The Ubiquitin Proteasome System (UPS)
In much the same way as kinases and phosphatases attach and remove, respectively, phosphate groups from proteins to modulate their activity, there are a series of enzymes (E1, E2, E3) that add one or more ubiquitins onto a protein, as well as enzymes that remove them (deubiquitinases), thereby regulating their activity, location and/or rate of degradation 1. Ubiquitin is a 76 amino acid protein that is added onto lysines in the target protein through the C-terminal residue of ubiquitin: one can be added (monoubiquitination) or as many as ten can be added (polyubiquitination). For polyubiquitinated proteins these can be linear or branched chains of ubiquitin, with the complexity of branching reminiscent of the complexity of protein glycosylation 1, 2. Ubiquitin contains seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63), and the N-terminal methionine that can be ubiquitinated. Common linkages include isopeptide bonds through the K48 and K63 on ubiquitin. K48 polyubiquitinated proteins are often targeted to the proteasome for protein degradation and recycling of the ubiquitin 1. Ubiquitination of a protein can also control its activity/function, such as K63 linkages that regulate DNA damage response or cell signaling 1, 3.
Interest in the ubiquitin-proteasome system (UPS) as a target for the treatment of disease, such as cancer, neurodegeneration and autoimmune disease, has increased steadily since the approval of the proteasome inhibitors bortezomib and carfilzomib 4. These drugs are used to treat hematological malignancies, such as multiple myeloma and mantle cell lymphoma. As yet this drug class has not been approved for solid tumors. Over time, resistance has begun to be observed for this class as well as side effect concerns, raising interest in targeting enzymes upstream of the UPS, such as the deubiquitinases and the E3 ligases, which may offer the possibility of more selectivity and fewer side effects 5.
Deubiquitinases (DUBs)
Deubiquitinases (DUBs) are upstream of the proteasome and have drawn interest as drug targets. The approximately 100 DUB enzymes can be grouped into five main classes, comprising the cysteine proteases ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), ovarian tumor proteases (OTUs), and Machado-Joseph domain proteases (MJDs) and the metalloproteases JAB1/MPN/MOV34 (JAMM) 6. The USPs are the largest family of DUBs, with approximately 56 members in humans, and are the focus of this review. In addition to the study of USPs as targets for drug discovery, there is much basic biology yet to be uncovered for this class of enzymes. Questions of substrate specificity, DUB redundancy and linkage selectivity have yet to be fully addressed for the majority of this enzyme class. To date both linkage selective DUBs, such as Cezanne which is specific for Lys11 linkages 7, and non-selective DUBs, such as USP2 which can cleave K48, K63 and linear, have been identified 6, 8–11. As has been seen for the kinase field 12, there is likely room for both selective and nonselective inhibitors as drugs and tool compounds.
The catalytic site of USPs contain a triad with a catalytic cysteine and nearby histidine and asparagine/aspartate to help poise the cysteine for nucleophilic attack. In addition to a USP domain various USPs have additional domains, such as ubiquitin-like domains and zinc-finger domains 6. Additionally, several of the USPs function as complexes, such as USP1/UAF1, USP12/UAF1/WDR20 and USP46/UAF1/WDR20 13, 14. Several USPs have crystal structures reported in the PDB, including USP2 (PDB ID 2HD5), USP5 (PDB ID 3IHP), USP7 (PDB ID 4M5W), USP14 (PDB ID 2AYN), CYLD (PDB ID 2VHF), and USP21 (2Y5B). USPs in an analogous way to kinases also seem to have active and inactive conformations with active conformations observed upon ubiquitin binding, although also like kinases not every USP has been observed in both conformations 6, 15.
