Abstract
The weighted histogram analysis method (WHAM) including its binless extension has been developed independently in several different contexts, and widely used in chemistry, physics, and statistics, for computing free energies and expectations from multiple ensembles. However, this method, while statistically efficient, is computationally costly or even infeasible when a large number, hundreds or more, of distributions are studied. We develop a locally WHAM (local WHAM) from the perspective of simulations of simulations (SOS), using generalized serial tempering (GST) to resample simulated data from multiple ensembles. The local WHAM equations based on one jump attempt per GST cycle can be solved by optimization algorithms orders of magnitude faster than standard implementations of global WHAM, but yield similarly accurate estimates of free energies to global WHAM estimates. Moreover, we propose an adaptive SOS procedure for solving local WHAM equations stochastically when multiple jump attempts are performed per GST cycle. Such a stochastic procedure can lead to more accurate estimates of equilibrium distributions than local WHAM with one jump attempt per cycle. The proposed methods are broadly applicable when the original data to be “WHAMMED” are obtained properly by any sampling algorithm including serial tempering and parallel tempering (replica exchange). To illustrate the methods, we estimated absolute binding free energies and binding energy distributions using the binding energy distribution analysis method from one and two dimensional replica exchange molecular dynamics simulations for the beta-cyclodextrin-heptanoate host-guest system. In addition to the computational advantage of handling large datasets, our two dimensional WHAM analysis also demonstrates that accurate results similar to those from well-converged data can be obtained from simulations for which sampling is limited and not fully equilibrated.
I. INTRODUCTION
To observe rare events in traditional molecular simulations, enhanced sampling techniques are generally applied to speed up, sometimes by many orders of magnitude, conformational interconversions and slow intrinsic dynamics of biomolecules and other condensed phase systems, based on the imposition of thermodynamic or alchemical biasing forces on the relevant chemical reaction coordinates or order parameter space.1–8 Typically true (unbiased) thermodynamic quantities need to be extracted from biased simulation results via postprocessing using reweighting techniques.9–17 The weighted histogram analysis method (WHAM)9 and the binless extension, multi-state Bennet acceptance ratio (MBAR)13 or unbinned WHAM (UWHAM),14 have been widely used for computing free energies and expectations from multiple ensembles, for example, at different temperatures or with different biasing potentials.9–16 In fact, this methodology can be seen to not only generalize the Bennett acceptance ratio (BAR)18 method from two to multiple ensembles but also remove the need of discretizing observations into bins in the original WHAM. Moreover, the same methodology has been extensively developed for statistical computation involving simulations from multiple distributions with intractable normalizing constants. Examples include reverse logistic regression,19 multiple bridge sampling,20 and the global likelihood method.21,22 For convenience, the methodology will be referred to as WHAM in short, even though the binless version will be discussed.
Generalized ensemble methods such as (parallel) replica exchange (RE) algorithms23–29 and (serial) simulated tempering (ST)30–39 are popular among many enhanced conformational sampling methods. These algorithms produce a random walk, not only in conformational space but also in thermodynamic or Hamiltonian parameter spaces of the simulated system, such as the temperature of the system treated as a random variable in the serial tempering method. Recently, there has been increasing interest in extending replica exchange simulations to a large number, hundreds or more, of thermodynamic states in a multidimensional parameter space (e.g., temperatures and biasing potentials along two or more coordinates), using national high performance clusters such as XSEDE17,40–44 or distributed networks such as campus grids or world-wide volunteer grids.45,46 However, there are serious computational difficulties in using WHAM for large-scale free energy estimation. For simulated data from m ensembles with n data points per ensemble (for example, m parallel simulations each of size n), WHAM involves, for each individual data point, evaluation and manipulation of m unnormalized density functions at each of the m thermodynamic states, not just at the thermodynamic state observed to be associated with the data point in the simulations. The number of computer operations, including taking exponentials, is then of order O(m2n), in addition to the fact that a total of m2n unnormalized density values need to be either computed during every step of solving the WHAM equations or pre-computed and stored in memory. Therefore, the computational cost of WHAM grows quadratically as the number of thermodynamic states under study increases. For this reason, the standard implementations of WHAM are ineffective or infeasible for analyzing simulations from a large number of ensembles.
To tackle this problem of WHAM, we develop a locally WHAM (local WHAM) and a stochastic solution, using serial tempering30–32 and its extensions38,47 to resample data obtained from multi-ensemble simulations, which is called simulations of simulations (SOS).48–53 To facilitate the derivation of local WHAM, serial tempering algorithms are presented in a generalized formulation, referred to as generalized serial tempering (GST), where the single state-dependent biasing factors used in the usual implementation of serial tempering are explicitly decomposed into two parameters, corresponding to hypothesized free energy values and, possibly non-uniform, target proportions of states. Although serial tempering was originally designed for the purpose of sampling from multiple complex ensembles using Markov chain Monte Carlo (MCMC) or molecular dynamics (MD), the SOS procedure based on GST (SOS-GST) involves performing GST on simulated data that are already obtained from real simulations for the purpose of analyzing such data, i.e., estimating free energies and equilibrium distributions. The real simulations from which the data are obtained for analysis by SOS-GST are allowed to be done by either GST or any other sampling algorithm such as replica exchange.
The proposed methods described in this work provide an alternative but closely related approach to a SOS-RE procedure53 we have recently proposed for solving the (global) WHAM equations stochastically by combining simulations of simulations and parallel tempering (replica exchange). Both approaches can be employed to analyze large datasets generated by multicanonical simulations, to obtain estimates of free energies and equilibrium distributions with substantially less computation time and memory than standard implementations of global WHAM. But as discussed later, local WHAM allows for a numerical implementation using optimization algorithms, which is faster than a stochastic solution and can still lead to accurate estimates of free energies, albeit the local WHAM solution is only an approximation to the global WHAM solution.
For SOS-GST, a neighborhood of states are specified for each thermodynamic state under study, similarly as in real GST simulations.30–32 For example, if the thermodynamic states are parameterized to lie on a regular grid, then the neighborhood of a state can consist of the nearest neighbors on the grid. At each cycle, the SOS-GST procedure consists of a move process sampling a data point from the simulated data given the current thermodynamic state and a jump process generating a new state in the neighborhood of the current state. The jump probability is determined by the Metropolis rule, depending on both hypothesized free energy values and target proportions of states, which are chosen for SOS-GST to be the observed proportions of thermodynamic states in the actual simulated data. As mentioned above, while serial tempering is not often implemented so as to include parameters specifying target proportions of states, it is convenient to make explicit the choice of target proportions so that hypothesized free energies can be identified as a vector of directly adjustable parameters to be determined in a self-consistent manner as follows. The local WHAM estimates of free energies are those values for hypothesized free energies such that the long-term (stationary) proportions of time spent on all states in SOS-GST are identical to the target proportions of states, which are taken to be the observed proportions of states in the actual simulated data. This is analogous to the fact that the true free energies are those values such that the long-term proportions of time spent on all states are identical to the target proportions of states in GST for real simulations.30–32
For SOS-GST with a single jump allowed at each cycle, the local WHAM equations for free energy estimates can be expressed in an analytically simple form and hence be solved directly by fast and reliable optimization algorithms similarly as the global WHAM equations,14,15 without running the SOS-GST procedure. The computational cost of local WHAM is of order O(smn), depending only linearly on the number of thermodynamic states for a fixed neighborhood size s, because the unnormalized density functions for each data point only need to be locally evaluated at the neighboring states to its observed thermodynamic state for reweighting. Hence, local WHAM can be executed orders of magnitude faster than global WHAM, when the number of states is large but the neighborhood sizes are small. On the other hand, the variability in free energy estimation is, to a large extent, affected by the degree of overlaps between different ensembles.20 Then, local WHAM is expected to yield similar statistical efficiency to that of global WHAM, because local WHAM is constructed to effectively exploit the overlaps between all pairs of neighboring ensembles, which often represent the majority of overlaps between all ensembles considered.
For SOS-GST with multiple jump attempts performed per cycle, the local WHAM equations become analytically too complicated to be handled by optimization methods. To address this issue, we propose an adaptive SOS-GST procedure, based on self-adjusted GST algorithms used in real simulations,47 for stochastically solving the local WHAM equations. At each cycle of this procedure, the hypothesized free energy values are adjusted after the usual move and jump processes by the theory of stochastic approximation,54–57 such that the adjusted values converge in probability to the local WHAM estimates of free energies, as the number of cycles increases to infinity. A potential advantage of the adaptive SOS-GST procedure using multiple jump attempts per cycle is that it can lead to estimates of equilibrium distributions matching the global WHAM estimates more closely than the local WHAM estimates using a single jump attempt per cycle, even though the free energy estimates from the three methods are similar to each other. As might be expected, estimation of equilibrium distributions is, in general, more sensitive to the presence of limited equilibration in simulated data than is free energy estimation.
To illustrate our new methods, we calculated the binding free energies of β-cyclodextrin-heptanoate host-guest system at different temperatures from 1D and 2D replica exchange molecular dynamics (REMD) simulations using the Binding Energy Distribution Analysis Method (BEDAM).58 The model complex is one of the host-guest systems well studied in our previous work45,59,60 and serves as a testbed for different methods.
II. THEORY AND METHODS
A. Setup
Consider a collection of m thermodynamic states associated with m probability distributions, (P1, …, Pm), with the density functions
| (1) |
where qj(x) is an unnormalized density function of the jth state, and Zj is the partition function of the jth state (i.e., the normalizing constant), for j = 1, …, m. Typically, qj(x) is defined from a Boltzmann distribution in the form
| (2) |
where h(x; θj) is a temperature-scaled potential energy function including the biasing potential, and θj is a parameter vector including the jth temperature Tj and alchemical parameters. For example, if generalized ensemble simulations are performed with respect to temperature, θj = βj = 1/(kBTj) is the jth inverse temperature and h(x; θj) = βjH(x), where H(x) is the original potential energy function. When we calculate free energy as a function of an alchemical parameter λ at a fixed temperature, θj = λj and h(x; θj) = βH(x; λj), where H(x; λj) is the modified potential energy at the jth state. It is also possible to construct a generalized ensemble using a vector of multiple parameters. For example, θj = (βl, λk) with two indices (l, k), and h(x; θj) is defined as βlH(x; λk) = βl{H0(x) + λkb(x)} in our 2D BEDAM for binding free energy calculations in Section III. For simplicity, we refer to each distribution Pj with a specific parameter vector θj of thermodynamic and alchemical parameters as a thermodynamic state.
