]. It became clear that tumorigenesis in this model, which occurs with a latency of about 11 months, required additional chromosomal abnormalities. For instance, five of the eight tumors studied also revealed whole chromosome losses of mouse chromosome 4. In addition, we observed copy number increases and translocations involving chromosome 11. These two chromosomes were of particular interest because the orthologous regions in the human genome are also involved in human breast cancers. Specifically, the distal region of human chromosome 1 p (orthologous to mouse chromosome 4) is commonly deleted in breast cancer and amplifications on human chromosome 17q (orthologous to mouse chromosome 11) are common as well [13]. It therefore seems reasonable to assume that both of these chromosomes and chromosomal regions contain genes that cooperate with myc in the induction of mouse mammary gland adenocarcinomas.
Table 1.
List of live different breast cancer models analyzed by CGH and SKY
| Mouse model | Number of tumors | Publication |
|---|---|---|
| MMTV-myc | 8 | Genes Chromosomes Cancer. 1999 25(3):251–260 |
| C3(1)SV40 TAg | 9 | Oncogene. 1998 17(18):2403–2411 |
| Brca1 conditional ko | 15 | Oncogene. 2002 21(33):5097–5107 |
| Her2/neu (endog. promoter) | 12 | Oncogene. 2002 21(6):890–898 |
| PyV-mT | 26 | Cancer Res. 2003 63(9):2179–2187 |
Amplification of the her2/neu oncogene occurs in some 25% of human breast cancers [14]. The expression of an activated form of her2/neu under its endogenous promotor results in the emergence of mammary gland adenocarcinomas in the mouse [15]. In a follow up study to the MMTV-myc transgenics, we applied CGH and SKY to 12 cases of murine tumors induced by expression of a mutated her/2neu gene under it’s endogenous promotor. We were particularly interested in comparing the patterns of genomic imbalances between these two models. Similar to the myc-induced tumors, frequent deletions that map to mouse chromosome 4 were observed. These deletions could comprise the entire chromosome, however, the minimally deleted region maps to the distal portion of this chromosome. CGH analysis also revealed common amplification of distal mouse chromosome 11. SKY clearly indicated that this amplification was in most instances due to double minute chromosomes containing numerous copies of the her2/neu gene. One possible interpretation of the frequent amplification of her2/neu would be that activation by mutation is not sufficient for tumorigenesis, but that increased copy numbers of this gene acquired via genomic amplification has to occur as well. The concurrent analysis of these tumors by both CGH and SKY suggests that the other observed genomic imbalances are mainly due to unbalanced chromosomal translocations and copy number imbalances of entire chromosomes (chromosomal aneuploidies). High-resolution deletion mapping with genomic clones that contain p53 and BRCA1 indicated that inactivation of these tumor suppressor genes is not required in this model system.
Tumor initiation in the mouse can be induced by mechanisms other than tissue specific overexpression of oncogenes. For instance, several models have been established in which the mammary gland specific expression of viral genes, e.g., polyoma virus middle T antigen (PyV-mT) or SV40 large T antigen (SV40Tag), resulted in tumor formation. Both transgenes compromise the function of the tumor suppressor genes p53 and Rb solely in the tissue of interest (as opposed to p53 or Rb knockouts) [16, 17]. In addition, the conditional KO of BRCA1 using CreLox technology results in blunted ductal morphogenesis and tumor formation [18]. We therefore extended our analyses to these model systems asking specifically whether tumorigenesis via oncogene overexpression compared to tumor suppressor gene inactivation resulted in different patterns of genomic imbalances. We first analyzed 15 tumors from BRCA1 conditional KO mice [19]. As in her2/neu and myc induced carcinomas, we observed a non-random distribution of genomic imbalances that revealed a predilection for certain chromosomes or chromosomal regions [19]. For instance, copy number increases on chromosome 11 were again identified in nine of 15 tumors and either partial or entire mouse chromosome 15 gains were observed in eight cases. In summary, regions of genomic copy number decrease were consistently found on the proximal part of mouse chromosome 11 and on distal chromosome 14.