Assay Technologies to Interrogate DUBs
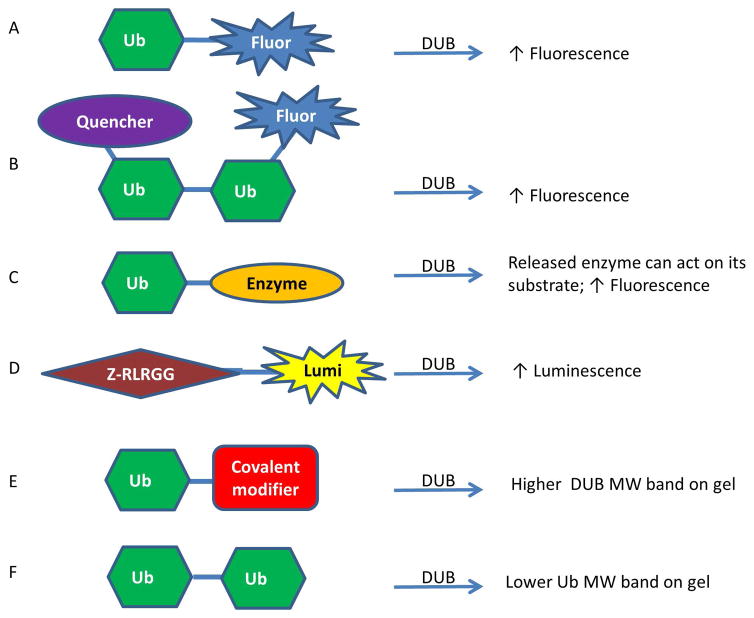
In order to identify DUB inhibitors, DUB substrates and DUB inhibitor selectivity, a variety of assay reagents have been identified and utilized in high-throughput screening (HTS) campaigns as well as lower throughput gel and western blot experiments (Figure 1) 16–18. The higher throughput methods generally involve an increase in luminescence or fluorescence upon cleavage that can be monitored on a plate reader 16. Commonly used reagents are ubiquitin linked to a fluorophore through a linear linkage, such as Ub-AMC (Ub-7-amino-4-methylcoumarin) and Ub-Rhodamine110 (Figure 1A), which have been used for screening various USPs including USP1 (PubChem Assay Identifier (AID) 504865), USP2 (PubChem AID 493170) and USP14 (PubChem AID 449747). More recently, reagents have been created that contain an isopeptide linkage between a di-ubiquitin (Di-Ub) to more closely mimic the most common in vivo Ub linkage. One example of this type of assay involves using an internally quenched fluorescent reagent in which one Ub has a fluorophore and the other has a quencher that quenches the fluorophore when the two are in close proximity but not once the Di-Ub is cleaved (Figure 1B) 8. Another method that has been utilized represents a coupled enzyme system. In one format, called Ub-Chop2, the ubiquitin is linked to an enzyme that is only active when released and thereby can produce a fluorescence enzyme product (Figure 1C); Ub-Chop2 has been used to identify inhibitors of USP2 in a large-scale screen (PubChem AID 463254). Additionally, having these reagents with different fluorescence excitation and emission wavelengths can help mitigate compound interference, which can be particularly problematic in the blue spectral region 19, such as for Ub-AMC, by allowing for orthogonal assay development. Lastly, when aminoluciferin is tethered to the DUB substrate Z-RLRGG it is not luminescent but upon cleavage by a DUB, its luminescence can be measured by a luminometer (Figure 1D), a technique that was used for USP8 20. All of these reagents can be read out in kinetic mode which can help minimize assay interference from compound fluorescence. Additional methods for identifying DUB substrates, DUB inhibitors and DUB linkage preferences include mass spectrometry based methods and protein microarray methods 17, 18, 21.
Figure 1. Representative assay formats used to study DUBs.
A) Ubiquitin linked to fluorophore that fluoresces upon cleavage. B) Internally quenched fluorescence Di-Ub that fluoresces upon cleavage. C) When cleaved, the enzyme becomes active and can act on its substrate and form a fluorescent product. D) A generic DUB substrate can be tethered to a proluminescent molecule that becomes luminescent only when cleaved. E) Upon binding a DUB, the Ub is covalently attached and the MW shift can be monitored by gel assay. F) Di-Ub cleavage can be monitored by gel assay. Fluor=fluorophore; Lumi=luminescent compound; Ub = ubiquitin.