In this report, we are primarily interested in analyzing data obtained for the m thermodynamic states from actual simulations, either independent or multicanonical, in order to extract accurate estimates of free energies and equilibrium distributions using all the data. Assume that real simulations are conducted, such that an approximate sample of size nj is obtained from the jth thermodynamic state (i.e., drawn from the jth distribution Pj) for j = 1, …, m. The pooled data are denoted by {(Li, Xi) : i = 1, …, N}, where Xi is the ith observation, Li stores the state label for Xi (i.e., set to j if Xi is drawn from the jth state), and is the total sample size.
The simulations from which the data are obtained can be carried out in a variety of ways. For example, it is possible to run m Metropolis chains independently targeting equilibrium distributions (P1, …, Pm) or to introduce interactions between different samples for the purpose of improving convergence, using specialized algorithms such as RE (also known as parallel tempering)23–29 and ST.30–39,47 For replica exchange, but not for serial tempering, the individual sample sizes are typically equal to each other (i.e., n1 = … = nm = N/m) except for the recently introduced asynchronous form of replica exchange.45,46 To extract accurate estimates of free energies and expectations from these possibly biased simulations, however, various reweighting techniques have to be applied to the pooled data.9–17
B. Globally weighted histogram analysis method
We describe a commonly used method for free energy estimation using observations simulated from multiple thermodynamic states. Although the method can be traced to the Bennett acceptance ratio in the case of only two states,18 the binned version of the method is known as the WHAM9,10 in physics, based on discretizing the observations into a finite number of bins in order to construct proper histograms. On the other hand, the unbinned version of the method, using the actual data instead of their discretizations, has been independently developed in statistics.19–21 To better deal with sparse distributions at certain thermodynamic states, the unbinned method has been (re-)introduced to physics as the MBAR13 method and also directly presented as a binless extension of WHAM, called UWHAM.14 In the following, the unbinned version of the method is discussed and, for simplicity, still referred to as WHAM.
The WHAM estimates of free energies at different thermodynamic states, log(Z1, …, Zm) (the constant multiple −kBT is reduced to 1 in Section II), are defined as a solution, , to the system of estimating equations,13,14
| (3) |
where πk = nk/N is the observed proportion of kth thermodynamic state for k = 1, …, m. In fact, are determined only up to an additive constant, because if satisfy Eq. (3), then also satisfy Eq. (3) for an arbitrary constant c. It is customary to pick a reference value, for example, logZ1, and then estimate the free energy differences, log(Z2/Z1, …, Zm/Z1), by the solution , with fixed, from Eq. (3). Once the free energy estimates are obtained, the WHAM estimate of the equilibrium distribution Pj for jth thermodynamic state is a discrete distribution , supported on the pooled data with the probabilities
| (4) |
In other words, Pj is approximated by attaching weight (4) to each observation Xi in the pooled data, where the weights sum up to 1 by Eq. (3). Moreover, let ϕ(x) be a scalar function of x. The marginal distribution of ϕ(x) under Pj is approximated by attaching the same weight (4) to ϕ(Xi) for each Xi in the pooled data. Then, the expectation of ϕ(x) under Pj (i.e., the ensemble average of the observable ϕ(x) from the jth state), denoted by 〈ϕ〉j, is estimated by the weighted average
| (5) |
The above WHAM estimates of free energies and expectations are shown to be statistically optimal with smallest possible asymptotic variances as N → ∞, in the case where (X1, …, XN) are independent.22 On the other hand, by Eq. (4), the WHAM probabilities are reweighted from all N data points Xi, depending on q1(Xi), …, qm(Xi). Namely, WHAM requires evaluating m functions q1(Xi), …, qm(Xi) at each observation Xi for i = 1, …, N. The total number of function evaluations is of order , where is the average sample size per distribution. Such high computational cost presents a serious limitation on the use of WHAM for the situation where the number of distributions, m, is large, for example, in the hundreds or more and the total number of data points, N, to be reweighted is also large (N = 3.5 × 107 and m = 240 in our application).
To reduce computational cost while preserving statistical efficiency, we adopt and further develop a locally WHAM (local WHAM),47 for estimating free energies and equilibrium distributions from large-scale simulations. Following our recent work,53 we reformulate the method from the perspective of SOS,48,50,53 but here, we use serial tempering30,32 instead of replica exchange to resample the data obtained from real simulations for the purpose of estimating free energies and expectation values from the original data.
C. Generalized serial tempering
We present a generalized formulation of serial tempering, referred to as GST, in the statistical framework of labeled mixture sampling47 which makes explicit the mixture structure, in particular, the relationship between target mixture weights and hypothesized free energies in serial tempering and its extensions.30–39 The main motivation for introducing GST here is to facilitate the derivation of local WHAM equations in the context of simulations of simulations in Section II D. Nevertheless, the GST formulation can also be useful in real simulations for allocating non-uniform amounts of time (or resources) on different thermodynamic states.61,62 For example, if the energy landscape of state j is much rougher than that of state k, it can be more efficient to bias the serial tempering simulations such that more time is spent at state j than at k.
For a standard formulation of ST, a single chain is generated by alternately updating a pair of thermodynamic state (“label” L) and coordinate configuration (“observation” X) such that the joint stationary distribution is
| (6) |
where are state-dependent biasing (or weight) factors, and Z is a weighted sum of the normalizing constants of the m canonical ensembles,
| (7) |
with and Zj defined in Eq. (1). By integration over the configuration space, the marginal stationary probability of the jth thermodynamic state can be found as62
| (8) |
Taking logarithms of both sides of Eq. (8) indicates that, up to an additive constant,
| (9) |
As shown by Eq. (9), a uniform distribution over thermodynamic states (i.e., p(j; ζB) = 1/m for j = 1, …, m) will be obtained if and only if the biasing factors are set equal to the true free energies , up to an additive constant.
For GST, we reformulate joint distribution (6) as
| (10) |
where two parameter vectors ζ and π0 are introduced with the following interpretation: ζ = (ζ1, …, ζm) with ζ1 ≡ 0 encode hypothesized (i.e., guessed) values of the true free energy differences with , and encode target mixture weights (i.e., proportions of states) that would be obtained if ζ were specified as identical to ζ∗. In fact, the marginal stationary distribution of X is
| (11) |
which is a mixture distribution with the mixture weight for the jth thermodynamic state equal to the corresponding marginal stationary probability
| (12) |
Similarly, as Eq. (8) leads to (9), Eq. (12) implies that, up to an additive constant,
| (13) |
In parallel to Eq. (9), Eq. (13) shows that the stationary mixture weights p(j; ζ, π0) are equal to for all thermodynamic states j = 1, …, m if and only if the guessed vector ζ coincides with the truth ζ∗ for free energy differences.
The relationship between ST and GST can be understood as follows. On one hand, the joint density function p(j, x; ζB) in ST can be seen as a special case of p(j, x; ζ, π0) in GST, by setting (i.e., the target proportions of states are uniform) and for j = 1, …, m (i.e., the hypothesized free energies ζj are the biasing factors up to an additive constant). On the other hand, the joint density function p(j, x; ζ, π0) can be equivalently put in the form of p(j, x; ζB), and Eqs. (12) and (13) can also be derived from Eqs. (8) and (9), by defining single biasing factors as
| (14) |
By Eq. (14), the biasing factors in the standard ST formulation are decomposed into two parameters, ζj and , corresponding to hypothesized free energies and target mixture weights in GST. Hence, the biasing factors , in general, differ from hypothesized free energies ζj unless the target proportions of states are uniform (). Although ST and GST can be made equivalent to each other by relationship (14), we find that the GST formulation allows an analytically more direct derivation of local WHAM equations, because hypothesized free energies ζj are identified in GST as a vector of directly adjustable parameters, separately from target proportions ; otherwise ζj would need to be identified as by Eq. (14). See further discussion in Section II D.
Under joint distribution (10) of thermodynamic state L and coordinate configuration X, the two conditional distributions can be easily shown to be
| (15) |
| (16) |
That is, p(x|L = j) corresponds to the jth distribution Pj, regardless of ζ and π0, whereas p(L = j|x; ζ, π0) is a discrete distribution on {1, …, m}, depending on ζ and π0. We notice that expression of p(L = j|x; ζ, π0) is similar to the individual terms in WHAM equation (3), with ζ and π0 corresponding to ζ and π, respectively.
A typical GST algorithm proceeds as follows: for some fixed choices of hypothesized free energies ζ and target proportions of states π0, generate a single Markov chain, {(Lt, Xt) : t = 1, 2, …}, with p(j, x; ζ, π0) as the stationary joint distribution. Each GST cycle consists of a move and a jump process for updating Xt and Lt, respectively.
GST:
-
•
Move: Generate Xt given (Xt−1, Lt−1 = j), leaving Pj invariant (namely, update X in the configuration space for the current thermodynamic state j);
-
•
Jump: Generate Lt given (Lt−1, Xt = x), leaving p(L = ⋅ |x; ζ, π0) invariant (namely, jump to another thermodynamic state but stay in the same configuration X).
