Abstract
Interval censored outcomes arise when a silent event of interest is known to have occurred within a specific time period determined by the times of the last negative and first positive diagnostic tests. There is a rich literature on parametric and non-parametric approaches for the analysis of interval-censored outcomes. A commonly used strategy is to use a proportional hazards (PH) model with the baseline hazard function parameterized. The proportional hazards assumption can be relaxed in stratified models by allowing the baseline hazard function to vary across strata defined by a subset of explanatory variables. In this paper, we describe and implement a new R package straweib, for fitting a stratified Weibull model appropriate for interval censored outcomes. We illustrate the R package straweib by analyzing data from a longitudinal oral health study on the timing of the emergence of permanent teeth in 4430 children.
Introduction
In many clinical studies, the time to a silent event is known only up to an interval defined by the times of the last negative and first positive diagnostic test. Event times arising from such studies are referred to as ’interval-censored’ data. For example, in pediatric HIV clinical studies, the timing of HIV infection is known only up to the interval from the last negative to the first positive HIV diagnostic test (Dunn et al., 2000). Examples of interval-censored outcomes can also be found in many other medical studies (Gomez et al., 2009).
A rich literature exists on the analysis of interval-censored outcomes. Non-parametric approaches include the self-consistency algorithm for the estimation of the survival function (Turnbull, 1976). A semi-parametric approach based on the proportional hazards model has been developed for interval-censored data (Finkelstein, 1986; Goetghebeur and Ryan, 2000). A variety of parametric models can also be used to estimate the distribution of the time to the event of interest, in the presence of interval-censoring (Lindsey and Ryan, 1998). An often used parametric approach for the analysis of interval-censored data is based on the assumption of a Weibull distribution for the event times (Lindsey and Ryan, 1998). The Weibull distribution is appropriate for modeling event times when the hazard function can be reliably assumed to be monotone. Covariate effects can be modeled through the assumption of proportional hazards (PH), which assumes that the ratio of hazard functions when comparing individuals in different strata defined by explanatory variables is time-invariant. The article by Gomez et al. (2009) presents a comprehensive review of the state-of-the-art techniques available for the analysis of interval-censored data.
In this paper, we implement a parametric approach for modeling covariates applicable to interval-censored outcomes, but where the assumption of proportional hazards may be questionable for a certain subset of explanatory variables. For this setting, we implement a stratified Weibull model by relaxing the PH assumption across levels of a subset of explanatory variables. We compare the proposed model to an alternative stratified Weibull regression model that is currently implemented in the R package survival (Therneau, 2012). We illustrate the difference between these two models analytically and through simulation.
The paper is organized as follows: In Section 2, we present and compare two models for relaxing the PH assumption, based on the assumption of a Weibull distribution for the time to event of interest. In this section, we discuss estimation of the unknown parameters of interest, hazard ratios comparing different groups of subjects based on specific values of explanatory covariates and tests of the PH assumption. These methods are implemented in a new R package, straweib (Gu and Balasubramanian, 2013). In Section 3, we perform simulation studies to compare two stratified Weibull models implemented in R packages straweib and survival. In Section 4, we illustrate the use of the R package straweib by analyzing data from a longitudinal oral health study on the timing of the emergence of permanent teeth in 4430 children in Belgium (Leroy et al., 2003; Gomez et al., 2009). In Section 5, we discuss the models implemented in this paper and present concluding remarks.
Weibull regression models
Let T denote the continuous, non-negative random variable corresponding to the time to event of interest, with corresponding probability distribution function (pdf) and cumulative distribution function (cdf), denoted by f(t) and F(t), respectively. We let S(t) = 1 − F(t) to denote the corresponding survival function and to denote the hazard function. We let Z denote the p × 1 vector of explanatory variables or covariates.
We assume that the random variable T | Z = 0 is distributed according to a Weibull distribution, with scale and shape parameters denoted by λ and γ, respectively. The well known PH model to accommodate the effect of covariates on T is expressed as:
where β denotes the p × 1 vector of regression coefficients corresponding to the vector of explanatory variables, Z.
Thus, under the Weibull PH model, the survival and hazard functions corresponding to T can be expressed as
| (1) |
| (2) |
where, λ > 0 and γ > 0 correspond to the scale and shape parameters corresponding to T when Z = 0. The hazard ratio comparing two individuals with covariate vectors Z and Z* is equal to exp(β′(Z − Z*)).
Stratified Weibull regression model implemented in the R package survival
In this section, we describe the stratified Weibull PH regression model implemented in the the R package survival (Therneau, 2012).