Lower throughput methods that are important for confirmation of hits rely primarily on a change in mass upon Di-Ub cleavage or DUB binding. Ubiquitin aldehyde or ubiquitin vinyl sulfone are reagents that form covalent irreversible linkages to DUBs upon binding, leading to observable mass changes to the DUBs (Figure 1E). These have been visualized in western blots with antibodies to the individual DUB, such as USP1, or to ubiquitin, or to a tag, such as HA, placed on the ubiquitin, HA-ubiquitin vinyl sulfone 22. Additionally, Di-Ub can be cleaved by a DUB and this new product can be visualized on a gel as a band with half the molecular weight of the substrate (Figure 1F), a technique that has been used for USP1 23. Tetra-ubiquitin molecules are becoming increasingly available that can also be used for this purpose. Having a diversity of assay technologies is important for identifying and validating inhibitors and studying the USP family.
USPs as Drug Targets
While the USP family as a whole is still widely unexplored, selected USPs have been the focus of intense research efforts both to uncover their physiological roles and to identify inhibitors 24, 25, with some of the highlights to date described below.
USP1 deubiquitinates PCNA and FANCD2, which are proteins important for DNA repair pathways and the Fanconi Anemia pathway, respectively 3. USP1, which is active as a complex with UAF1 13, can deubiquitinate PCNA, which upon DNA damage is ubiquitinated and thereby recruits translesion DNA polymerase eta. ML323 was developed as an inhibitor of USP1/UAF1 after a high-throughput screen using Ub-Rhodamine110 assay 26. Additional compounds, pimozide, GW7647 and C527 were found to be sub μM inhibitors 22, 23.
USP2, found in vivo as the alternately spliced USP2a and USP2b, has a large number of identified substrates important in cancer including cyclin D1, MDM2, and fatty acid synthase (FAS) 27–29. Cyclin D1, which can be overexpressed in cancer, was degraded in response to USP2-targeting siRNA leading to inhibition of cell cycle progression in cancer cells but not normal cells 28. USP2 can also deubiquitinate MDM2, thereby destabilizing p53 29. USP2a was shown to help protect prostate cancer cells from apoptosis by deubiquitinating the antiapoptotic proteins MDM2 and FAS, which can be overexpressed in cancer 27, 30. Indeed 44% of prostate tumors tested had overexpression of USP2a. 2-cyano-pyrimidines and -triazines have been identified as inhibitors of USP2 and UCH-L3 31.
USP5 has been implicated in suppression of both p53 and FAS levels in melanoma cells. An inhibitor EOAI3402143, which had been optimized from WP1130 32, was shown to recapitulate USP5 knockdown results and block melanoma growth in a mouse model 33. Inhibition was able to overcome resistance to BRAF-targeting kinase inhibitors in melanoma 33. This compound also inhibits USP9x, a drug target discussed below 34–36.
USP7/HAUSP (herpes virus-associated ubiquitin-specific protease) deubiquitinates MDM2 (an E3 ligase; also called HDM2), thereby destabilizing p53, and can be targeted by small molecule inhibitors 37. Additional roles for USP7/HAUSP include deubiquitination of the tumor suppressors PTEN (phosphatase and tensin homologue deleted in chromosome 10) and FOXO4 (Forkhead box O), which favors their localization to the cytoplasm versus the nucleus limiting their transcriptional activity 38, 39. It should be noted that PTEN is also deubiquitinated by USP13 40. USP7/HAUSP is overexpressed in cancer, such as prostate cancer. Inhibitors of USP7 include HBX 41,108 and P22077, which were identified from HTS campaigns 37, 41, 42.