A major challenge for serial tempering simulations to achieve reasonable convergence is that the hypothesized free energies ζ should be chosen to be roughly near the true ζ∗ (that is, the biasing factors ζB should be near ζ∗ for uniform sampling over different thermodynamic states).29 Various techniques have been proposed to tackle this issue. A common strategy is to estimate free energies from short trial RE simulations using WHAM28,40 or other approximate methods.30,34,36 The choice of ζ can also be adjusted periodically during the subsequent simulation.33,35,37,39 In Section II D, we will make use of GST with a freely specified but fixed choice of ζ in SOS as an analytical tool (without running SOS-GST) to derive local WHAM equations for free energy estimation with data already generated from real simulations. In Section II E, we will then implement SOS-GST with an adaptive scheme for sequentially adjusting the choice of hypothesized free energies ζ to obtain a stochastic solution of the local WHAM equations.
The move process above is, in practice, accomplished by MCMC or MD in the coordinate configuration space. For implementing the jump process we describe a non-weighted, local jump scheme, where the jump is restricted to a neighborhood of the current state Lt−1 and the proposal probabilities of jump are not weighted depending on the current configuration Xt. Such jump schemes are commonly used and central to the computational advantage of local WHAM developed in Sections II C and D. For k = 1, …, m, let be a neighborhood of thermodynamic states (excluding k itself) to the state k, such that if and only if . For example, if the thermodynamic states are arranged on a regular grid, then the neighborhood of a state can be defined as the nearest neighbors on the grid. Moreover, let Γ(k, ⋅ ) be a proposal distribution for jumping from the state k to another state such that Γ(k, j) = 0 if , where . A typical example is to set Γ(k, j) = 1/s(k) (uniform distribution) if and 0 otherwise (including j = k), where s(k) is the size of . With one jump attempt, the resulting jump scheme is as follows.
Non-weighted jump scheme with one jump attempt:
-
•
Proposal: Sample j ∼ Γ(Lt−1, ⋅ ) (namely, select state j with, e.g., uniform probabilities from the neighborhood of Lt−1);
-
•Acceptance-rejection: Set Lt = j (namely, accept j as the new state) with the Metropolis probability
and, with the remaining probability, set Lt = Lt−1.(17)
In general, multiple (say r) jump attempts can be successively performed for the jump process in each GST cycle, leading to the following iterated jump scheme.
Non-weighted jump scheme with multiple attempts:
-
•
For i = 1, …, r, generate Lt,i from Lt,i−1 using the non-weighted jump scheme above with the same Xt, where Lt,0 = Lt−1.
-
•
Set Lt = Lt,r.
As the number of jump attempts r increases to ∞, the non-weighted jump scheme can be shown to yield a weighted global-jump scheme, so called Gibbs sampling,38 where the proposal probabilities depend on the current configuration Xt and all proposals are accepted.
Weighted global-jump scheme:
-
•
Proposal: Sample j ∈ {1, …, m} with probability p(j|Xt; ζ, π0);
-
•
Acceptance: Set Lt = j.
In Appendix A, we show that the non-weighted jump scheme with one jump attempt can be extended to a weighted jump scheme which, in the extreme case where consists of all m states for each k, also reduces to the weighted global-jump scheme above.
D. SOS-GST and derivation of local WHAM equations
In this section, we use GST in SOS48–53 as an analytical tool to derive local WHAM equations.47 Suppose that simulations have been completed so that an approximate sample of size nj is obtained from Pj for j = 1, …, m, with the total sample size . Let π = (π1, …, πm), where πj = nj/N is the observed proportion of the jth state. As emphasized above, the real simulations are allowed to be done by serial tempering, replica exchange, or other sampling algorithms including independent sampling of different thermodynamic states. The interest is then centered on how such samples can be combined to estimate free energies and equilibrium distributions.
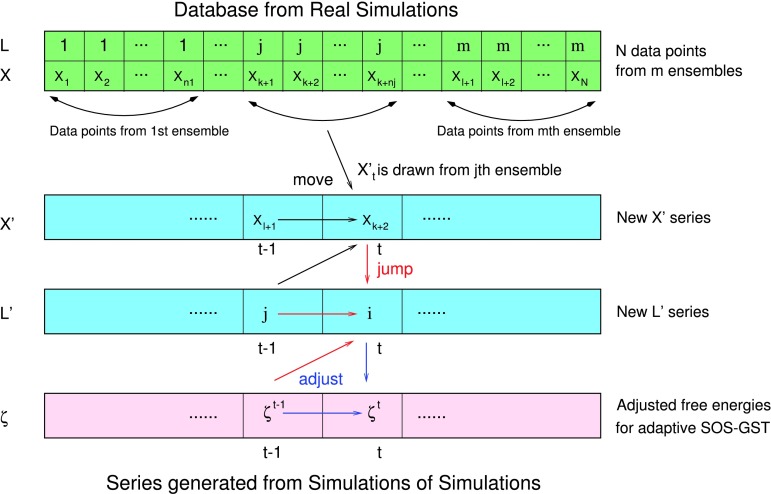
As illustrated in Fig. 1, a general idea of SOS is to apply a multicanonical sampling algorithm to resample datasets (such as binding energies in our application examples) obtained from simulations at multiple thermodynamic states.48,50,53 Specifically, consider a SOS procedure using GST described in Section II C, with two important choices: replacing the move process by simple random sampling without replacement from the individual sample datasets, and setting the target mixture weights identical to πj, i.e., , the proportion of the jth state contained in the original data. If the real simulations are done by synchronous replica exchange, then n1/N = ⋯ = nm/N = m−1. But (n1/N, …, nm/N) may in general differ from m−1, for example, when the real simulations are done by serial tempering or asynchronous replica exchange.45,46
FIG. 1.
Schematic representation of SOS-GST (first two steps) and adaptive SOS-GST (three steps): (1) black arrows show the move process in the configuration space; (2) red arrows denote the jump process in the thermodynamic state space; and (3) blue arrow represents the additional operation of adjusting the hypothesized free energies in the adaptive SOS-GST procedure.
SOS based on GST (SOS-GST as shown in Fig. 1):
-
•
Move: Given (in the jth state), resample from all data belonging to the jth state, {Xi : Li = j, i = 1, …, N}, with uniform probabilities ;
-
•
Jump: Generate given , by a non-weighted jump scheme possibly with multiple attempts (Section II C) or a weighted jump scheme (Appendix A), except that the target mixture weights π0 are set to π.
The foregoing procedure yields a valid Markov chain , with a stationary distribution, p′(j, x; ζ, π) (similar to p(j, x; ζ, π0) in Eq. (10) for real GST simulations), depending on the choice of hypothesized free energies ζ. The target mixture weights π0 are fixed as the observed proportions of states π in the simulated data. Throughout the discussion, we use superscripts ′ to indicate variables and functions defined in SOS. In Appendix B, we demonstrate several properties for the joint distribution p′(j, x; ζ, π). In particular, the marginal stationary probabilities p′(j; ζ, π), or in short , satisfy the following equations by detailed balance:
| (18) |
where and uj(l, x; ζ, π) denotes the jump probability from the lth to jth state given configuration x. It can be shown that gives the transition probability from the lth to jth state in the SOS chain. For the non-weighted jump scheme with one jump attempt, the jump probability uj(l, x; ζ, π) can be computed in a simple closed form by Eq. (A4), with π0 replaced by π. But the expression of uj(l, x; ζ, π) becomes analytically complicated when multiple jump attempts are performed per cycle. Local WHAM Eq. (22) will be derived regardless of the number of jump attempts per cycle, although numerical strategies for solving Eq. (22) and statistical properties may depend on whether one or multiple jump attempts are involved per cycle.
The marginal stationary probabilities , in general, differ from the target mixture weights πj = nj/N for the SOS procedure. Although there is no closed-form expression such as Eq. (12) for the marginal probabilities of states in real simulations, we show in Appendix B that can be related to the true free energies ζ∗, approximately by Eq. (12),
| (19) |
The approximation in Eq. (19) holds in an asymptotic sense as the original sample sizes nj increase to infinity for all thermodynamic states j = 1, …, m such that the time averages from the original data are close to the thermodynamic-ensemble averages. Similarly, as Eq. (12) leads to (13), taking logarithms of both sides of Eq. (19) shows that up to an additive constant, the true free energies can be approximated by
| (20) |
Eq. (20) is a SOS counterpart of Eq. (13) with π0 = π in real simulations. But an important difference is that serves only as an estimate of given the data, rather than itself.
Eq. (20) can be seen as a mapping to provide updated free energy estimates from initial guessed free energies ζ. Different choices of ζ, in general, lead to different estimates . By the idea of self-consistency, we require
| (21) |
That is, we require ζ to be such that the SOS stationary probabilities of states agree with the target mixture weights πj used in SOS-GST. By substituting Eq. (21) into Eq. (18), we then define the local WHAM free energy estimates as a solution, , to the system of estimating equations,
| (22) |
Similar ideas of self-consistency are used to derive global WHAM equations.9,14 In general, self-consistency can be applied and local WHAM equations can be extended in the case where the target mixture weights π0 in SOS-GST can be set different from the observed proportions π in the original data, even though we focus on the case of setting π0 identical to π. See Appendix C for such an extension of Eqs. (21) and (22).
Local WHAM Eq. (22) is derived above in the GST formulation using two separate parameters, ζ and π0 = π. The derivation can also be described in the standard ST formulation, where single biasing factors are defined as by Eq. (14) with set to πj. In fact, Eq. (20) can be recast as a SOS counterpart of Eq. (9),
| (23) |
where, by abuse of notation, is the same as but re-expressed to depend on only the biasing factors ζB. Moreover, self-consistency Eq. (21) can be recast as for j = 1, …, m. Hence, the local WHAM free energy estimates can be equivalently obtained (up to an additive constant) as
| (24) |
where is determined as a solution to
| (25) |
and, by abuse of notation, uj(Li, Xi; ζB) is the same as uj(Li, Xi; ζ, π) but re-expressed to depend on only the biasing factors ζB. Although Eq. (25) is expressed using only the biasing factors ζB, the solution of these equations, in general, gives free energy estimates only indirectly after the transformation by Eq. (24).