The commonly amplified region on chromosome 11 was close to the map location of the her2/neu oncogene. Despite the consistent amplification of this region of chromosome 11, only a fraction of the cases actually displayed amplification of her2/neu by FISH and immunohistochemical and Western blot analysis revealed less than 25% of tumors showing overexpression of the oncogene. In other cases, the amplicon comprised a region distal to the her2/neu locus, thereby pointing to the involvement of another potential oncogene(s). This finding corresponds to human breast cancer, because amplification of the orthologous region in the human genome (i.e., distal chromosome 17q) has been shown to be independent of amplification of her2/neu [13]. Our finding is also in concordance with results that expression of her2/neu is less frequently found in BRCA1 associated human cancer compared to sporadic carcinomas [14, 20].
p53 plays a crucial role in BRCA1 induced response to DNA damage [21]. In the tumors studied here, we could show that the chromosomal mapping position of p53 on mouse chromosome 11 is frequently involved in translocations and deletions. High-resolution FISH mapping with a p53-specific genomic clone and Western blot analysis showed that p53 is indeed frequently altered in the BRCA1-associated mammary gland adenocarcinomas. However, it also became evident that normal levels of the protein were identified in some tumors, indicating that p53 inactivation might promote, but is not required for BRCA1-associated tumorigenesis. In addition, amplification of myc has been observed in BRCA1 tumors [22], a result that was supported by the common gain of mouse chromosome 15 in eight of our tumors. In conclusion, the similarity of BRCA1-associated mammary gland adenocarcinomas and BRCA1-associated human cancers is supported by a similar distribution of genomic imbalances (Figs 2 and 3). The involvement of such specific chromosomal aneuploidies is therefore required for tumorigenesis across species boundaries, at least in this particular model. Another notable difference between the BRCA1-associated mouse tumors and the her2/neu and myc induced carcinomas was the degree of genomic instability as established by the average number of chromosome copy alterations (ANCA). The ANCA index is derived by dividing the total number of genomic imbalances by the number of cases studied [10]. BRCA1-associated tumors revealed an ANCA value more than twice as high as the oncogene-driven carcinomas (Table 2). One could speculate that the relatively few genomic imbalances detected in the her2/neu and myc induced carcinomas are a reflection of the fact that a strong oncogenic stimulus overrides the requirement for acquiring multiple aneuploidies, such as those commonly observed in human carcinomas.
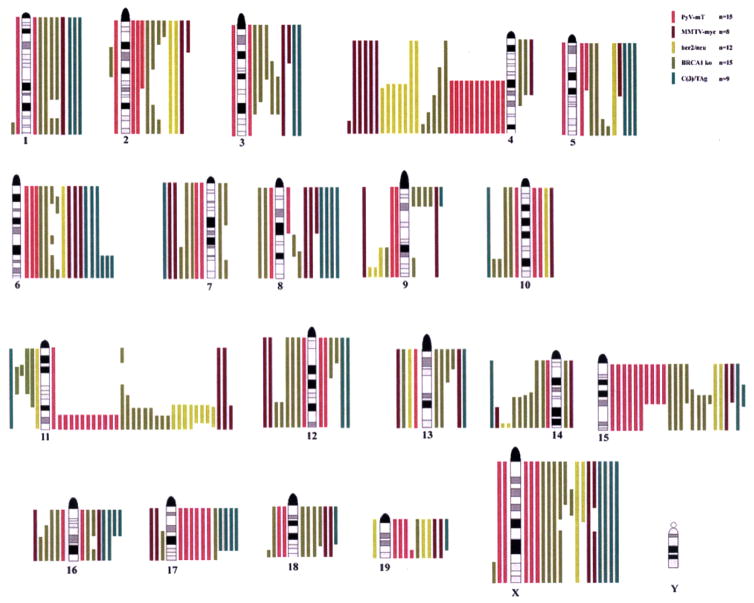
Fig. 2.