USP8 (UBPY) knockdown in gefitinib-resistant NSCLC but not gefitinib-sensitive NSCLC leads to cell death, potentially providing an avenue to pursue when resistance to EGFR receptor kinase inhibitors develops 20. This knockdown effect could be recapitulated with a USP8 inhibitor (9-ethyloxyimino-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile) 43 that showed efficacy in a mouse xenograft model. USP8 had previously been shown to be involved in EGFR degradation 44. Additionally, USP8 mutations cause Cushing’s Disease, a disease caused by pituitary corticotroph adenomas hypersecting adenocorticotropin (ACTH) 45. The mutations cause a decrease in binding to 14-3-3 protein, increased cleavage of USP8 and an increase in DUB activity, which leads to increased recycling of EGFR increasing the levels of ACTH 45.
USP9x plays an important role in stabilizing beta-catenin 35, MCL1 36 and SMAD4 34, a component of the TGFβ signaling pathway. Each of these proteins is in its own right a target for cancer therapy. USP9x is one of the DUBs inhibited by WP1130 32.
USP12 is active as a complex USP12/UAF1/WDR20 and has several substrates identified to date including the androgen receptor, which is important in prostate and some breast cancers 14, 46. To date only one compound has been reported to inhibit USP12/UAF1/WDR20 and that is GW7647, which also inhibits USP1 23, 47.
USP14 is associated with the proteasome and is one of the DUBs responsible for ensuring that ubiquitin is recycled rather than degraded by the proteasome, contributing to ubiquitin homeostasis. An inhibitor of USP14 was identified and shown to enhance the activity of the proteasome 48. Inhibition of USP14 lead to accelerated proteasomal degradation of proteins involved in neurodegenerative diseases.
USP28 has a role in stabilizing the oncoprotein c-myc as discovered in a shRNA screen 49, but also is implicated in other functions that may lead to side effects 5. For example, USP28 is important for the DNA damage response 50. USP28 is highly expressed in colon and breast carcinomas and has been implicated in conferring stem-cell-like traits to breast cancer cells 51.
Perspective
Deubiquitinases (DUBs) have been a subject of increased interest of late as a potential novel drug target class, in turn spurring efforts to develop relevant activity assays. Most reagents used for DUB assays contain one Ub linked to a fluorophore or luminescent compound by a linear linkage or two Ubs linked by an isopeptide linkage but polyubiquitinated proteins can have as many as 10 Ubs linked together in linear or branched conformations 1, 9. Having additional assay reagents, such as tetraubiquitin, with > 2 ubiquitins as well as branched Ubs may be important for further understanding Dub linkage selectivity. It will be interesting to see the impact on HTS campaigns of utilizing the different available assay reagents to understand whether specific inhibitors exist targeting the different linkages and poly-ubiquitin reagents.
Screening a large panel size was important to obtain an accurate assessment of inhibitor selectivity for kinases 12, and it will be interesting to see whether the same will hold for DUB families, such as the USPs, or whether a small panel will accurately represent overall selectivity. There is a need for the development and execution of HTS assays and the discovery of molecule probes for all of the ~56 USPs in order to understand their biology. Once a critical mass of inhibitors are available, a broad selectivity profile will be important in order to understand whether highly selective modulators across the whole DUB class can be obtained as is the case for some kinase inhibitors, such as lapatinib for EGFR, which is selective across the whole kinome 12. A given DUB, such as USP2, can have multiple substrates that it can deubiquitinate, so in addition to polypharmacology from inhibitors that hit more than one DUB, even a DUB-selective inhibitor may impact multiple cellular proteins and processes due to multiple substrates acted on in vivo. Additionally, redundancy within the USP family is already being observed with multiple DUBs impacting MDM2, p53, FAS and PTEN 27, 29, 33, 37, 38. Thus, the DUB field seems poised for rapid development: in much the same way as the approval of Gleevec sparked fervor into the kinase field 52, having a DUB inhibitor approved would be expected to do the same for the deubiquitinase field.