Local WHAM Eq. (22) depends on the choice of a jump scheme through jump probability uj(l, x; ζ, π) in SOS-GST. It is easy to see that if the weighted global-jump scheme is used with jump probability uj(l, x; ζ, π) defined as (A3) with π0 replaced by π, then local WHAM Eq. (22) reduces to global WHAM Eq. (3). In Appendix D, we show that for the non-weighted jump scheme with one jump attempt per cycle based on Metropolis acceptance probability (17), local WHAM Eq. (22) reduces to
| (26) |
for j = 1, …, m. As further discussed in Appendix D, solving Eq. (26) is equivalent to minimizing a convex function, similarly to the minimization method for solving global WHAM equations (3) 14,15 and local WHAM equations (22) with the jump probability based on Barker’s63 acceptance probability.47 In our numerical work, Eq. (26) is solved to obtain the local WHAM estimates , using a fast and reliable optimization algorithm which incorporates backtracking into Newton steps.
Similarly as global WHAM, the local WHAM method can be used to estimate equilibrium distributions, in addition to free energies. From the perspective of SOS, we define the local WHAM estimate of the jth equilibrium distribution Pj as the conditional distribution , induced from the joint stationary distribution for the SOS chain. As shown in Appendix B, this estimate of Pj is a discrete distribution , supported on the data with the probabilities
| (27) |
where nj is the number of data points Xi associated with Li = j. For the non-weighted jump scheme with one jump attempt per cycle, the local WHAM estimate is supported on the locally pooled data from the jth and its neighboring states, because is 0 if (i.e., Li is not included in the jth or neighboring states). In other words, Pj is approximated by attaching weight (27) to each observation Xi in the locally pooled data, where the weights sum up to 1 because satisfies Eq. (22). The global WHAM estimate in Eq. (4), giving a weight to each observation Xi from the pooled data from all thermodynamic states, corresponds to a special case of , where the jump probability uj(l, x; ζ, π) is defined as (A3) under the weighted global-jump scheme. This relationship justifies the names, local and global WHAMs.
The comparison between local and global WHAMs can be seen as follows in terms of computational cost and statistical efficiency. The local method is, by design, computationally far less costly than the global method. In fact, the local method based on the non-weighted jump scheme with one jump attempt per cycle requires, for each observation Xi, evaluating only the unnormalized density functions qj(Xi) for (j equal to the state of Xi in the simulated data or its neighboring states). The computational savings are substantial when the sizes of the neighborhoods are much smaller than the total number of distributions, m. On the other hand, statistical efficiency of the local method can be similar to that of the global method, because the accuracy in free energy estimation is, to a large extent, affected by the degree of overlaps between the distributions (P1, …, Pm),20 and each distribution Pj typically overlaps more with the neighboring distributions than with other distributions. Therefore, local WHAM can be much faster but almost as accurate as global WHAM, especially for handling a large number of distributions.
Finally, we point out that local WHAM equations (22), in particular Eq. (26), can also be justified as combining estimating equations by the BAR method over all pairs of neighboring states.47 By direct calculation, Eq. (26) can be re-expressed as an average, over , of equations of the form
| (28) |
or, with Eq. (16) substituted for , equivalently as
| (29) |
where αjl(x) is a scalar function of x, possibly depending on , but such dependency is suppressed in the notation. Eq. (29) is a sample analog of the BAR identity18 or equivalently the bridge-sampling identity20 for estimating the free energy difference, , between two thermodynamic states. Therefore, large-sample statistical theory can be established for the local WHAM estimators similarly as done for the global WHAM estimators and extended bridge sampling estimators.19,22 Such development will be reported elsewhere to further study statistical properties of local WHAM including the derivation of analytical formulas of asymptotic variances for error analysis.
E. Adaptive SOS-GST and stochastic solution for local WHAM
In Section II D, we derived local WHAM equations (22) by working with analytic formulas of stationary distributions resulting from SOS-GST with a freely specified but fixed choice of ζ. If the jump probability uj(l, x; ζ, π) is analytically tractable, for example, under the non-weighted jump scheme with one jump attempt per cycle, then Eq. (22) reduces to Eq. (26) and can be solved directly by a deterministic optimization algorithm. In this section, we develop an adaptive SOS-GST procedure to provide a stochastic solution of local WHAM equations (22), which is easily applicable even when multiple jump attempts are performed per cycle, and hence, the jump probability uj(l, x; ζ, π) is analytically complicated. Compared with the optimization method for local WHAM with one jump attempt per cycle, a potential advantage of the stochastic method using multiple jump attempts is to provide a better approximation to global WHAM, which can be obtained as the limit of local WHAM as the number of jump attempts per cycle increases to infinity.
As illustrated in Fig. 1, a stochastic solution of local WHAM Eq. (22) involves literarly running SOS-GST, but with an adaptive scheme for successively tuning the choice of hypothesized free energies ζ until self-consistency Eq. (21) is satisfied, i.e., the stationary probabilities of states for the SOS chain match the target proportions of states π in the original data. Specifically, consider the following adaptive SOS-GST procedure, which is similar to the non-adaptive SOS-GST procedure (Section II D) in generating , but incorporating an additional operation of adjusting the choice of hypothesized free energies ζ, denoted by , in each cycle t.
Adaptive SOS-GST (as shown in Fig. 1):
-
•
Move and Jump: Generate and then , as in the SOS-GST procedure in Section II D but with the hypothesized free energies ζ set to ζ(t−1).
-
•Update: For j = 1, …, m, set if or 0 otherwise, and update the hypothesized free energies ζ(t) as follows: for j = 1, …, m,
(30)
Eq. (31) is to ensure , and the coefficient γt is a function of t, decreasing to 0 as t → ∞, defined in two stages as(31)
where π∗ = min(π1, …, πm), t0 is a burn-in size, and 1/2 < α < 1 is an initial decay rate (discussed below).(32)
Adaptive scheme (30) for updating hypothesized free energies is constructed with a self-adjusting mechanism as follows to ensure that the sequence ζ(t) converges as t → ∞ to the local WHAM estimate . If is smaller (or greater) than for some state j, then the stationary distribution of the sequence will, on average over time, take value j more likely (or less likely) than with probability πj. This can be understood by analogy with the fact that by Eq. (12) in real GST simulations, a smaller (or greater) ζj leads to a greater (or smaller) marginal probability for L = j. By the update rule (30), will then be increased (or decreased) from to get close to . Theoretically, the adaptive SOS-GST procedure can be justified as a stochastic approximation algorithm54–57 to find the local WHAM estimate as a solution to Eq. (22). In fact, this procedure can be seen as a SOS application of a self-adjusted GST algorithm developed in Ref. 47 for real simulations, where the move process is implemented by Markov chain Monte Carlo.
Eq. (30) is qualitatively similar to the schemes previously used for adjusting free energy estimates in ST simulations,33,35 based on the Wang–Landau recursive scheme.64 However, the update coefficient γt in Eq. (30) is specified in two stages as in Ref. 47 by theory and practice of stochastic approximation. The minimum with π∗ is taken in Eq. (32) to ensure that the adjustment, , in update rule (30) is bounded between 0 and 1 for all t ≥ 1, even when the number of states m is large and some πj is small. On one hand, the particular choice of γt in the second stage is asymptotically equivalent to t−1 as t → ∞. Update scheme (30) with γt = t−1 is shown to be optimal,47 resulting in the minimum asymptotic variance for ζ(t), in spite of the fact that such optimal schemes are generally infeasible.55,56 On the other hand, a slow-decaying coefficient of order t−α is used in the first stage for 1/2 < α < 1, to introduce larger adjustments than with the coefficient t−1 and hence drive ζ(t) to fall faster into a neighborhood of the target .47
The adaptive SOS-GST procedure is particularly useful for solving local WHAM Eq. (22) stochastically when a jump scheme with multiple jump attempts is used per cycle. In such situations, the jump probability uj(l, x; ζ, π) is analytically complicated so that it is difficult to solve Eq. (22) by standard numerical methods. The adaptive SOS-GST procedure, however, does not require evaluation of jump probabilities uj(Li, Xi; ζ, π) appearing in Eq. (22). In effect, the jump probability uj(Li, Xi; ζ, π) is implicitly approximated by time-averages of the state indicators in update scheme (30).47
The output from adaptive SOS-GST, , can be used to approximate the local WHAM estimates of equilibrium distributions, in addition to free energies. After a large number (say T) of cycles of adaptive SOS-GST, the free energy estimate can be approximated by ζ(T). For equilibrium distribution Pj, the local WHAM estimate, defined as the conditional distribution , can be approximated by the empirical distribution (or histogram) of the jth SOS sample, . For a jump scheme with one jump attempt per cycle, each observation with is contained in the locally pooled data , and hence, the approximate local WHAM estimate of Pj amounts to weighting all observations Xi in the locally pooled data, similarly as the exact local WHAM estimate by Eq. (27). If a jump scheme with multiple (say r) jump attempts is used per cycle, then an observation with may be obtained as Xi with Li = j, or (i.e., Li belongs to the first-order neighborhood of j), or (i.e., Li belongs to the second-order neighborhood of j), etc., depending on the choice of r. Then, the approximate local WHAM estimate of Pj involves properly weighting observations Xi with the original labels Li not only in the first-order neighborhood of j but also in up to rth-order neighborhood of j. Such an estimate of Pj can be very close to global WHAM estimate (4) for a large number of jump attempts r per cycle and approaches the latter as r increases to infinity.