Consider the following log-linear model for the random variable T:
where, α1, ⋯, αp denote unknown regression coefficients corresponding to the p dimensional vector of explanatory variables, μ denotes the intercept, and σ denotes the scale parameter. The random variable ε captures the random deviation of event times on the natural logarithm scale (i. e. log(T)) from the linear model as a function of the covariate vector Z. In general, the log-linear form of the model for T can be shown to be equivalent to the accelerated failure time (AFT) model (Collett, 2003).
The assumption of a standard Gumbel distribution with location and scale parameters equal to 0 and 1, respectively, implies that the random variable T follows a Weibull distribution. Moreover, in this case, both the PH and AFT assumptions (or equivalently, the log-linear model) lead to identical models with different parameterizations (Collett, 2003). The survival and hazard functions can be expressed as:
| (3) |
| (4) |
The coefficients for the explanatory variables (β) in the hazard function (h(t | Z)) are equal to . Moreover, there is a one-one correspondence between the parameters λ, γ, β in equations (1)–(2) and the parameters μ, σ, α in equations (3)–(4), where , γ = σ−1 and (Collett, 2003).
The log-linear form of the Weibull model can be generalized to allow arbitrary baseline hazard functions within subgroups defined by a stratum indicator S = 1, ⋯, s. Thus, the stratified Weibull regression model for an individual in the jth stratum is expressed as:
where μj and σj denote stratum specific intercept and scale parameters. This model is implemented in the R package survival (Therneau, 2012). In this model, the regression coefficients α on the AFT scale are assumed to be stratum independent.
However, the hazard ratio comparing two individuals with covariate vectors and stratum indicators denoted by (Z, S = j) and (Z*, S = k) is stratum specific and is given by:
For j ≠ k, the hazard ratio varies with time t. However, when j = k, the hazard ratio comparing two individuals within the same stratum S = j is invariant with respect to time t but is stratum-dependent and reduces to:
| (5) |
Stratified Weibull regression model implemented in R package straweib
In this section, we describe the stratified Weibull regression model that is implemented in the new R package, straweib (Gu and Balasubramanian, 2013).
To relax the proportional hazards assumption in the Weibull regression model, we propose the following model for an individual in the stratum S = j:
| (6) |
Equivalently, the model can be stated in terms of the survival function as:
Here, we assume that the scale and shape parameters (λ, γ) are stratum specific - however, the regression coefficients β are assumed to be constant across strata (S). The hazard ratio comparing two individuals with covariate vectors and stratum indicators denoted by (Z, S = j) and (Z*, S = k) is given by:
For j ≠ k, the hazard ratio varies with time t and thus relaxes the PH assumption. However, for j = k, the hazard ratio comparing two individuals within the same stratum S = j reduces to:
| (7) |
This hazard ratio is invariant with respect to time t and stratum S, as in the stratified Cox model (Collett, 2003).
Estimation
Let uj = log(λj) and υj = log(γj). Let nj denote the number of subjects in stratum S = j. For the kth subject in stratum j, let Zjk denote the p dimensional vector of covariates and let ajk and bjk denote the left and right endpoints of the censoring interval. That is, ajk denotes the time of the last negative test and bjk denotes the time of the first positive test for the event of interest. Then the log-likelihood function can be expressed as:
The unknown parameters to be estimated are υ, u, and β. The log-likelihood function can be optimized using the optim function in R. The shape and scale parameters can be estimated from the estimates of υ and u. The covariance matrix of the estimates of these unknown parameters can be obtained by inverting the negative Hessian matrix that is output from the optimization routine (Cox and Hinkley, 1979).
Test of the PH assumption
One can test whether or not the baseline hazard functions of each strata are proportional to each other, by testing the equality of shape parameters across strata S = 1, ⋯, s. That is,
or equivalently,
The null hypothesis H0 can be tested using a likelihood ratio test, by comparing a reduced model that assumes that γ1 = γ2 == ⋯ = γs to the full model in (6) assuming stratum specific shape parameters. We note that the reduced model is equivalent to the Weibull PH model that includes the stratum indicator S as an explanatory variable. Thus the reduced model has s − 1 fewer parameters than the stratified model, or the full model. Let lF and lR denote the log-likelihoods of the full and reduced models evaluated at their MLE. Then the test statistic T = −2(lR − lF) follows a distribution under H0. In addition to the likelihood ratio test, one can also use a Wald test to test the null hypothesis H0. The R package straweib illustrated in Section 3 outputs both the Wald and Likelihood Ratio test statistics.