References
- 1.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 2.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448–62. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacq X, Kemp M, Martin NM, Jackson SP. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem Biophys. 2013 Sep;67(1):25–43. doi: 10.1007/s12013-013-9635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dou QP, Zonder JA. Overview of proteasome inhibitor-based anti-cancer therapies: perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr Cancer Drug Targets. 2014;14(6):517–36. doi: 10.2174/1568009614666140804154511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi D, Grossman SR. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol Ther. 2010 Oct 15;10(8):737–47. doi: 10.4161/cbt.10.8.13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009 Aug;10(8):550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 7.Bremm A, Freund SMV, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17(8):939–47. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Progenra, Inc., Lifesensors, Inc, assignee. Di- and poly-ubiquitin deubiquitinase substrates and uses there of 2013. [Google Scholar]
- 9.Faesen AC, Luna-Vargas MP, Geurink PP, Clerici M, Merkx R, van Dijk WJ, et al. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol. 2011 Dec 23;18(12):1550–61. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Reports. 2009;10(5):466–73. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005 Dec 2;123(5):773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011 Nov;29(11):1046–51. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 13.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Molecular cell. 2007 Dec 14;28(5):786–97. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 14.Kee Y, Yang K, Cohn MA, Haas W, Gygi SP, D’Andrea AD. WDR20 Regulates Activity of the USP12 ·UAF1 Deubiquitinating Enzyme Complex. Journal of Biological Chemistry. 2010 Apr 9;285(15):11252–7. doi: 10.1074/jbc.M109.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolen B, Taylor S, Ghosh G. Regulation of Protein Kinases: Controlling Activity through Activation Segment Conformation. Molecular Cell. 2004;15(5):661–75. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 16.Kramer HB, Nicholson B, Kessler BM, Altun M. Detection of ubiquitin–proteasome enzymatic activities in cells: Application of activity-based probes to inhibitor development. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823(11):2029–37. doi: 10.1016/j.bbamcr.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loch CM, Strickler JE. A microarray of ubiquitylated proteins for profiling deubiquitylase activity reveals the critical roles of both chain and substrate. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823(11):2069–78. doi: 10.1016/j.bbamcr.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritorto MS, Ewan R, Perez-Oliva AB, Knebel A, Buhrlage SJ, Wightman M, et al. Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat Commun. 2014;5:4763. doi: 10.1038/ncomms5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008 Apr 24;51(8):2363–71. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 20.Byun S, Lee SY, Lee J, Jeong CH, Farrand L, Lim S, et al. USP8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin Cancer Res. 2013 Jul 15;19(14):3894–904. doi: 10.1158/1078-0432.CCR-12-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGouran JF, Gaertner SR, Altun M, Kramer HB, Kessler BM. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem Biol. 2013 Dec 19;20(12):1447–55. doi: 10.1016/j.chembiol.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mistry H, Hsieh G, Buhrlage SJ, Huang M, Park E, Cuny GD, et al. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther. 2013 Dec;12(12):2651–62. doi: 10.1158/1535-7163.MCT-13-0103-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, et al. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol. 2011 Nov 23;18(11):1390–400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edelmann MJ, Nicholson B, Kessler BM. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Rev Mol Med. 2011;13:e35. doi: 10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farshi P, Deshmukh RR, Nwankwo JO, Arkwright RT, Cvek B, Liu J, et al. Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opin Ther Pat. 2015 Jun;16:1–18. doi: 10.1517/13543776.2015.1056737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, et al. A selective USP1–UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10(4):298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5(3):253–61. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 28.Shan J, Zhao W, Gu W. Suppression of Cancer Cell Growth by Promoting Cyclin D1 Degradation. Molecular Cell. 2009;36(3):469–76. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. The EMBO Journal. 2007;26(4):976–86. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Priolo C, Tang D, Brahamandan M, Benassi B, Sicinska E, Ogino S, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 2006 Sep 1;66(17):8625–32. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- 31.2-cyano-pyrimidines and -triazines as cysteine protease inhibitors. 2007. [Google Scholar]
- 32.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010 Nov 15;70(22):9265–76. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 33.Potu H, Peterson LF, Pal A, Verhaegen M, Cao J, Talpaz M, et al. Usp5 links suppression of p53 and FAS levels in melanoma to the BRAF pathway. Oncotarget. 2014 Jul 30;5(14):5559–69. doi: 10.18632/oncotarget.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. FAM/USP9x, a Deubiquitinating Enzyme Essential for TGFβ Signaling, Controls Smad4 Monoubiquitination. Cell. 2009;136(1):123–35. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 35.Murray RZ, Jolly LA, Wood SA. The FAM Deubiquitylating Enzyme Localizes to Multiple Points of Protein Trafficking in Epithelia, where It Associates with E-cadherin and β-catenin. Molecular Biology of the Cell. 2004 Apr 1;15(4):1591–9. doi: 10.1091/mbc.E03-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463(7277):103–7. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 37.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009 Aug;8(8):2286–95. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 38.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455(7214):813–7. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Horst A, de Vries-Smits AMM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8(10):1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, et al. Deubiquitylation and stabilization of PTEN by USP13. Nature cell biology. 2013 Dec;15(12):1486–94. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012 Apr 20;19(4):467–77. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, et al. Selective Dual Inhibitors of the Cancer-Related Deubiquitylating Proteases USP7 and USP47. ACS Med Chem Lett. 2012 Oct 11;3(10):789–92. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colombo M, Vallese S, Peretto I, Jacq X, Rain JC, Colland F, et al. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem. 2010 Apr 6;5(4):552–8. doi: 10.1002/cmdc.200900409. [DOI] [PubMed] [Google Scholar]
- 44.Alwan HAJ, van Leeuwen JEM. UBPY-mediated Epidermal Growth Factor Receptor (EGFR) De-ubiquitination Promotes EGFR Degradation. Journal of Biological Chemistry. 2007;282(3):1658–69. doi: 10.1074/jbc.M604711200. [DOI] [PubMed] [Google Scholar]
- 45.Reincke M, Sbiera S, Hayakawa A, Theodoropoulou M, Osswald A, Beuschlein F, et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat Genet. 2015;47(1):31–8. doi: 10.1038/ng.3166. [DOI] [PubMed] [Google Scholar]
- 46.Burska UL, Harle VJ, Coffey K, Darby S, Ramsey H, O’Neill D, et al. Deubiquitinating Enzyme Usp12 Is a Novel Co-activator of the Androgen Receptor. Journal of Biological Chemistry. 2013 Nov 8;288(45):32641–50. doi: 10.1074/jbc.M113.485912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClurg UL, Summerscales EE, Harle VJ, Gaughan L, Robson CN. Deubiquitinating enzyme Usp12 regulates the interaction between the androgen receptor and the Akt pathway. Oncotarget. 2014 Aug 30;5(16):7081–92. doi: 10.18632/oncotarget.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010 Sep 9;467(7312):179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9(7):765–74. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 50.Zhang D, Zaugg K, Mak TW, Elledge SJ. A Role for the Deubiquitinating Enzyme USP28 in Control of the DNA-Damage Response. Cell. 2006;126(3):529–42. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 51.Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, et al. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013 Oct 17;5(1):224–36. doi: 10.1016/j.celrep.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen MH, Williams G, Johnson JR, Duan J, Gobburu J, Rahman A, et al. Approval Summary for Imatinib Mesylate Capsules in the Treatment of Chronic Myelogenous Leukemia. Clinical Cancer Research 2002. 2002 May 1;8(5):935–42. [PubMed] [Google Scholar]