The adaptive SOS-GST procedure with multiple jump attempts per cycle is, in general, computationally more intensive than the optimization method for local WHAM with one jump attempt per cycle, although it remains much faster than standard implementations of global WHAM.13,14 On the other hand, there is a potential advantage of using the stochastic method with multiple jump attempts, as opposed to the optimization method for local WHAM. As mentioned in Section II C, a non-weighted jump scheme with infinitely many jump attempts yields the weighted global-jump scheme, for which local WHAM Eq. (22) reduces to global WHAM Eq. (3). Hence, local WHAM with multiple jump attempts can provide a better approximation to global WHAM than with only one jump attempt per cycle. In our application (Section III), the two methods of local WHAM yield similar estimates to global WHAM estimates for both free energies and equilibrium distributions in the analysis of well converged datasets. When applied to poorly converged datasets, the two local WHAM estimates are still similar to global WHAM estimates for free energies, but the estimates of equilibrium distributions from the stochastic method based on multiple jump attempts per cycle agree with global WHAM estimates noticeably better than the local WHAM estimates based on one jump attempt per cycle.
Finally, we briefly discuss the relationship between adaptive SOS-GST and the SOS-RE procedures which we recently proposed53 for a stochastic solution to the global WHAM equations. Both procedures are developed using the idea of SOS: SOS-RE is based on running RE simulations whereas (adaptive) SOS-GST is based on running (adaptive) GST simulations, such that the “move” process is replaced by simple random sampling from individual sample datasets. But there are subtle differences between the two procedures in addition to RE versus GST. The SOS-RE procedure involves updating the individual datasets when an exchange attempt between two configurations from two states is accepted. As a result, the output histogram of SOS-RE for jth state can provide an approximation of the global WHAM estimate of jth equilibrium distribution, where all observations from the m states are associated with positive weights. In contrast, adaptive SOS-GST involves updating hypothesized free energies, without changing individual datasets. The output histogram of adaptive SOS-GST for jth state is, in general, an approximation of the local WHAM estimate of jth equilibrium distribution, where only observations from up to rth-order neighborhood of jth state are assigned positive weights. A systematic comparison of the performances of these procedures will be pursued in future work.
III. APPLICATION
A. BEDAM method
The BEDAM58–60,65,66 is a novel approach developed recently in our group to calculate absolute binding free energies for receptor-ligand systems. The BEDAM method is based on a sound statistical mechanics theory67 of molecular association, efficient computational strategies built upon parallel Hamiltonian replica exchange sampling (λ hopping),68,69 and thermodynamic reweighting to recover unbiased results.10,12–14 In this method, the total potential energy of the receptor-ligand complex can be reduced to the dimensionless form as
| (33) |
where β = 1/(kBT), and λ is an alchemical parameter ranging from 0, corresponding to the uncoupled state of the complex, to 1, corresponding to the fully coupled state of the complex. H0(x) is the potential energy of the complex when receptor and ligand are uncoupled, that is, as if they were separated at infinite distance from each other. The binding energy, b(x), is defined as the change in effective potential energy of the complex for bringing the receptor and ligand from infinite separation to the given conformation x of the complex.
The BEDAM method calculates the binding free energy between a receptor A and a ligand B using the AGBNP implicit solvation model70,71 as
| (34) |
where C∘ (=1M) is the standard concentration of ligand molecules, Vsite is the volume of the binding site, and p0(b) is the probability distribution of binding energy (b(x) in Eqs. (33) and (34)) collected in an appropriate decoupled ensemble of conformations in which the ligand is confined in the binding site while the receptor and the ligand are not interacting with each other but both only with the solvent continuum. For the subsequent calculation, the binding free energy is simplified as △Gb = − kBTlog[∫p0(b) e−βbdb], ignoring the additive constant −kBTln(C∘Vsite).
After introducing the intermediate states with different λ values, the BEDAM method58 was implemented in the IMPACT72 molecular simulation package using the synchronous nearest-neighbor exchange scheme and performing Hamiltonian replica exchange in the λ space and more recently in the ASyncRE python package using asynchronous pairwise independence exchange scheme45,46 for a multidimensional parameter space including both λ and temperature. The purpose of sampling along λ is to enhance mixing of conformations along the alchemical pathway while high temperatures enhance sampling of internal molecular degrees of freedom at each alchemical state. In this work, we employed different reweighting methods to estimate binding energy distributions p0(b) and binding free energies △Gb from binding energy samples obtained from 1D and 2D REMD simulations.
B. REMD simulations of a model system
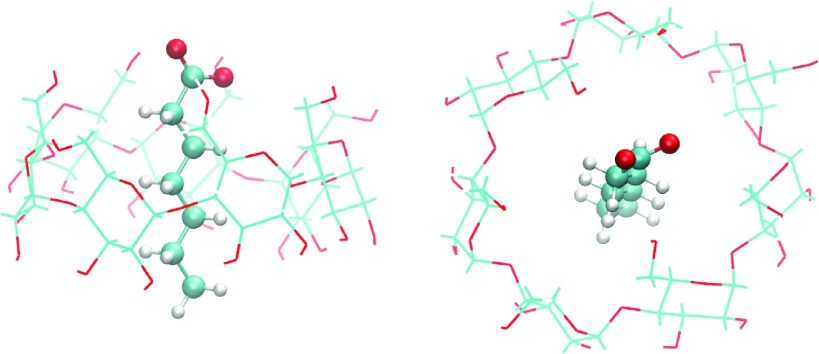
The model complex, β-cyclodextrin-heptanoate, as depicted in Fig. 2, is one of the host-guest systems investigated in our previous work.59,60 We performed standard 1D synchronous BEDAM simulations at 16 λ values (0.0, 0.001, 0.002, 0.004, 0.01, 0.04, 0.07, 0.1, 0.2, 0.4, 0.6, 0.7, 0.8, 0.9, 0.95, and 1.0) with a MD period of 0.5 ps per RE cycle, using the OPSL-AA force field73,74 and the AGBNP2 implicit solvent model.71 In total, we obtained 15 independent 1D BEDAM simulations at 15 different temperatures (200, 206, 212, 218, 225, 231, 238, 245, 252, 260, 267, 275, 283, 291, and 300 K). Each has 1.152 μs aggregated 1D BEDAM simulations with 16 replicas (72 ns for each replica). The binding energies were saved every 0.5 ps, resulting in a huge number of data points (144 000 × 240 = 34.56 × 106) from 16 × 15 = 240 states. Using this large set of data, we applied different WHAM methods to calculate binding free energies and binding free energy distributions. We will show below that even though the data are not fully converged at some thermodynamic states, our 2D WHAM methods can obtain results comparable with converged datasets.
FIG. 2.
Side and top view of β-cyclodextrin-heptonoate complex.
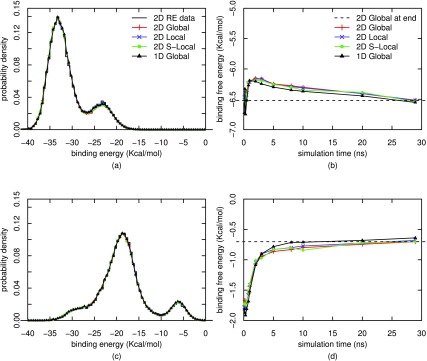
To serve as the benchmark for the comparisons between different methods, we adopted a well-converged dataset from our recent 2D asynchronous REMD simulations45 with the same 16 λ values and the same 15 temperatures resulting in a total of 16 × 15 = 240 replicas associated with 240 pairs of (λ, β) values. The binding energies were recorded every 1 ps during the simulation time of 30 ns per replica, resulting in a total of 7.2 × 106 data points (30 000 × 240). The binding energy distributions at λ = 1.0 are displayed in Fig. 3 for two temperatures (200 K and 300 K) after the UWHAM reweighting.14 It is evident that multiple peaks appear in the binding energy distributions. From our previous studies, we know that those peaks correspond to different orientations of heptanoate in the host cavity, each characterized by different hydrogen bonding patterns with β-cyclodextrin.59,60 The binding free energies estimated from these two simulations are −0.70 and −6.52 kcal/mol for 300 and 200 K, respectively (including the constant multiple −kBT).
FIG. 3.
Free energies and binding energy distributions for λ = 1 at two different temperatures, calculated by four different reweighting methods from the well-converged 2D Async REMD simulations. T = 200 K for (a) and (b), and T = 300 K for (c) and (d).
C. Comparison of different WHAMs
Figure 3 exhibits the binding free energies and binding energy distributions for λ = 1.0 at temperatures 200 and 300 K (see Figs. S1 and S2 in the supplementary material75 for all 15 temperatures), calculated from the well-converged 2D Async REMD simulations using the following WHAM methods.
-
•
2D Global: the minimization method of global WHAM14 applied to all data from 240 states, defined as a 16 × 15 grid of (λ, β) values;
-
•
2D Local: the minimization method of local WHAM, based on the non-weighted, nearest-neighbor jump scheme with one jump attempt per cycle;
-
•
2D S-Local: the stochastic method of local WHAM (i.e., the adaptive SOS-GST procedure), based on the non-weighted nearest-neighbor jump scheme with 10 jump attempts per cycle;
-
•
1D Global: global WHAM applied separately to 15 datasets, each obtained from 16 λ values at one of the 15 temperatures.
The adaptive SOS-GST procedure is run for a total of T = t0 + 4.8 × 107 cycles, with the burn-in size t0 = 4.8 × 106 and initial decay rate α = 0.6 in Eq. (32).
As shown in Figs. 3, S1, and S2,75 the free energy estimates from different WHAM methods are similar over the entire range of simulation time (i.e., converging as fast as each other). Moreover, the distributions of binding energies from different WHAM reweightings agree closely with those from the raw data, suggesting that the 2D Async REMD simulations are well equilibrated at all thermodynamic states. The convergence of binding energy distributions to the same distribution also ensures the correctness of our implementations of different reweighting methods. Hence, there are no significant differences except the Central Processing Unit (CPU) time (see further discussion below) when the four different reweighting methods are applied to the well-converged data from 2D Async REMD simulations. But such large-scale 2D simulations are computationally intensive.45,46 For the rest of this section, we will focus on the analysis of another dataset from 15 independent 1D REMD simulations (which are easier to implement than 2D simulations) and find that the differences of reweighting methods become more pronounced as the raw data are not fully converged at some thermodynamic states (see Fig. S375 for the binding energy distributions from the raw data).