Estimating hazard ratios
The log hazard ratio comparing two individuals with covariate vectors and stratum indicators denoted by (Z, S = j) and (Z*, S = j*) at time t can be expressed as:
Let υ̂, û and β̂ denote the maximum likelihood estimates for υ, u and β, then rtjj* can be estimated by
Let w = (υ, u, β) = (υ1, υ2, ⋯, υs, u1, u2, ⋯, us, β1, ⋯, βp). Let Σ̂ denote the estimate of the covariance matrix of ŵ. Let Jtjj* denote the Jacobian vector, . Thus, the estimate of the variance of r̂tjj* is obtained by:
We obtain a 95% confidence interval for rtjj* as . We exponentiate r̂tjj* and its corresponding 95% confidence interval to obtain the estimate and the 95% confidence interval for the hazard ratio, Rtjj*. We illustrate the use of the straweib R package for obtaining hazard ratios and corresponding confidence intervals in Section 4.
Comparison of models implemented in packages survival and straweib
In this section, we compare the stratified Weibull regression model implemented in the survival package to that implemented in our package, straweib.
In the absence of stratification, both models are identical and reduce to the Weibull PH model. However, in the presence of a stratification factor, the models implemented by survival and straweib correspond to different models, resulting in different likelihood functions and inference. As we discussed in Section 2, the hazard ratio between two subjects with different covariate values within same stratum depends on their stratum in the model implemented in the R package survival (Equation (5)), whereas the hazard ratio comparing two individuals within the same stratum is invariant to stratum in the model implemented in the R package straweib (Equation (7)). In particular, the Weibull model implemented in the straweib shares similarities with the semi-parametric, stratified Cox model for right censored data.
To illustrate the difference between the models implemented in the R packages survival and straweib, we conducted a simulation study in which 1000 datasets were simulated under the model assumed in the straweib package (Equation (6)). For each simulated dataset, since both models have the same number of unknown parameters, we compare the values of the log-likelihood evaluated at the MLEs. Datasets were simulated based on the assumptions that there are 3 strata, each with a 100 subjects; the shape parameters (γ) in the three strata were set to 1.5, 2, and 1, respectively; the baseline scale parameters in the three strata (λ) were set to 0.01, 0.015, and 0.02, respectively. We assumed that there are two independent explanatory variables available for each subject, randomly drawn from N(0, 1) random variables. The coefficients corresponding to each of the two covariates were set to 0.5 and 1, respectively. To simulate interval censored outcomes, we first simulated the true event time for each subject by sampling from a Weibull distribution with the appropriate parameters. We assumed that each subject has 20 equally spaced diagnostic tests, at which the true event status is observed. Each test has a probability of 70% being missing. To obtain the maximum likelihood estimates under each model, we used the survreg function in the R package survival and the icweib function in the straweib package.
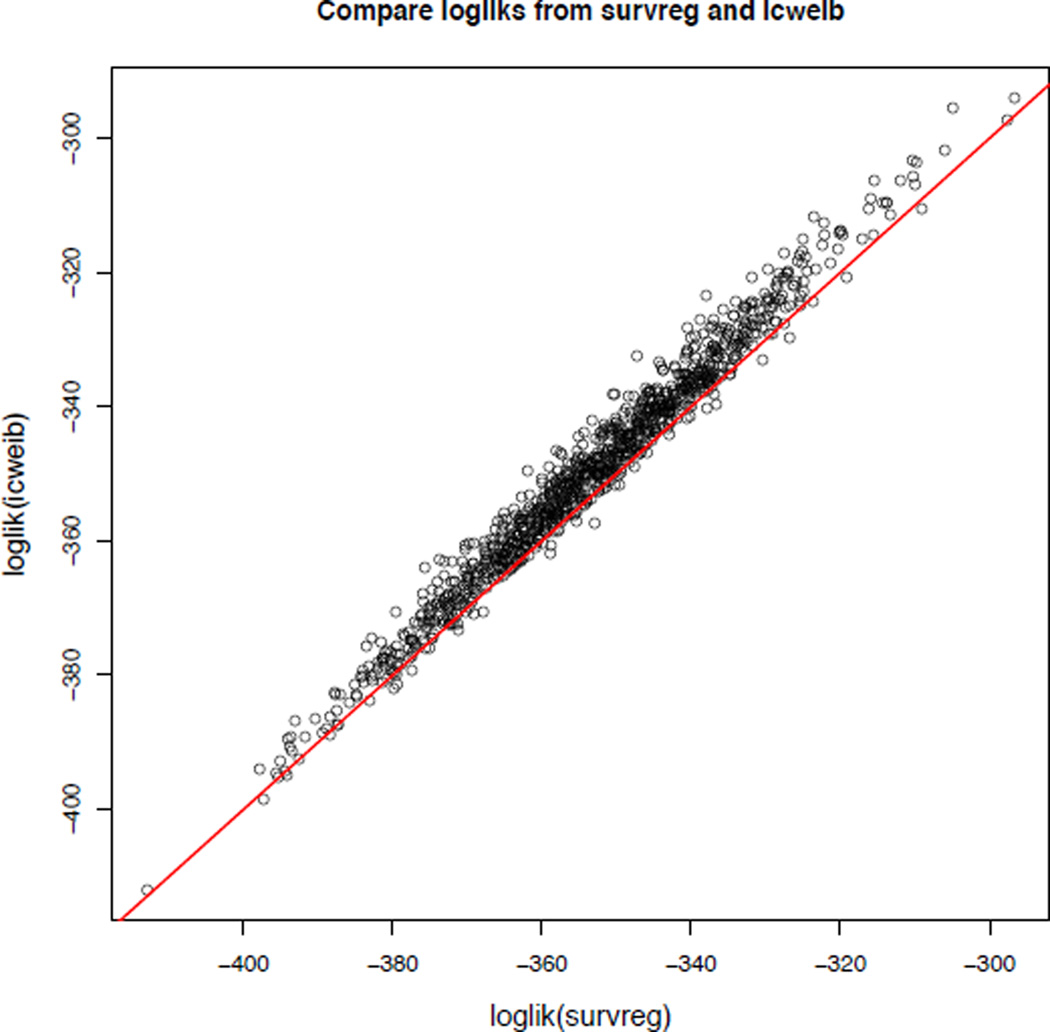