Summary of CGH analyses of five different mouse models of breast cancer. A non-random distribution of genomic imbalances, e.g. recurrent losses on chromosome 4 and gains of distal chromosome 11 and chromosome 15 is evident. Note that the number of cases per models is different.
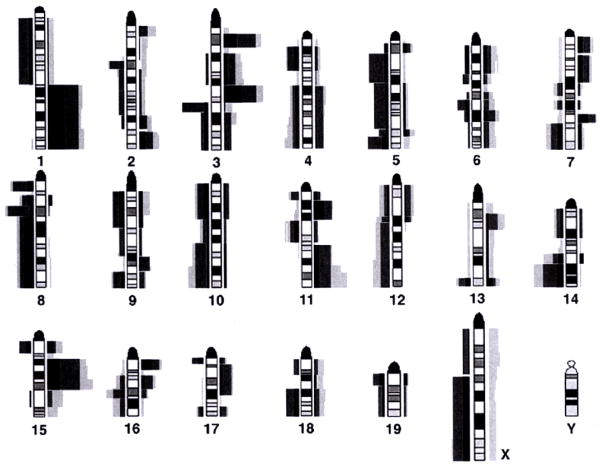
Fig. 3.
Projection of CGH results of human sporadic and BRCA1-associated breast cancers on mouse chromosomes. The dark gray indicated the relative involvement of chromosomal regions in gains and losses in sporadic human breast carcinomas, black the same data for BRCA1-associated tumors. The light gray bars reflect the relative involvement of mouse chromosomal regions in the BRCA1-associated murine tumors. With the exception of mouse chromosomes 4 and 18, similar genomic loci are involved in human and mouse tumors.
Table 2.
Comparison of the ANCA-values of different mouse models of breast cancer. The ANCA-value is derived by dividing the total number of copy alterations by the case number. Note that only the BRCA1-associated mouse tumors reach ANCA values comparable to human breast cancers
| HUMAN CANCERS | ANCA |
|---|---|
| Colon | 7.6 |
| Cervix uteri | 8.2 |
| Colon, liver metastases | 11.7 |
| Brain | 10.5 |
| Breast | 10.6 |
| Lung | 13.0 |
|
| |
| MOUSE MODELS | ANCA |
|
| |
| ErbB2 (e.p.) | 2.7 |
| MMTV-PyMT | 4 |
| C3(1)SV40TAg | 5.5 |
| MMTV-myc | 5.7 |
| BRCA1ko | 14.2 |
If this holds true, one would also expect that tumors induced by compromising the function of the tumor suppressor genes p53 and Rb via SV40 or PyV-mT would be more similar to the BRCA1 associated tumors, at least with respect to the ANCA value. We therefore analyzed by SKY and CGH 26 PyV-mT induced tumors and eight SV40Tag induced tumors [23, 24] and Difilippantonio, personal communication. The results showed that, contrary to the initial prediction, both models have an ANCA value that is more similar to the one in carcinomas induced by oncogene overexpression. However, this conundrum can potentially be explained: in both models induced by mammary gland specific expression of viral genes, the cytogenetic profiles suggested strong and localized genomic amplifications. In the SV40Tag model, these amplifications mapped to distal mouse chromosome 6, and were shown to result in genomic amplification and overexpression of the k-ras oncogene, which was commonly present in the form of double minute chromosome [24]. This was possibly attributable to the integration of the SV40 large T-Antigen transgene in proximity to k-ras.