Table I displays the CPU time for different reweighting methods we implemented, using the dataset from the 15 independent 1D REMD simulations. Each method is successively applied to the data up to the following simulation time (in ns per state): 0.05, 0.10, 0.15, 0.25, 0.4, 0.5, 1.0, 1.5, 2.5, 4, 5, 10, 15, 25, 40, 50, and 72. The initial values of free energies are set to the estimates from the previous period, except for the first period where all initial values are set to 0. As might be broadly used, this stagewise scheme leads to much faster implementation for the optimization methods 2D Global, 2D Local, and 1D Global than a naive application where the initial values of free energies are set to 0 regardless of data sizes. (In practice, initial values can also be obtained from some inexpensive method as discussed in Ref. 13.) For 2D S-Local, the stagewise scheme is generally less effective than a direct application with the initial values set to 0 or inexpensive estimates. As discussed in Section II E, the free energy estimates ζ(t) can fall fast near the target values in the first stage of SA recursion (30) using a slow-decaying coefficient γt.47 Nevertheless, the comparison in Table I is valid in terms of CPU time for individual stages.
TABLE I.
CPU time for different WHAM methods.
| 2D Global | 2D Local | 2D S-Local | 1D Global | |
|---|---|---|---|---|
| Data size | N = 1.2 × 107 (25 ns per state) | |||
| CPU timea | 19510.1 s | 39.7 | 436.4 | 159.7 |
| Improvement rate | 1 | 491.8 | 44.7 | 122.1 |
| Data size | N = 3.5 × 107 (72 ns per state) | |||
| CPU time | 56411.6 s | 114.0 | 453.3 | 458.3 |
| Improvement rate | 1 | 494.8 | 124.5 | 123.1 |
It is evident that 2D local WHAM using the minimization method with one jump attempt per cycle is the fastest, with the improvement rate more than 490 times over 2D global WHAM and even 40 times over 1D global WHAM for which the data at each temperature are processed as one independent dataset. The 2D stochastic local WHAM with 10 jump attempts per cycle is slower than 2D local WHAM using the minimization method, but the improvement rate is still more than 120 times over 2D global WHAM when all data points are included. However, the stochastic method with multiple jump attempts per cycle can lead to more accurate estimates of binding energy distributions than local WHAM with one jump attempt per cycle when applied to data not fully converged as shown below.
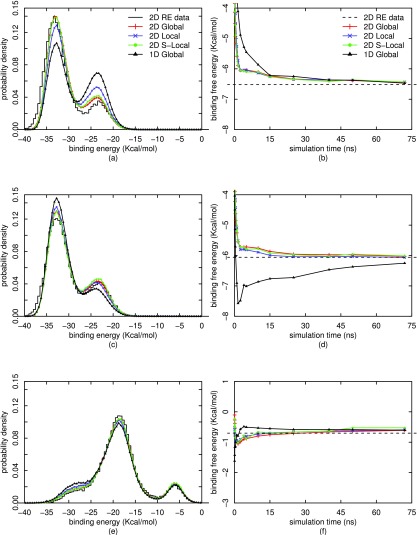
Figure 4 shows the binding free energies as a function of simulation time, as well as the final binding energy distributions for λ = 1.0 at three temperatures 200, 206, and 300 K (see Figs. S4 and S575 for all 15 temperatures), calculated from 15 independent 1D REMD simulations at 15 temperatures using different reweighting methods. We find that the two methods of 2D local WHAM, based on either one or multiple jump attempts per cycle, give similar estimates of free energies to those from 2D global WHAM, and all three estimates appear to converge much faster over the simulation time than 1D global WHAM to the true values (estimated by 2D global WHAM from the benchmark dataset of 2D Async REMD simulations). Even though the 2D local and global WHAM estimates of free energies may, in some cases, be similar to the 1D global WHAM estimates in point values, the standard errors associated with 2D local and global WHAM (computed by block bootstrap as in Ref. 14) are smaller than those from 1D global WHAM (see Fig. S6).75 The underlying rationale for the faster convergence and the smaller variance (fluctuation) of 2D WHAMs than 1D WHAM is that 2D WHAMs include the contributions of data from multiple other temperatures instead of only one target temperature in the 1D WHAM.
FIG. 4.
Free energies and binding energy distributions for λ = 1 at three different temperatures, calculated by four different reweighting methods from 15 independent 1D REMD simulations. T = 200 K for (a) and (b), T = 206 K for (c) and (d), and T = 300 K for (e) and (f). The bar lines in the distribution plots are raw histograms from 2D Async REMD simulations, and the dashed lines in the free energy plots are the global WHAM estimates from the 2D simulations.
The slower convergence of 1D global WHAM can also be observed from the estimates of binding energy distributions in Figs. 4(a) and S5:75 these estimates are not yet converged after 72 ns 1D REMD simulations and appear to be biased at low temperatures as evaluated against the benchmark estimates from the 2D Async REMD simulations. In contrast, the 2D local and global WHAM estimates show improvement over those from 1D global WHAM. The better performance of 2D WHAMs can be confirmed not only from smaller differences from the benchmark histograms from the 2D Async REMD simulations but also directly from the improvement of the left peak in Figs. 4(a) and S575 since the left peak should become more pronounced as the temperature is decreased. This corresponds to a full equilibration of the two major heptonoate guest orientations within the host.
From Figs. 4(a) and S5,75 we also see that 2D local WHAM based on one jump attempt per cycle shows some discrepancy in the estimates of binding energy distributions (although not in free energy estimates) from the 2D global WHAM estimates at low temperatures. Compared with these local WHAM estimates, the 2D stochastic local WHAM based on multiple jump attempts per cycle gives estimates of binding energy distributions more closely matching the 2D global WHAM estimates and the benchmark histograms from 2D Async REMD simulations. Hence, although both 2D local WHAMs achieve better convergence than 1D global WHAM, the stochastic method based on multiple jump attempts per cycle is superior to the minimization method based on one jump attempt per cycle in more accurately estimating binding energy distributions, although the stochastic method takes longer CPU time. The advantage of the stochastic method lies in the use of multiple jump attempts per cycle, such that the data points are redistributed from a large subset of thermodynamic states through the jump process according to detailed balance, and hence, a better approximation to the global equilibrium can be obtained for each thermodynamic state.
IV. DISCUSSION AND CONCLUSION
In this report, we develop local WHAM and a stochastic solution to analyze data from multi-ensemble simulations from the perspective of simulations of simulations using generalized serial tempering. The local WHAM involves reweighting data points only across neighboring thermodynamic states instead of all states under study, and hence has computational cost which scales linearly as the number of thermodynamic states, instead of quadratically as in standard implementations of global WHAM.13,14 The improvement rate of local over global WHAM in CPU time is about 490 times for the minimization method with one jump attempt per cycle, and 120 times for the stochastic method with multiple jump attempts per cycle, in our application of analyzing a large dataset obtained from REMD simulations with 240 thermodynamic states.
All of the four reweighting methods (2D Global, Local, S-Local, and 1D Global WHAM) lead to similar estimates of free energies and equilibrium distributions, and the 2D local WHAM using the minimization method is the fastest, for a well-converged dataset from 2D Async REMD simulations. But such large-scale 2D simulations are computationally intensive.45,46 On the other hand, these methods show substantial differences in their estimates of free energies and equilibrium distributions for a dataset with limited equilibration from independent 1D REMD simulations, which are easier to implement than 2D simulations. The two methods of 2D local WHAM, either deterministic with one jump attempt per cycle or stochastic with multiple jump attempts per cycle, result in estimates of free energies similar to 2D global WHAM estimates, but are computationally orders of magnitude less costly than 2D global WHAM. Moreover, the 2D local WHAM estimates, like the 2D global WHAM estimates, converge faster over simulation time than 1D global WHAM estimates to the benchmark estimates computed from well-converged 2D Async REMD simulations. The stochastic method for local WHAM with multiple jump attempts per cycle, however, appears to outperform the minimization method with one jump attempt per cycle, in more accurately estimating the binding energy distributions, at some increased computational cost but still substantially faster than global WHAM.
In summary, these results demonstrate the benefit of using local WHAM (including the stochastic solution) as a fast and accurate method to incorporate information from multiple temperatures even for estimating binding free energies along the λ dimension at individual temperatures in our application. More generally, the proposed methods can be used to effectively extract information from all the thermodynamic states for free energy estimation in large-scale multi-state simulations, especially when some of the simulations are not fully equilibrated. Recently, alternative methods76–78 to WHAM have been proposed to estimate kinetic and equilibrium properties by working with transition matrices in Markov state models for multi-state simulations. These methods appear to be useful for estimating kinetic properties. Their performance of estimating equilibrium properties compared with WHAM remains a question of interest for future research.
Acknowledgments
This work has been supported in part by the National Science Foundation (CDI type II 1125332) and the National Institutes of Health (Nos. GM30580 and P50 GM103368) awarded to R.M.L. We thank Emilio Gallicchio and Peng He for helpful discussions about the BEDAM simulations of the host-guest binding system used as an example in this paper. REMD simulations were carried out on the Gordon, Trestles, Stampede, Comet, and SuperMIC clusters of XSEDE resources (supported by No. TG-MCB100145).
APPENDIX A: WEIGHTED JUMP SCHEME AND JUMP PROBABILITIES
The jump scheme described in Section II C is referred to as non-weighted, because the proposal probabilities Γ(Lt−1, ⋅ ) remain fixed, regardless of the current observation Xt. Alternatively, the proposal probabilities of jumping from Lt−1 to j can be chosen depending on Xt, for example, in proportion to the conditional probabilities p(j|Xt; ζ, π0) for . This leads to the following jump scheme.