Figure 1 compares the maximized value of the log-likelihoods under both models, when the data are generated using a simulation mechanism that corresponds to the model implemented in the R package straweib. The maximized value of the log-likelihood from the R package survival is lower than that from the R package straweib for 93.1% of simulated datasets. This is expected as in this simulation study the data generating mechanism is identical to the model implemented in the R package straweib. In applications where the proportional hazards assumption is questionable, we recommend fitting both models and comparing the resulting maximized values of the log likelihood. Whether one model is better than another depends on the data.
Figure 1.
Comparing the maximized values of the log-likelihood obtained from the models implemented in the R package survival (X axis) to that from the R package straweib (Y axis), when the data is simulated under the model implemented in the R package straweib
Example
We illustrate the R package straweib with data from a study on the timing of emergence of permanent teeth in Flemish children in Belgium (Leroy et al., 2003). The data analyzed were from the Signal-Tandmobiel project (Vanobbergen et al., 2000), a longitudinal oral health study in a sample of 4430 children conducted between 1996 and 2001. Dental examinations were conducted annually for a period of 6 years and tooth emergence was recorded based on visual inspection. As in Gomez et al. (2009), we will illustrate our R package by analyzing the timing of emergence of the permanent upper left first premolars. As dental exams were conducted annually, for each child, the timing of tooth emergence is known up to the interval from the last negative to the first positive dental examination.
data(tooth24) head(tooth24) id left right sex dmf 1 1 2.7 3.5 1 1 2 2 2.4 3.4 0 1 3 3 4.5 5.5 1 0 4 4 5.9 Inf 1 0 5 5 4.1 5.0 1 1 6 6 3.7 4.5 0 1
The dataset is formatted to include 1 row per child. The variable denoted id corresponds to the ID of the child, left and right correspond to the left and right endpoints of the censoring interval in years, sex denotes the gender of the child (0 = boy, and 1 = girl), and dmf denotes the status of primary predecessor of the tooth (0 = sound, and 1 = decayed or missing due to caries or filled). Right censored observations are denoted by setting the variable right to "Inf".
In our analysis below, we use the function icweib in the package straweib, to fit a stratified Weibull regression model, where the variable dmf is the stratum indicator (S) and the variable sex is an explanatory variable (Z).
fit <- icweib(L = left, R = right, data = tooth24, strata = dmf, covariates = ~sex)
fit
Total observations used: 4386. Model Convergence: TRUE
Coefficients:
coefficient SE z p.value
sex 0.331 0.0387 8.55 0
Weibull parameters - gamma(shape), lambda(scale):
straname strata gamma lambda
dmf 0 5.99 1.63e-05
dmf 1 4.85 1.76e-04
Test of proportional hazards for strata (H0: all strata's shape parameters are equal):
test TestStat df p.value
Wald 44.2 1 2.96e-11
Likelihood Ratio 44.2 1 3.00e-11
Loglik(model)= −5501.781 Loglik(reduced)= −5523.87
Loglik(null)= −5538.309 Chisq= 73.05611 df= 1 p.value= 0
The likelihood ratio test of the PH assumption results in a p value of 3.00e-11, indicating that the PH model is not appropriate for this dataset. Or in other words, the data suggest that the hazard functions corresponding to the strata defined by dmf = 0 and dmf = 1 are not proportional. From the stratified Weibull regression model, the estimated regression coefficient for sex is 0.331, corresponding to a hazard ratio of 1.39 (95% CI: 1.29 – 1.50). In the output above, the maximized value of the log likelihood of the null model corresponds to the model stratified by covariate dmf but excluding the explanatory variable sex.