In the PyV-mT induced tumors, we observed recurrent amplification of distal chromosome 11, caused again by multiple double minute chromosomes. High-resolution mapping in this chromosomal region led to the identification of a single BAC clone that labeled the double minute chromosomes (Fig. 1b). This BAC clone contained sequences for the septin 9 gene. Quantitative RT-PCR established highly increased expression levels of this gene (see Fig. 1d for presentation of results), a finding that was confirmed not only in other mouse models but also in cell lines derived from human breast cancer that showed genomic amplification of 17q23 (orthologous to mouse 11) [23]. The family of septin genes is involved in a plethora of cellular processes, including cytokinesis in yeast and vesicle transport, and possesses GTPase activity [25]. Septin 9 is also activated by way of chromosomal translocations in acute myeloid leukemia [26, 27]. We therefore propose that the SV40 induced genomic amplification and overexpression of k-ras and early amplification and overexpression of septin 9 in these models generates enormous amounts of transforming genes, and hence a genetic environment that is similar to the her2/neu and myc-induced cancers. As in these models, the acquisition of multiple genomic imbalances is less important than in the BRCA1-associated mouse tumors. Another contributing factor to the elevated degree of genomic instability in the BRCA1-associated mammary tumors could relate to the role of BRCA1 in DNA damage repair. BRCA1-associated human breast cancers also display increased numbers of genomic imbalances when compared to sporadic breast cancers [28]. Figure 2 summarizes the CGH profiles of the mouse tumors discussed here. The projection of genomic imabalances in sporadic and BRCA1-associated human breast cancers on the mouse genome, and the results of CGH analysis of mouse BRCA1-associated mammary gland adenocarcinomas are shown in Fig. 3.
COMPARISON TO HUMAN CANCERS
Human carcinomas are defined by a non-random, tumor specific distribution of chromosomal aneuploidies and resulting genomic imbalances. In breast cancers, for instance, copy number increases of chromosome arms 1q, 8q, 17q and 20q, accompanied by losses of chromosome arms 8p, 13q and 17p are common [13, 29, 30]. These aneuploidies are acquired sequentially during tumorigenesis and can emerge in an otherwise stable, diploid genome [31]. The first such aneuploidies can be detected in premalignant dysplastic lesions. Crude aneuploidy, as evidenced by heterogeneity of the cellular DNA content, occurs at later stages of tumorigenesis, and so do high-level amplification of genomic loci that contain oncogenes [10]. The acquisition of these early genomic imbalances can be associated, and is probably facilitated, by the impairment of surveillance mechanisms of genomic integrity, for instance by the infection of cervical epithelial cells with high-risk papilloma viruses (HPV), which are necessary, but not sufficient requirements for tumorigenesis. In the absence of oncogene amplification at early steps of cellular transformation and tumorigenesis, emerging cancer cells therefore need to acquire multiple genetic insults, that are reflected in increasing numbers of chromosomal aneuploidies and gene mutations. An a priori strong oncogenic stimulus, such as the overexpression of her2/neu and myc under tissue specific promoters, or the SV40 induced genomic amplification and overexpression of k-ras that are used to induce mammary gland tumorigenesis in transgenic mice, is usually not observed. This in turn, could explain the scarcity of genomic imbalances in these models. These observations are corroborated by recently published evidence that strong oncogene expression can cause cellular transformation [32]. Suppression of oncogene expression in these models systems then results in reversion of the tumorigenic phenotype, which persists unless additional secondary karyotypic changes are acquired [33].