Weighted jump scheme with one jump attempt:
-
•Proposal: Sample (select) with probability
(A1) -
•Acceptance-rejection: Set (accept) Lt = j with probability
and, with the remaining probability, set Lt = Lt−1.(A2)
In the extreme case where consists of all m states for each k, the weighted jump scheme yields a weighted global-jump scheme with no rejection of proposed jumps as mentioned in Section II C. It should be noted that, in this extreme case, the non-weighted jump scheme will lead to a different global-jump scheme, which involves a non-vanishing rejection step and does not seem to be of interest in the subsequent development.
Let uj(l, x; ζ, π0) be the jump probability from l to j given the current observation x. Under the weighted global-jump scheme, we have
| (A3) |
which is in fact independent of l. In general, uj(l, x; ζ, π0) can be computed as follows for the non-weighted and weighted jump schemes with one jump attempt per cycle.
Under the non-weighted jump scheme, we have
| (A4) |
and uj(l, x; ζ, π0) = 0 if . Alternatively, Metropolis acceptance probability (17) can be replaced by Barker’s acceptance probability63 to obtain the following jump probability used in Ref. 47:
| (A5) |
and uj(l, x; ζ, π0) = 0 if . Finally, under the weighted jump scheme, we have
| (A6) |
and uj(l, x; ζ, π0) = 0 if .
APPENDIX B: STATIONARY DISTRIBUTION OF SOS CHAIN
The SOS chain is constructed in the following order: . That is, depends on the history only through , and depends on the history only through . Then, the sequence is also a valid Markov chain and has a joint stationary distribution with the density , where as before, and denotes the empirical distribution, placing probability at each observation in the jth sample of size nj, {Xi : Li = j, i = 1, …, N}. More precisely, is the density function of such a discrete distribution with respect to the counting measure on the pooled data . All elements Xi in are treated as distinct, even though one element may be numerically equal to another element in .
From the joint stationary distribution of , the marginal stationary distribution of is , i.e., a mixture distribution with component densities and weights . Moreover, the joint stationary distribution of is
| (B1) |
By summation of the above expression over , the marginal stationary distribution of satisfies Eq. (18).
To demonstrate the approximation of by Eq. (19), assume that are uniquely determined by Eq. (18); otherwise the Markov chain would admit multiple stationary distributions. Then, it suffices to show that Eq. (19) holds, with replaced by and replaced by the equilibrium expectation 〈uj(l, x; ζ, π)〉l under state l,
| (B2) |
But Eq. (B2) follows by summing over (l, x) the detailed balance equation
| (B3) |
which is satisfied by the construction of the jump schemes.
By definition, the local WHAM estimate satisfies for j = 1, …, m. Note that if x represents an observation Xi with Li = l, and otherwise. Then, the marginal stationary distribution of is for all . The joint stationary distribution of is
| (B4) |
Dividing Eq. (B4) by the marginal stationary probability shows that the conditional stationary distribution is given by Eq. (27).
APPENDIX C: GENERALIZED GLOBAL AND LOCAL WHAMS
Suppose that for SOS-GST in Section II C, the target mixture weights π0 are set possibly different from the observed proportions of states π. Nevertheless, the proof in Appendix B remains valid, with π replaced by π0. Then, the stationary marginal probabilities of the SOS chain satisfy the equations, similar to Eq. (19),
| (C1) |
where .
By self-consistency, we require ζ to be such that the SOS stationary probabilities of states agree with the target mixture weights for j = 1, …, m. Hence, we define the generalized local WHAM estimates of free energies as a solution, , to the system of estimating equations (similar to Eq. (22)),
| (C2) |
Setting π0 = π in Eq. (C2) leads back to local WHAM equation (22).
If the jump probability uj(l, x; ζ, π0) is defined as (A3) under the weighted global-jump scheme, then Eq. (C2) reduces to
| (C3) |
This generalizes global WHAM equation (3) in allowing π0 possibly different from π. The mixture weights π0 can be interpreted as the proportions of effective sample sizes of states, accounting for possible autocorrelations in the simulated data. If the simulated data are independent from each state, then π0 should be identical to π. For dependent data, use of the effective proportions π0 may lead to better estimation of free energies. This topic can be investigated for both global and local WHAMs in future work.
APPENDIX D: LOCAL WHAM EQUATIONS
For the jump probability uj(l, x; ζ, π), Tan47 suggested choice Eq. (A5) based on Barker’s acceptance probability63 under the non-weighted jump scheme with one jump attempt per cycle and demonstrated several useful properties of this choice. Solving Eq. (22) for the choice (A5) is equivalent to minimizing a convex function in ζ,
| (D1) |
where hbar(r) = log(1 + r). In the following, we discuss related properties of choices (A4) and (A6) for the jump probability uj(l, x; ζ, π).
First, we show that Eq. (22) reduces to (26) for choice (A4) based on Metropolis acceptance probability (17) under the non-weighted jump scheme with one jump attempt per cycle. If Li = l for some l such that or equivalently , then
| (D2) |
If Li = j, then
| (D3) |
| (D4) |
Substituting these expressions in Eq. (22) and noticing that yield Eq. (26).
Second, we show that solving Eq. (26) for choice (A4) is equivalent to minimizing a function similar to κbar(ζ) above,
| (D5) |
where hmet(r) = r if r ≤ 1 and 1 + logr if r > 1. For any fixed 1 ≤ j ≤ m, rewrite κmet(ζ) as
| (D6) |
where does not depend on ζj. By directly taking derivatives by the chain rule and noticing that (d/dr) hmet(r) = min(1, r−1), we have
| (D7) |
and
| (D8) |
Substituting these expressions in (∂/∂ζj) κmet(ζ) = 0 yields Eq. (26).
Third, we show that κmet(ζ) is convex in ζ. Rewrite κmet(ζ) as
| (D9) |
which is a sum of functions of ζ, each in the form for some constant c ≥ 0 free of ζ. The convexity of κmet(ζ) follows because is convex in ζ.
Finally, it can be shown, by similar calculation as in obtaining Eq. (26), that for the jump probability uj(l, x; ζ, π) given by Eq. (A6) under the weighted jump scheme, local WHAM Eq. (22) reduces to the following equations:
| (D10) |
for j = 1, …, m, where . In contrast with the global WHAM and the local WHAM under the non-weighted jump scheme, solving Eq. (D10) cannot be made equivalent to minimizing a scalar function (much less a convex function), because the derivative matrix with respect to ζ of the vector defined by the right hand side of (D10) for j = 1, …, m is, in general, asymmetric.
REFERENCES
- 1.Dellago C., Bolhuis P. G., and Chandler D., J. Chem. Phys. 108, 9236 (1998). 10.1063/1.476378 [DOI] [Google Scholar]
- 2.Zuckerman D. M. and Woolf T. B., J. Chem. Phys. 111, 9475 (1999). 10.1063/1.480278 [DOI] [Google Scholar]
- 3.Laio A. and Parrinello M., Proc. Natl. Acad. Sci. U. S. A. 99, 12562 (2002). 10.1073/pnas.202427399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamelberg D., Mongan J., and McCammon J. A., J. Chem. Phys. 120, 11919 (2004). 10.1063/1.1755656 [DOI] [PubMed] [Google Scholar]
- 5.Zhang B. W., Jasnow D., and Zuckerman D. M., Proc. Natl. Acad. Sci. U. S. A. 104, 18043 (2007). 10.1073/pnas.0706349104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan A. C., Sezer D., and Roux B., J. Phys. Chem. B 112, 3432 (2008). 10.1021/jp0777059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawk A. T., Konda S. S. M., and Makarov D. E., J. Chem. Phys. 139, 064101 (2013). 10.1063/1.4817200 [DOI] [PubMed] [Google Scholar]
- 8.Arrar M., de Oliveira C. A. F., Fajer M., Sinko W., and McCammon J. A., J. Chem. Theory Comput. 9, 18 (2013). 10.1021/ct300896h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrenberg A. and Swendsen R., Phys. Rev. Lett. 63, 1195 (1989). 10.1103/PhysRevLett.63.1195 [DOI] [PubMed] [Google Scholar]
- 10.Kumar S., Rosenberg J. M., Bouzida D., Swendsen R. H., and Kollman P. A., J. Comput. Chem. 13, 1011 (1992). 10.1002/jcc.540130812 [DOI] [Google Scholar]
- 11.Bartels C. and Karplus M., J. Comput. Chem. 18, 1450 (1997). [DOI] [Google Scholar]
- 12.Gallicchio E., Andrec M., Felts A. K., and Levy R. M., J. Phys. Chem. B 109, 6722 (2005). 10.1021/jp045294f [DOI] [PubMed] [Google Scholar]
- 13.Shirts M. R. and Chodera J. D., J. Chem. Phys. 129, 124105 (2008). 10.1063/1.2978177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Z., Gallicchio E., Lapelosa M., and Levy R. M., J. Chem. Phys. 136, 144102 (2012). 10.1063/1.3701175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu F. and Hummer G., J. Comput. Chem. 33, 453 (2012). 10.1002/jcc.21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C., Lai C.-L., and Pettitt B. M., “Accelerating the weighted histogram analysis method by direct inversion in the iterative subspace,” Mol. Simul. (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng Y. and Roux B., J. Chem. Theory Comput. 11, 3523 (2015). 10.1021/ct501130r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett C. H., J. Comput. Phys. 22, 245 (1976). 10.1016/0021-9991(76)90078-4 [DOI] [Google Scholar]