The p value from the Wald test of the null hypothesis of no effect of gender results in a p value of approximately 0 (p < 10−16), which indicates that the timing of emergence of teeth is significantly different between girls and boys.
To test the global null hypothesis that both covariates sex and dmf are not associated with the outcome (time to teeth emergence), we obtain the log-likelihood for global null model, as shown below.
fit0 <- icweib(L = left, R = right, data = tooth24) fit0 Total observations used: 4386. Model Convergence: TRUE Weibull parameters - gamma(shape), lambda(scale): straname strata gamma lambda strata ALL 5.3 7.78e-05 Loglik(model)= −5596.986 Loglik(null)= −5596.986
The likelihood ratio test testing the global null hypothesis results in a test statistic T = −2(lR − lF) = −2(−5596.986 + 5501.781) = 190.41, which follows a distribution under H0, resulting in a p value of approximately 0 (p < 10−16).
We illustrate the HRatio function in the straweib package to estimate the hazard ratio and corresponding 95% confidence intervals for comparing boys without tooth decay (dmf = 0) to boys with evidence of tooth decay (dmf = 1), where the hazard ratio is evaluated at various time points from 1 through 7 years.
HRatio(fit, times = 1:7, NumStra = 0, NumZ = 0, DemStra = 1, DemZ = 0) time NumStra DemStra beta*(Z1–Z2) HR low95 high95 1 1 0 1 0 0.1143698 0.06596383 0.1982972 2 2 0 1 0 0.2520248 0.18308361 0.3469262 3 3 0 1 0 0.4000946 0.33112219 0.4834339 4 4 0 1 0 0.5553610 0.49863912 0.6185351 5 5 0 1 0 0.7162080 0.66319999 0.7734529 6 6 0 1 0 0.8816470 0.79879884 0.9730878 7 7 0 1 0 1.0510048 0.91593721 1.2059899
The output indicates that the hazard ratio for boys comparing the stratum dmf = 0 to stratum dmf = 1 is small initially (e.g. 0.11 at 1 year) but tends to 1 in later years (e.g. 0.88 at 6 years and 1.05 at 7 years). Prior to 6 years, the hazard ratio is significantly less than 1, indicating that the timing of teeth emergence is delayed in children with tooth decay (dmf = 1) when compared to children without tooth decay (dmf = 0).
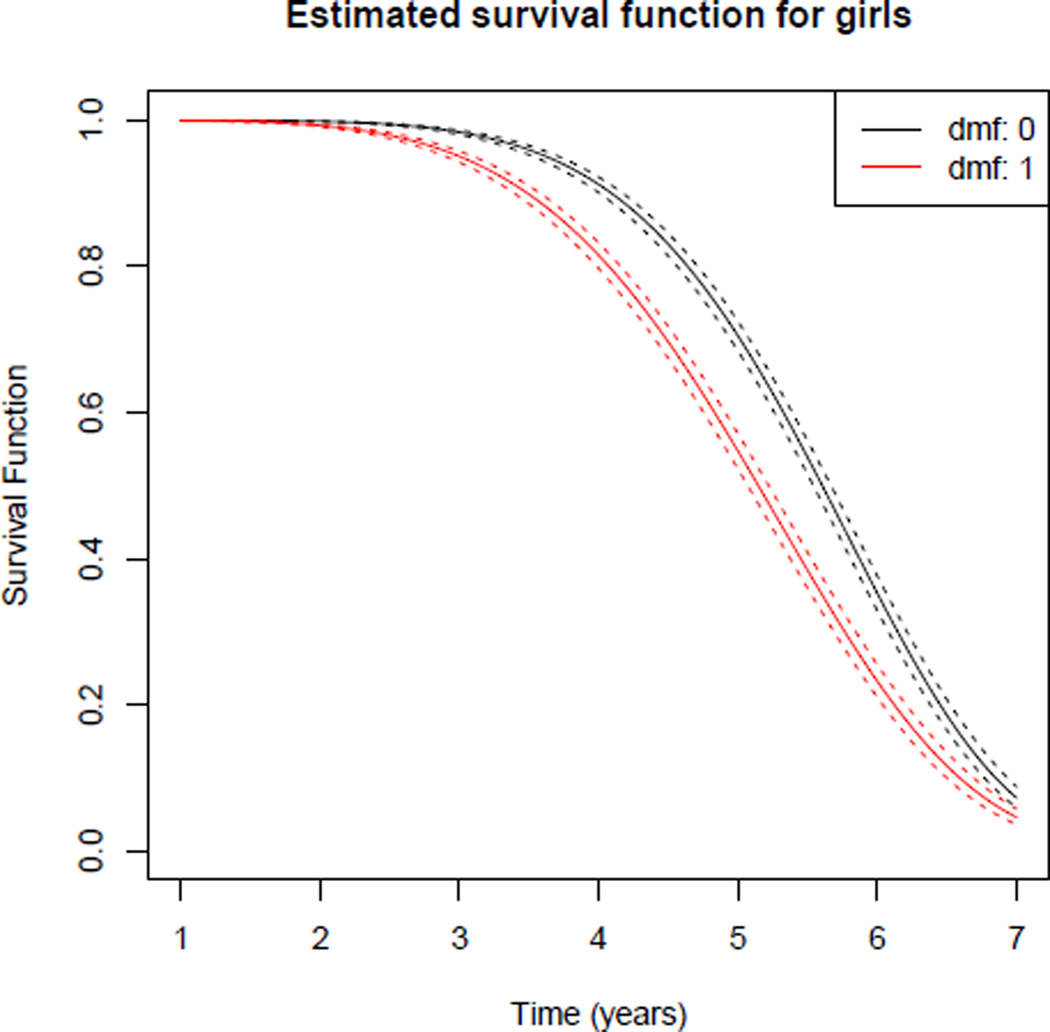
We illustrate estimation of the survival function in Figure 2 by plotting the survival functions and corresponding 95% point wise confidence intervals for girls (Z = 1), with and without tooth decay.
plot(fit, Z = 1, tRange = c(1, 7), xlab = ”Time (years)”, ylab = ”Survival Function”,
main = ”Estimated survival function for girls”)
Figure 2.
Estimated survival functions for girls, comparing the subgroup with sound primary predecessor of the tooth (dmf = 0) to the subgroup with unsound primary predecessor of the tooth (dmf = 1).
We compare our results from the straweib package to that obtained from the survival package.
library(survival)
tooth24.survreg <- tooth24