The strict conservation of the pattern of genomic imbalances in human cancers could also suggest that tumorigenesis in the respective tissue context in the mouse would require the acquisition of corresponding genomic imbalances. The projection of imbalances observed in human breast cancers onto the shuffled mouse chromosome would therefore reveal a similar distribution. For example, both sporadic breast cancers and those associated with BRCA1 mutations, very commonly carry extra copies of the long arms of chromosomes 1 and 8 [34]. See Fig. 3 for details. It is a yet unresolved fundamental scientific question whether breast carcinomas (or other tumors) require the low level transcriptional gain of all or most genes on these chromosomes, or whether the acquisition of these tissue specific chromosomal aneuploidies merely serves as the vehicle for the low copy number increase of one or a few tumor promoting genes, with the remainder of the genes residing on these chromosome merely coming along for the ride. If the first hypothesis applies, than one would expect that all or most of the orthologous regions of chromosomes gained in human cancers, would be subject to copy alterations in the mouse models as well. Alternatively, only a few corresponding regions (i.e., those containing the relevant oncogenes) would be affected. Based on the analyses discussed here, we cannot endorse of refute one or the other hypothesis with certainty. However, using human chromosome 8 as an example, it seems clear that the region that contains the c-myc oncogene is the most important target for gain of this chromosome. The murine orthologous region (bands 15D) is the most commonoly gained in mouse mammary gland adenocarcinomas. Other regions on human chromosome 8 appear to be less important.
Our results suggest that the distribution of genomic imbalances in human breast cancer and the mouse models analyzed here is most similar in the mammary gland adenocarcinomas induced by conditional inactivation of BRCA1. The models induced by oncogene overexpression reveal fewer of the human breast cancer defining chromosomal aneuploidies. It will be revealing in this respect to comprehensively study the respective consequences of chromosomal aneuploidies on global gene expression patterns in both human cancer and mouse models. Gene expression profiling allows discrimination of some murine models of breast cancer [35]. Future studies will have to integrate both maps of genomic imbalances and global gene expression profiles to help understand to which extent these differences are attributable to the different patterns of chromosomal aberrations in the respective mouse models. In addition to the comprehensive molecular cytogenetic analysis of mouse models of breast cancer, we have also extensively mapped cytogenetic aberrations in mouse models of hematological malignancies. The comparative mapping of these aberrations is more straightforward. For instance, elimination of genes involved in the repair of DNA damage, such as Ku80 and ATM result in lymphomas that faithfully recapitulate the genetic events in human disease: dysregulation of c-myc via translocation to the IgH locus [36] or T-cell receptor rearrangements [37] occur in both human lymphomas and mouse models thereof [1]. This might be a reflection of the relative “simplicity” of karyotypic changes in some hematological malignancies, in which recurrent chromosomal translocations are sufficient to trigger malignant transformation.
In summary, the comprehensive molecular cytogenetic analysis of mouse models of breast cancers has shown that tumorigenesis requires, as in human cancers, the acquisition of distinct genomic imbalances. Some of these imbalances are common to all analyzed mouse models, such as frequent copy number losses on chromosome 4. However, distinct differences exist as well. The projection of the maps of genomic imbalances identified in human breast carcinomas onto the mouse genome shows that a considerable proportion of genomic imbalances occur in both species, however, the comparative maps do not result in “mirror” images of genomic imbalances. That being said, the BRCA1-associated mouse tumors reveal a distribution of genomic imbalances and a level of genomic instability that is most similar to human breast cancers.