- 19.Geyer C. J., “Estimating normalizing constants and reweighting mixtures in Markov chain Monte Carlo,” Technical Report, University of Minnesota, School of Statistics, 1994. [Google Scholar]
- 20.Meng X.-L. and Wong W. H., STAT SINICA 6, 831 (1996). [Google Scholar]
- 21.Kong A., McCullagh P., Meng X.-L., Nicolae D., and Tan Z., J. R. Statist. Soc. Ser. B 65, 585 (2003). 10.1111/1467-9868.00404 [DOI] [Google Scholar]
- 22.Tan Z., J. Am. Stat. Assoc. 99, 1027 (2004). 10.1198/016214504000001664 [DOI] [Google Scholar]
- 23.Swendsen R. and Wang J.-S., Phys. Rev. Lett. 57, 2607 (1986). 10.1103/PhysRevLett.57.2607 [DOI] [PubMed] [Google Scholar]
- 24.Geyer C. J., Computing Science and Statistics: Proceedings of 23rd Symposium on the Interface (Interface Foundation of North America, 1991), p. 156. [Google Scholar]
- 25.Hansmann U. H., Chem. Phys. Lett. 281, 140 (1997). 10.1016/s0009-2614(97)01198-6 [DOI] [Google Scholar]
- 26.Sugita Y. and Okamoto Y., Chem. Phys. Lett. 314, 141 (1999). 10.1016/S0009-2614(99)01123-9 [DOI] [Google Scholar]
- 27.Sugita Y., Kitao A., and Okamoto Y., J. Chem. Phys. 113, 6042 (2000). 10.1063/1.1308516 [DOI] [Google Scholar]
- 28.Mitsutake A. and Okamoto Y., J. Chem. Phys. 121, 2491 (2004). 10.1063/1.1766015 [DOI] [PubMed] [Google Scholar]
- 29.Rauscher S., Neale C., and Pomes R., J. Chem. Theory Comput. 10, 2640 (2009). 10.1021/ct900302n [DOI] [PubMed] [Google Scholar]
- 30.Marinari E. and Parisi G., Europhys. Lett. 19, 451 (1992). 10.1209/0295-5075/19/6/002 [DOI] [Google Scholar]
- 31.Lyubartsev A. P., Martsinovski A. A., Shevkunov S. V., and Vorontsov-Velyaminov P. N., J. Chem. Phys. 96, 1776 (1992). 10.1063/1.462133 [DOI] [Google Scholar]
- 32.Geyer C. J. and Thompson E. A., J. Am. Stat. Assoc. 90, 909 (1995). 10.1080/01621459.1995.10476590 [DOI] [Google Scholar]
- 33.Li H., Fajer M., and Yang W., J. Chem. Phys. 126, 024106 (2007). 10.1063/1.2424700 [DOI] [PubMed] [Google Scholar]
- 34.Park S. and Pande V. S., Phys. Rev. E 76, 016703 (2007). 10.1103/PhysRevE.76.016703 [DOI] [PubMed] [Google Scholar]
- 35.Zhang C. and Ma J., Phys. Rev. E 76, 036708 (2007). 10.1103/PhysRevE.76.036708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X., Bowman G. R., and Pande V. S., J. Chem. Phys. 128, 205106 (2008). 10.1063/1.2908251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chelli R., J. Chem. Theory Comput. 6, 1935 (2010). 10.1021/ct100105z [DOI] [PubMed] [Google Scholar]
- 38.Chodera J. D. and Shirts M. R., J. Chem. Phys. 135, 194110 (2011). 10.1063/1.3660669 [DOI] [PubMed] [Google Scholar]
- 39.Nguyen P. H., Okamoto Y., and Derreumaux P., J. Chem. Phys. 138, 061102 (2013). 10.1063/1.4792046 [DOI] [PubMed] [Google Scholar]
- 40.Mitsutake A. and Okamoto Y., J. Chem. Phys. 130, 214105 (2009). 10.1063/1.3127783 [DOI] [PubMed] [Google Scholar]
- 41.Mitsutake A., J. Chem. Phys. 131, 094105 (2009). 10.1063/1.3204443 [DOI] [PubMed] [Google Scholar]
- 42.Jiang W. and Roux B., J. Chem. Theory Comput. 6, 2559 (2010). 10.1021/ct1001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang W., Luo Y., Maragliano L., and Roux B., J. Chem. Theory Comput. 8, 4672 (2012). 10.1021/ct300468g [DOI] [PubMed] [Google Scholar]
- 44.Kokubo H., Tanaka T., and Okamoto Y., J. Comput. Chem. 34, 2601 (2013). 10.1002/jcc.23427 [DOI] [PubMed] [Google Scholar]
- 45.Xia J., Flynn W. F., Gallicchio E., Zhang W., He B. P., Tan Z., and Levy R. M., J. Comput. Chem. 36, 1772 (2015). 10.1002/jcc.23996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallicchio E., Xia J., Flynn W. F., Zhang B., Samlalsingh S., Mentes A., and Levy R. M., Comput. Phys. Commun. 196, 236 (2015). 10.1016/j.cpc.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan Z., “Optimally adjusted mixture sampling and locally weighted histogram analysis,” J. Comput. Graphical Stat. (published online). 10.1080/10618600.2015.1113975 [DOI] [Google Scholar]
- 48.Zheng W., Andrec M., Gallicchio E., and Levy R. M., Proc. Natl. Acad. Sci. U. S. A. 104, 15340 (2007). 10.1073/pnas.0704418104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hritz J. and Oostenbrink C., J. Chem. Phys. 127, 204104 (2007). 10.1063/1.2790427 [DOI] [PubMed] [Google Scholar]
- 50.Zheng W., Andrec M., Gallicchio E., and Levy R. M., J. Phys. Chem. B 112, 6083 (2008). 10.1021/jp076377+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosta E. and Hummer G., J. Chem. Phys. 131, 165102 (2009). 10.1063/1.3249608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith D. B., Okur A., and Brooks B., Chem. Phys. Lett. 545, 118 (2012). 10.1016/j.cplett.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang B. W., Xia J., Tan Z., and Levy R. M., J. Phys. Chem. Lett. 6, 3834 (2015). 10.1021/acs.jpclett.5b01771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robbins H. and Monroe S., Ann. Stat. 22, 400 (1951). 10.1214/aoms/1177729586 [DOI] [Google Scholar]
- 55.Benveniste A., Métivier M., and Priouret P., Adaptive Algorithms and Stochastic Approximations (Springer, New York, 1990). [Google Scholar]
- 56.Chen H.-F., Stochastic Approximation and Its Applications (Kluwer Academic Publishers, Dordrecht, 2002). [Google Scholar]
- 57.Song Q., Wu M., and Liang F., Adv. Appl. Probab. 46, 1059 (2014). 10.1239/aap/1418396243 [DOI] [Google Scholar]
- 58.Gallicchio E., Lapelosa M., and Levy R. M., J. Chem. Theory Comput. 6, 2961 (2010). 10.1021/ct1002913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallicchio E. and Levy R. M., J. Comput.-Aided Mol. Des. 26, 505 (2012). 10.1007/s10822-012-9552-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickstrom L., He P., Gallicchio E., and Levy R. M., J. Chem. Theory Comput. 9, 3136 (2013). 10.1021/ct400003r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang C. and Ma J., J. Chem. Phys. 129, 134112 (2008). 10.1063/1.2988339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosta E. and Hummer G., J. Chem. Phys. 132, 034102 (2010). 10.1063/1.3290767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barker A. A., Aust. J. Phys. 18, 119 (1965). 10.1071/PH650119 [DOI] [Google Scholar]
- 64.Wang F. and Landau D. P., Phys. Rev. Lett. 86, 2050 (2001). 10.1103/PhysRevLett.86.2050 [DOI] [PubMed] [Google Scholar]
- 65.Lapelosa M., Gallicchio E., and Levy R. M., J. Chem. Theory Comput. 8, 47 (2012). 10.1021/ct200684b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallicchio E., Deng N., He P., Wickstrom L., Perryman A. L., Santiago D. N., Forli S., Olson A. J., and Levy R. M., J. Comput.-Aided Mol. Des. 28, 475 (2014). 10.1007/s10822-014-9711-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilson M. K., Given J. A., Bush B. L., and McCammon J. A., Biophys. J. 72, 1047 (1997). 10.1016/S0006-3495(97)78756-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukunishi H., Watanabe O., and Takada S., J. Chem. Phys. 116, 9058 (2002). 10.1063/1.1472510 [DOI] [Google Scholar]
- 69.Liu P., Kim B., Friesner R. A., and Berne B. J., Proc. Natl. Acad. Sci. U. S. A. 102, 13749 (2005). 10.1073/pnas.0506346102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallicchio E. and Levy R. M., J. Comput. Chem. 25, 479 (2004). 10.1002/jcc.10400 [DOI] [PubMed] [Google Scholar]
- 71.Gallicchio E., Paris K., and Levy R. M., J. Chem. Theory Comput. 5, 2544 (2009). 10.1021/ct900234u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banks J., Beard J., Cao Y., Cho A., Damm W., Farid R., Felts A., Halgren T., Mainz D., Maple J., Murphy R., Philipp D., Repasky M., Zhang L., Berne B., Friesner R., Gallicchio E., and Levy R. M., J. Comput. Chem. 26, 1752 (2005). 10.1002/jcc.20292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jorgensen W. L., Maxwell D. S., and Tirado-Rives J., J. Am. Chem. Soc. 118, 11225 (1996). 10.1021/ja9621760 [DOI] [Google Scholar]
- 74.Kaminski G. A., Friesner R. A., Tirado-Rives J., and Jorgensen W. L., J. Phys. Chem. B 105, 6474 (2001). 10.1021/jp003919d [DOI] [Google Scholar]
- 75.See supplementary material at http://dx.doi.org/10.1063/1.4939768 E-JCPSA6-144-025603 for additional results and cumulative CPU time in our numerical study.
- 76.Rosta E. and Hummer G., J. Chem. Theory Comput. 11, 276 (2014). 10.1021/ct500719p [DOI] [PubMed] [Google Scholar]
- 77.Wu H., Mey A. S. J. S., Rosta E., and Noe F., J. Chem. Phys. 141, 214106 (2014). 10.1063/1.4902240 [DOI] [PubMed] [Google Scholar]
- 78.Mey A. S. J. S., Wu H., and Noe F., Phys. Rev. X 4, 041018 (2014). 10.1103/physrevx.4.041018 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4939768 E-JCPSA6-144-025603 for additional results and cumulative CPU time in our numerical study.