tooth24.survreg$right <- with(tooth24, ifelse(is.finite(right), right, NA))
fit1 <- survreg(Surv(left, right, type=”interval2”) ~ sex + strata(dmf) + factor(dmf),
data = tooth24.survreg)
fit1
Call:
survreg(formula = Surv(left, right, type = ”interval2”) ~ sex +
strata(dmf) + factor(dmf), data = tooth24.survreg)
Coefficients:
(Intercept) sex factor(dmf)1
1.84389938 −0.06254599 −0.06491729
Scale:
dmf=Sound1 dmf=Sound2
0.1659477 0.2072465
Loglik(model)= −5499.3 Loglik(intercept only)= −5576.2
Chisq= 153.8 on 2 degrees of freedom, p= 0
n= 4386
The maximized value of the log-likelihood from the R package survival is −5499.3 (shown below), as compared to the maximized value of the log-likelihood of −5501.8 from the R package straweib.
To clarify the specific assumptions made by the models implemented in the survival and straweib packages, we carried out subgroup analyses in which we fit a Weibull PH model separately to each of the strata dmf = 0 and dmf = 1. The results from the Weibull PH model fit to the subgroup of children in the dmf = 0 stratum is shown below:
fit20 <- icweib(L= left, R=right, data=tooth24[tooth24$dmf==0, ], covariates = ~sex)
fit20 ### Partial results shown below
Coefficients:
coefficient SE z p.value
sex 0.448 0.0543 8.25 2.22e-16
The results from the Weibull PH model fit to the subgroup dmf = 1 is shown below:
fit21 <- icweib(L= left, R=right, data=tooth24[tooth24$dmf==1, ], covariates = ~sex)
fit21 ### Partial results shown below
Coefficients:
coefficient SE z p.value
sex 0.208 0.0554 3.76 0.000169
The model using the PH scale (implemented by straweib package) replaces the stratum specific hazard ratios for sex of e0.448 = 1.57 for the subgroup dmf = 0 and e0.208 = 1.23 for the subgroup dmf = 1 with a common value, e0.331 = 1.39.
Since the Weibull distribution has both the PH and accelerated failure time (AFT) property (Collett, 2003), the identical set of subgroup analyses can be fit using the survival package. Results from the fit using the survival package for the subgroup dmf = 0 are shown below:
fit20.survreg <- survreg(Surv(left, right, type=”interval2”) ~ sex,
data = tooth24.survreg[tooth24.survreg$dmf==0, ])
fit20.survreg ### Partial results shown below
Coefficients:
(Intercept) sex
1.85029150 −0.07453785
Similar results using the survival package for the subgroup dmf = 1 are shown below:
fit21.survreg <- survreg(Surv(left, right, type=”interval2”) ~ sex,
data = tooth24.survreg[tooth24.survreg$dmf==1, ])
fit21.survreg ### Partial results shown below
Coefficients:
(Intercept) sex
1.76931556 −0.04303767
In particular, the model assuming a common sex coefficient in the AFT scale (implemented by survival package) replaces the value of sex coefficient −0.075 for the subgroup with dmf = 0 and sex coefficient of −0.043 for the subgroup dmf = 1 with a shared common value, −0.063.