References
- 1.Heim S, Mitelman F. Cancer Cytogenetics. Wiloy-Liss; 1995. [Google Scholar]
- 2.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 3.Donehower LA, Godley LA, Aldaz CM, Pyle R, Shi YP, Pinkel D, Gray J, Bradley A, Medina D, Varmus HE. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 4.Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 5.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 6.Liyanage M, Coleman A, du Manoir S, Veldman T, McCormack S, Dickson RB, Barlow C, Wynshaw-Boris A, Janz S, Wienberg J, Ferguson-Smith MA, Schrock E, Ried T. Multicolour spectral karyotyping of mouse chromosomes. Nat Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- 7.Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 8.Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius VM, Vidgren V, Zhu Y. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- 9.Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius VM, Wolf M, Zhu Y. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Weaver ZA, McCormack SJ, Liyanage M, du Manoir S. A mors of MMTV-cmyc transgenic mice. Genes Chromosomes Cancer. 1999;25:251–260. [PubMed] [Google Scholar]
- 12.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 13.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 14.Johannsson OT, Idvall I, Anderson C, Borg A, Barkardottir RB, Egilsson V, Olsson H. Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur J Cancer. 1997;33:362–371. doi: 10.1016/s0959-8049(97)89007-7. [DOI] [PubMed] [Google Scholar]
- 15.Andrechek ER, Hardy WR, Siegel PM, Rudnicki MA, Cardiff RD, Muller WJ. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci USA. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci USA. 1994;91:11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 19.Weaver Z, Montagna C, Xu X, Howard T, Gadina M, Brodie SG, Deng CX, Ried T. Mammary tumors in mice conditionally mutant for Brca1 exhibit gross genomic instability and centrosome amplification yet display a recurring distribution of genomic imbalances that is similar to human breast cancer. Oncogene. 2002;21:5097–5107. doi: 10.1038/sj.onc.1205636. [DOI] [PubMed] [Google Scholar]
- 20.Robson M, Rajan P, Rosen PP, Gilewski T, Hirschaut Y, Pressman P, Haas B, Norton L, Offit K. BRCA-associated breast cancer: absence of a characteristic immunophenotype. Cancer Res. 1998;58:1839–1842. [PubMed] [Google Scholar]
- 21.Deng CX, Brodie SG. Roles of BRCA1 and its interacting proteins. Bioessays. 2000;22:728–737. doi: 10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 22.Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene. 2001;20:7514–7523. doi: 10.1038/sj.onc.1204929. [DOI] [PubMed] [Google Scholar]
- 23.Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z, Ried T. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63:2179–2187. [PubMed] [Google Scholar]
- 24.Liu ML, Von Lintig FC, Liyanage M, Shibata MA, Jorcyk CL, Ried T, Boss GR, Green JE. Amplification of Ki-ras and elevation of MAP kinase activity during mammary tumor progression in C3(1)/SV40 Tag transgenic mice. Oncogene. 1998;17:2403–2411. doi: 10.1038/sj.onc.1202456. [DOI] [PubMed] [Google Scholar]
- 25.Field CM, al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalikin LM, Sims HL, Petty EM. Genomic and expression analyses of alternatively spliced transcripts of the MLL loss in breast and ovarian tumors. Genomics. 2000;63:165–172. doi: 10.1006/geno.1999.6077. [DOI] [PubMed] [Google Scholar]
- 27.Osaka M, Rowley JD, Zeleznik-Le NJ. MSF (MLL septin-likc fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with at(11:17)(q23:q25) Proc Natl Acad Sci USA. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tirkkonen M, Johannsson O, Agnarsson BA, Olsson H, Ingvarsson S, Karhu R, Tanner M, Isola J, Barkardottir RB, Borg A, Kallioniemi OP. Distinct somatic genetic changes associated with tumor progression in carriers of BRCA1 and BRCA2 germ-line mutations. Cancer Res. 1997;57:1222–1227. [PubMed] [Google Scholar]
- 29.Ried T, Just KE, Holtgreve-Grez H, du Manoir S, Speicher MR, Schrock E, Latham C, Blegen H, Zetterberg A, Cremer T, Auer G. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55:5415–5423. [PubMed] [Google Scholar]
- 30.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heselmeyer-Haddad K, Janz V, Castle PE, Chaudhri N, White N, Wilber K, Morrison LE, Auer G, Burroughs FH, Sherman MF, Ried T. Detection of genomic amplification of the human telomerase gene (TERC) in cytologic specimens as a genetic test for the diagnosis of cervical dysplasia. Am J Pathol. 2003;163:1405–1416. doi: 10.1016/S0002-9440(10)63498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 33.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, Horner JW, 2nd, Cordon-Cardo C, Yancopoulos GD, DePinho RA. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 35.Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R, Simon R, Liu ET, Green JE. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci USA. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liyanage M, Weaver Z, Barlow C, Coleman A, Pankratz DG, Anderson S, Wynshaw-Boris A, Ried T. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]