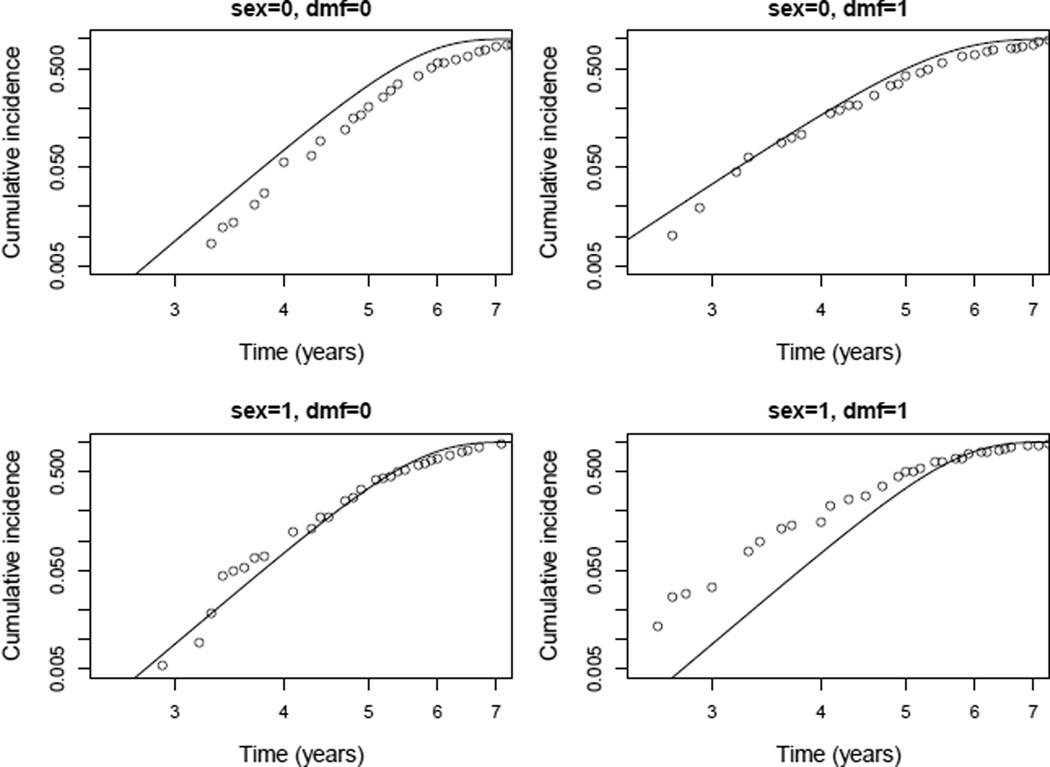
To assess the goodness of fit of the stratified Weibull model implemented by straweib, we created a multiple probability plot, as described in chapter 19 of Meeker and Escobar (1998). This diagnostic plot was created by splitting the dataset into 4 subgroups based on the values of sex and dmf. Within each group, we estimated the cumulative incidence at each visit time using a non-parametric procedure for interval censored data (Turnbull, 1976). The non-parametric estimates of cumulative incidence within each subgroup were compared to that obtained from the stratified Weibull model implemented by straweib package. We use the R package interval (Fay and Shaw, 2010) to obtain Turnbull’s NPMLE estimates and the R package straweib for the estimates from the stratified Weibull model (code available upon request). Figure 3 shows the diagnostic plot.
Figure 3.
Comparing non-parametric (points) and Weibull model (lines) based estimates of cumulative incidence within each group based on covariates sex and dmf.
Table 1 presents the estimates of hazard ratio for sex, within each of the strata defined by dmf = 0 and dmf = 1, comparing three different analyses - (1) Using the survival package to stratify on the variable dmf and including sex as an explanatory variable; (2) Using the straweib package to stratify on the variable dmf and including sex as an explanatory variable; (3) Fitting a Weibull PH model with sex as an explanatory variable, separately within each of the two subgroups defined by dmf = 0 and dmf = 1.
HR.straweib <- exp(fit$coef[1, 1]) HR.survreg <- exp(−fit1$coefficients['sex']/fit1$scale) HR.subgroup <- exp(c(fit20$coef[1, 1], fit21$coef[1, 1]))
Table 1.
Hazard ratio estimates for gender, comparing the models implemented in the R packages survival, straweib and subgroup analyses
| stratum | Package survival | Package straweib | Stratum specific subgroup analyses |
|---|---|---|---|
| dmf = 0 | 1.46 | 1.39 | 1.56 |
| dmf = 1 | 1.35 | 1.39 | 1.23 |
Concluding remarks
We have developed and illustrated an R package straweib for the analysis of interval-censored outcomes, based on a stratified Weibull regression model. The proposed model shares similarities with the semi-parametric stratified Cox model. We illustrated the R package straweib using data from a prospective study on the timing of emergence of permanent teeth in Flemish children in Belgium (Leroy et al., 2003).
Although the models and R package are illustrated for the analysis of interval-censored time-to-event outcomes, the methods proposed here are equally applicable for the analysis of right-censored outcomes. The syntax for the analysis of right-censored observations is explained in the manual accompanying the straweib package available on CRAN (Gu and Balasubramanian, 2013).
Acknowledgments
This research was supported by NICHD grant R21 HD072792.
Contributor Information
Xiangdong Gu, Division of Biostatistics and Epidemiology, University of Massachusetts, Amherst, MA, USA, xdgu@schoolph.umass.edu.
David Shapiro, Department of Biostatistics, Harvard School of Public Health, Boston, MA, USA, shapiro@sdac.harvard.edu.
Michael D. Hughes, Department of Biostatistics, Harvard School of Public Health, Boston, MA, USA, mhughes@hsph.harvard.edu
Raji Balasubramanian, Division of Biostatistics and Epidemiology. University of Massachusetts, Amherst, MA, USA, rbalasub@schoolph.umass.edu.
Bibliography
- Collett D. Modelling Survival Data in Medical Research. Second. Taylor & Francis: Texts in statistical science; 2003. ISBN 9781584883258. [Google Scholar]
- Cox DR, Hinkley DV. Theoretical Statistics. Chapman and Hall; 1979. [Google Scholar]
- Dunn DT, Simonds RJ, Bulterys M, Kalish LA, Moye J, de Maria A, Kind C, Rudin C, Denamur E, Krivine A, Loveday C, Newell ML. Interventions to prevent vertical transmission of HIV-1: effect on viral detection rate in early infant samples. AIDS. 2000;14(10):1421–1428. doi: 10.1097/00002030-200007070-00016. [DOI] [PubMed] [Google Scholar]
- Fay MP, Shaw PA. Exact and asymptotic weighted logrank tests for interval censored data: The interval R package. Journal of Statistical Software. 2010;36(2):1–34. doi: 10.18637/jss.v036.i02. URL http://www.jstatsoft.org/v36/i02/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein DM. A proportional hazards model for interval-censored failure time data. Biometrics. 1986;42(4):845–854. [PubMed] [Google Scholar]
- Goetghebeur E, Ryan L. Semiparametric regression analysis of interval-censored data. Biometrics. 2000;56(4):1139–1144. doi: 10.1111/j.0006-341x.2000.01139.x. [DOI] [PubMed] [Google Scholar]
- Gomez G, Calle ML, Oller R, Langohr K. Tutorial on methods for interval-censored data and their implementation in R. Statistical Modelling. 2009;9(4):259–297. [Google Scholar]
- Gu X, Balasubramanian R. straweib: Stratified Weibull Regression Model. 2013 URL http://CRAN.Rproject.org/package=straweib. R package version 1.0. [PMC free article] [PubMed] [Google Scholar]
- Leroy R, Bogaerts K, Lesaffre E, Declerck D. The emergence of permanent teeth in flemish children. Community Dentistry and Oral Epidemiology. 2003;31(1):30–39. doi: 10.1034/j.1600-0528.2003.00023.x. [DOI] [PubMed] [Google Scholar]
- Lindsey JC, Ryan LM. Tutorial in biostatistics - methods for interval-censored data. Statistics in Medicine. 1998;17(2):219–238. doi: 10.1002/(sici)1097-0258(19980130)17:2<219::aid-sim735>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Meeker W, Escobar L. Statistical Methods for Reliability Data. Wiley: Wiley Series in Probability and Statistics; 1998. ISBN 9780471673279. [Google Scholar]
- Therneau T. A Package for Survival Analysis in S. 2012 R package version 2.36-14. [Google Scholar]
- Turnbull BW. Empirical distribution function with arbitrarily grouped, censored and truncated data. Journal of the Royal Statistical Society Series B-Methodological. 1976;38(3):290–295. [Google Scholar]
- Vanobbergen J, Martens L, Lesaffre E, Declerck D. The Signal-Tandmobiel project a longitudinal intervention health promotion study in flanders (belgium): baseline and first year results. Eur J Paediatr Dent. 2000;2:87–96. [Google Scholar]